The Mechanosensing and Global DNA Methylation of Human Osteoblasts on MEW Fibers
Abstract
:1. Introduction
2. Materials and Methods
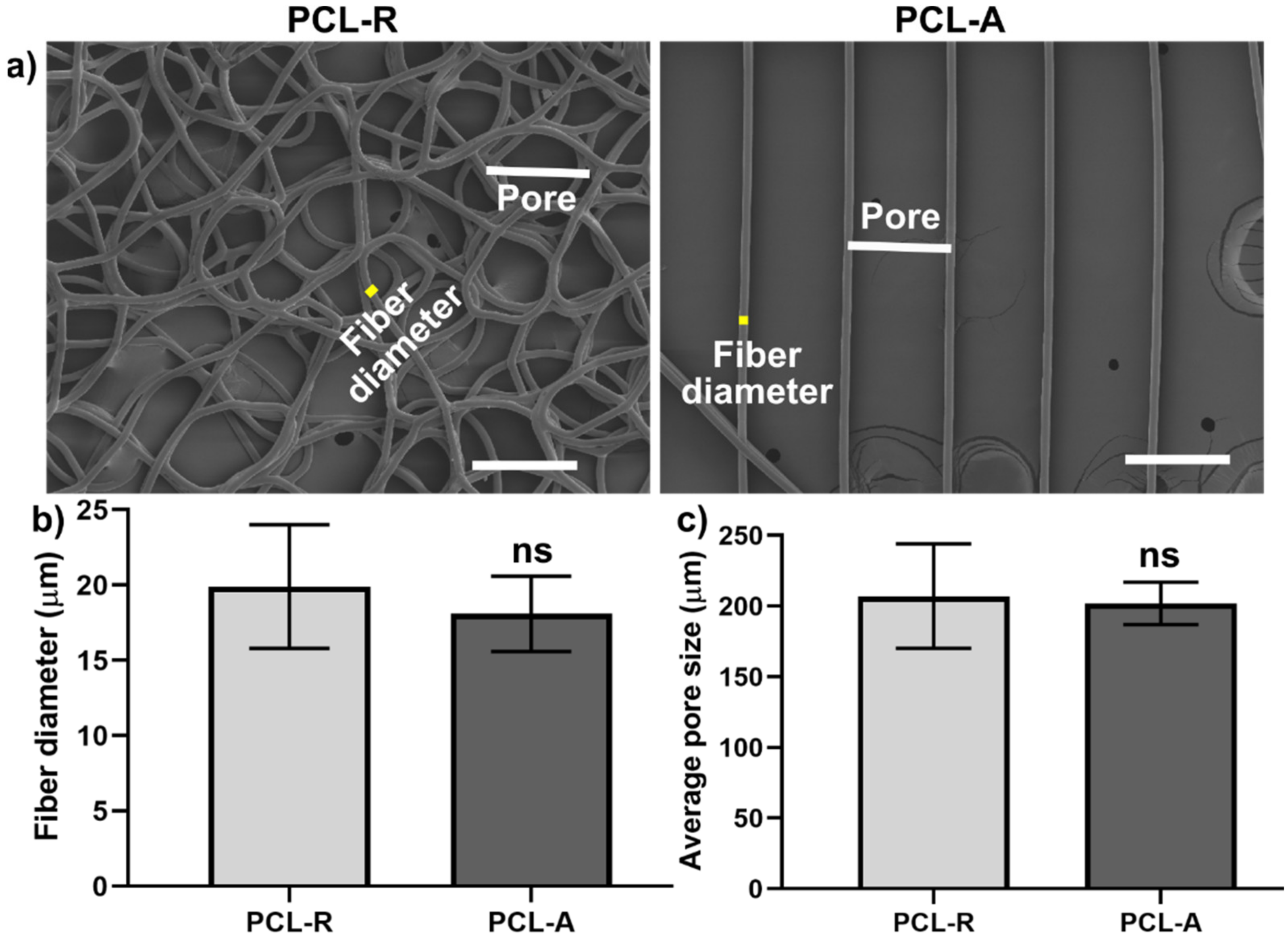
2.1. 3D PCL Porous Membranes Fabrication and Characterization
2.2. Human Primary Osteoblast Culture and Differentiation
2.3. Immunofluorescence Staining
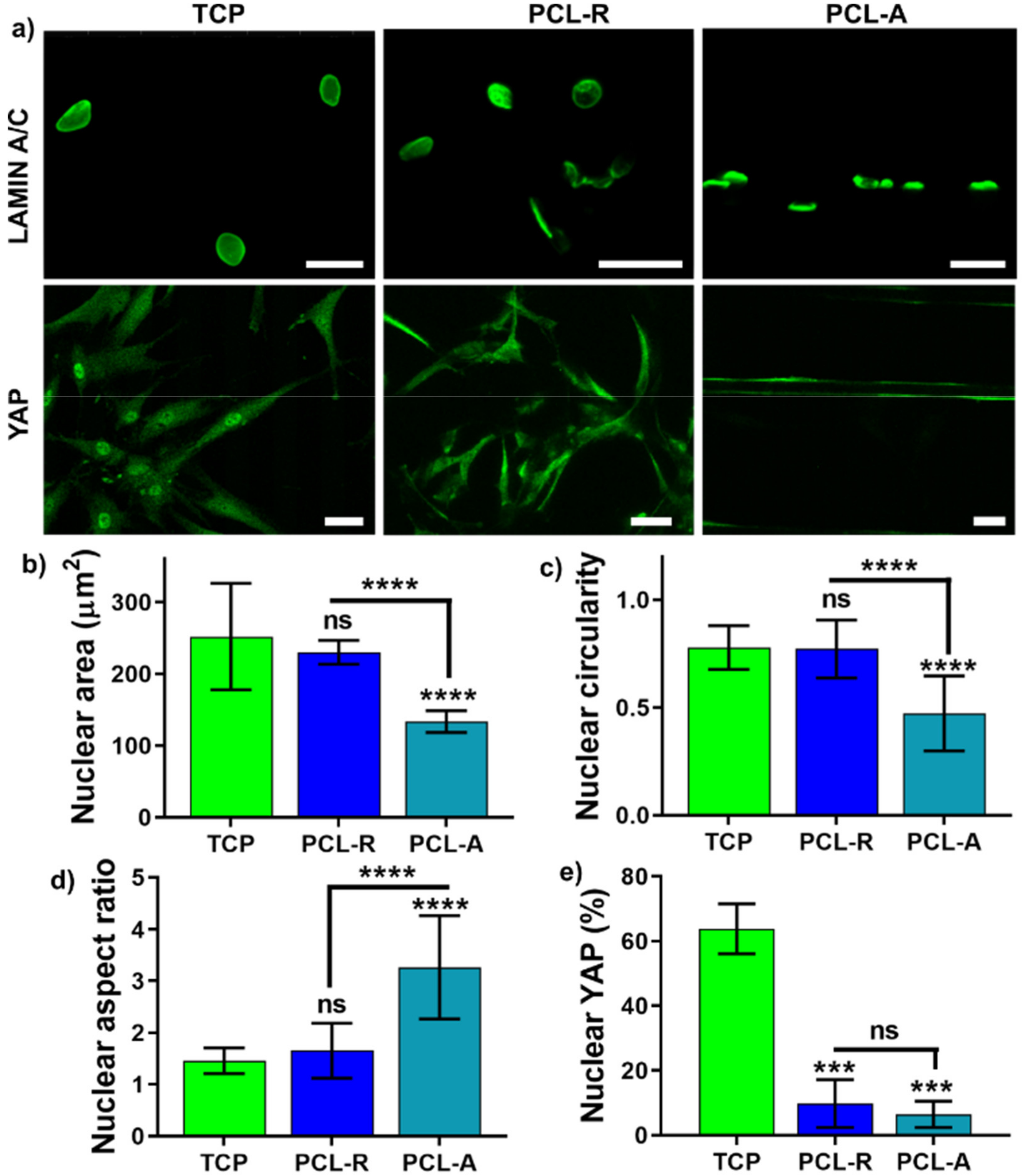
2.4. Focal Adhesion, Nucleus and Nuclear YAP Data Analysis
2.5. DNA Isolation and Global DNA Methylation Analysis
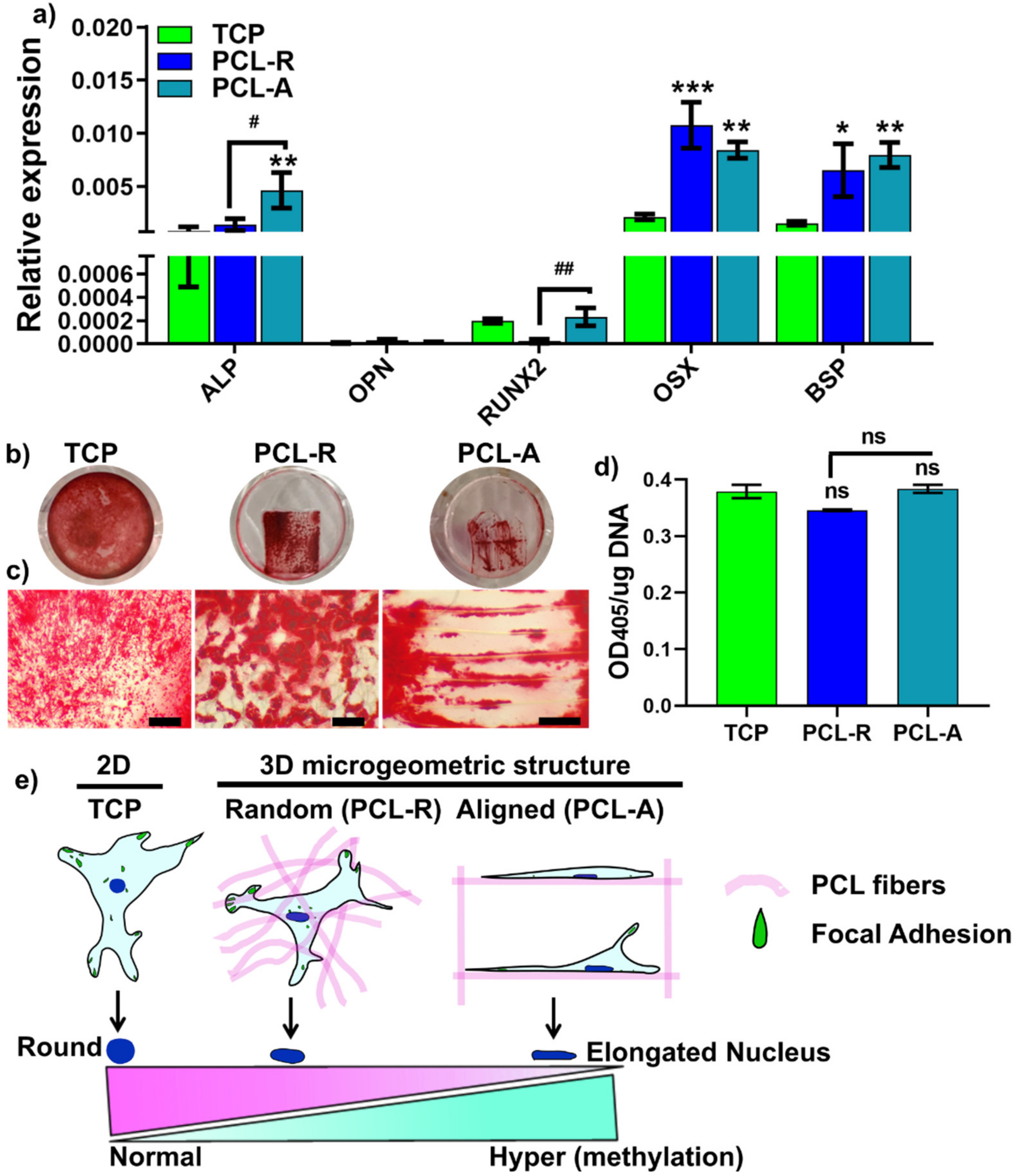
2.6. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.7. Alizarin Red S Staining
2.8. Statistical Analysis
3. Result and Discussion
3.1. Focal Adhesions Are Altered on Aligned Fibers on a PCL Porous Scaffold
3.2. The Effect of Fiber Alignment on Nucleus Mechanosensing
3.3. DNA Epigenetics Is Modulated by Fiber Alignment
3.4. Cell Differentiation on Different Fiber Aligned Porous Membranes
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lv, L.W.; Tang, Y.M.; Zhang, P.; Liu, Y.S.; Bai, X.S.; Zhou, Y.S. Biomaterial Cues Regulate Epigenetic State and Cell Functions—A Systematic Review. Tissue Eng. Part B Rev. 2018, 24, 112–132. [Google Scholar] [CrossRef] [PubMed]
- Matsugaki, A.; Aramoto, G.; Ninomiya, T.; Sawada, H.; Hata, S.; Nakano, T. Abnormal arrangement of a collagen/apatite extracellular matrix orthogonal to osteoblast alignment is constructed by a nanoscale periodic surface structure. Biomaterials 2015, 37, 134–143. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Matsugaki, A.; Kawahara, K.; Ninomiya, T.; Sawada, H.; Nakano, T. Unique arrangement of bone matrix orthogonal to osteoblast alignment controlled by Tspan11-mediated focal adhesion assembly. Biomaterials 2019, 209, 103–110. [Google Scholar] [CrossRef]
- Kubow, K.E.; Conrad, S.K.; Horwitz, A.R. Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr. Biol. 2013, 23, 1607–1619. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Kuzma, M.L.; Bai, X.; Yang, J. Biomaterial-Based Metabolic Regulation in Regenerative Engineering. Adv. Sci. 2019, 6, 1900819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amani, H.; Arzaghi, H.; Bayandori, M.; Dezfuli, A.S.; Pazoki-Toroudi, H.; Shafiee, A.; Moradi, L. Controlling Cell Behavior through the Design of Biomaterial Surfaces: A Focus on Surface Modification Techniques. Adv. Mater. Interfaces 2019, 6, 1900572. [Google Scholar] [CrossRef] [Green Version]
- Hynes, R.O. The extracellular matrix: Not just pretty fibrils. Science 2009, 326, 1216–1219. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; Frith, J.E.; Gomez, G.A.; Yap, A.S.; O’Neill, G.M.; Cooper-White, J.J. Five Piconewtons: The Difference between Osteogenic and Adipogenic Fate Choice in Human Mesenchymal Stem Cells. ACS Nano 2019, 13, 11129–11143. [Google Scholar] [CrossRef] [PubMed]
- Haupt, A.; Minc, N. How cells sense their own shape—Mechanisms to probe cell geometry and their implications in cellular organization and function. J. Cell Sci. 2018, 131, jcs214015. [Google Scholar] [CrossRef] [Green Version]
- Swaminathan, V.; Waterman, C.M. The molecular clutch model for mechanotransduction evolves. Nat. Cell Biol. 2016, 18, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Irianto, J.; Discher, D.E. Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol. 2017, 216, 305–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowder, S.W.; Leonardo, V.; Whittaker, T.; Papathanasiou, P.; Stevens, M.M. Material Cues as Potent Regulators of Epigenetics and Stem Cell Function. Cell Stem Cell 2016, 18, 39–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worley, K.; Certo, A.; Wan, L.Q. Geometry-Force Control of Stem Cell Fate. Bionanoscience 2013, 3, 43–51. [Google Scholar] [CrossRef]
- Larsson, L. Current Concepts of Epigenetics and Its Role in Periodontitis. Curr. Oral Health Rep. 2017, 4, 286–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [Green Version]
- Robertson, K.D.; Wolffe, A.P. DNA methylation in health and disease. Nat. Rev. Genet. 2000, 1, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Zhang, J.; Wang, X.; Zheng, X.; Qin, X.; Zhang, Q.; Li, W.; Hu, W.; Lin, J.; Chen, F. DNA methylation profile is associated with the osteogenic potential of three distinct human odontogenic stem cells. Signal Transduct. Target. Ther. 2018, 3, 1. [Google Scholar] [CrossRef]
- Xin, T.-Y.; Yu, T.-T.; Yang, R.-L. DNA methylation and demethylation link the properties of mesenchymal stem cells: Regeneration and immunomodulation. World J. Stem Cells 2020, 12, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Bartold, P.M.; Salomon, C.; Ivanovski, S. Salivary Outer Membrane Vesicles and DNA Methylation of Small Extracellular Vesicles as Biomarkers for Periodontal Status: A Pilot Study. Int. J. Mol. Sci. 2021, 22, 2423. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.; Pilipchuk, S.P.; Giannobile, W.V.; Castilho, R.M. When epigenetics meets bioengineering-A material characteristics and surface topography perspective. J. Biomed. Mater. Res. B Appl. Biomater. 2018, 106, 2065–2071. [Google Scholar] [CrossRef] [PubMed]
- Downing, T.L.; Soto, J.; Morez, C.; Houssin, T.; Fritz, A.; Yuan, F.; Chu, J.; Patel, S.; Schaffer, D.V.; Li, S. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 2013, 12, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, N.; Abdal-hay, A.; Hamlet, S.; Graham, E.; Ivanovski, S. Effects of Gradient and Offset Architectures on the Mechanical and Biological Properties of 3-D Melt Electrowritten (MEW) Scaffolds. ACS Biomater. Sci. Eng. 2019, 5, 3448–3461. [Google Scholar] [CrossRef] [PubMed]
- Abdal-hay, A.; Abbasi, N.; Gwiazda, M.; Hamlet, S.; Ivanovski, S. Novel polycaprolactone/hydroxyapatite nanocomposite fibrous scaffolds by direct melt-electrospinning writing. Eur. Polym. J. 2018, 105, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Barrows, T. Degradable implant materials: A review of synthetic absorbable polymers and their applications. Clin. Mater. 1986, 1, 233–257. [Google Scholar] [CrossRef]
- Abdal-hay, A.; Raveendran, N.T.; Fournier, B.; Ivanovski, S. Fabrication of biocompatible and bioabsorbable polycaprolactone/magnesium hydroxide 3D printed scaffolds: Degradation and in vitro osteoblasts interactions. Compos. Part B Eng. 2020, 197, 108158. [Google Scholar] [CrossRef]
- Abdal-hay, A.; Amna, T.; Lim, J.K. Biocorrosion and Osteoconductivity of PCL/nHAp Composite Porous Film-Based Coating of Magnesium Alloy. Solid State Sci. 2012, 18, 131–140. [Google Scholar] [CrossRef]
- Gwiazda, M.; Kumar, S.; Swieszkowski, W.; Ivanovski, S.; Vaquette, C. The effect of melt electrospun writing fiber orientation onto cellular organization and mechanical properties for application in Anterior Cruciate Ligament tissue engineering. J. Mech. Behav. Biomed. Mater. 2020, 104, 103631. [Google Scholar] [CrossRef] [PubMed]
- Dan, H.; Vaquette, C.; Fisher, A.G.; Hamlet, S.M.; Xiao, Y.; Hutmacher, D.W.; Ivanovski, S. The influence of cellular source on periodontal regeneration using calcium phosphate coated polycaprolactone scaffold supported cell sheets. Biomaterials 2014, 35, 113–122. [Google Scholar] [CrossRef] [Green Version]
- Haase, H.R.; Ivanovski, S.; Waters, M.J.; Bartold, P.M. Growth hormone regulates osteogenic marker mRNA expression in human periodontal fibroblasts and alveolar bone-derived cells. J. Periodontal Res. 2003, 38, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Ivanovski, S.; Crawford, R.; Xiao, Y. Activation of the Canonical Wnt Signaling Pathway Induces Cementum Regeneration. J. Bone Miner. Res. 2015, 30, 1160–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blaudez, F.; Ivanovski, S.; Ipe, D.; Vaquette, C. A comprehensive comparison of cell seeding methods using highly porous melt electrowriting scaffolds. Mater. Sci. Eng. C Mater. 2020, 117, 111282. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Han, S.; Xiao, L.; Han, P.; Wang, S.; He, J.; Chang, J.; Wu, C.; Xiao, Y. Accelerated host angiogenesis and immune responses by ion release from mesoporous bioactive glass. J. Mater. Chem. B 2018, 6, 3274–3284. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Lloyd, T.; Chen, Z.; Xiao, Y. Proinflammatory Cytokines Regulate Cementogenic Differentiation of Periodontal Ligament Cells by Wnt/Ca(2+) Signaling Pathway. J. Interferon Cytokine Res. 2016, 36, 328–337. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Xiao, Z.; Chen, T.; Wei, J.; Chen, L.; Liu, L.; Chen, B.; Wang, X.; Li, X.; Dai, J. The miR-7 identified from collagen biomaterial-based three-dimensional cultured cells regulates neural stem cell differentiation. Stem Cells Dev. 2014, 23, 393–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Beyec, J.; Xu, R.; Lee, S.Y.; Nelson, C.M.; Rizki, A.; Alcaraz, J.; Bissell, M.J. Cell shape regulates global histone acetylation in human mammary epithelial cells. Exp. Cell Res. 2007, 313, 3066–3075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, A.D.; Carvajal, N.; Jin, A.; Matsumoto, K.; Yamada, K.M. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nat. Commun. 2015, 6, 8720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Lian, M.; Wu, Q.; Qiao, Z.; Sun, B.; Dai, K. Effect of Pore Size on Cell Behavior Using Melt Electrowritten Scaffolds. Front. Bioeng. Biotechnol. 2021, 9, 495. [Google Scholar] [CrossRef] [PubMed]
- Lanaro, M.; McLaughlin, M.P.; Simpson, M.J.; Buenzli, P.R.; Wong, C.S.; Allenby, M.C.; Woodruff, M.A. A quantitative analysis of cell bridging kinetics on a scaffold using computer vision algorithms. Acta Biomater. 2021. [Google Scholar] [CrossRef] [PubMed]
- Cukierman, E.; Pankov, R.; Stevens, D.R.; Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 2001, 294, 1708–1712. [Google Scholar] [CrossRef] [PubMed]
- Grashoff, C.; Hoffman, B.D.; Brenner, M.D.; Zhou, R.; Parsons, M.; Yang, M.T.; McLean, M.A.; Sligar, S.G.; Chen, C.S.; Ha, T.; et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 2010, 466, 263–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hakkinen, K.M.; Harunaga, J.S.; Doyle, A.D.; Yamada, K.M. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng. Part A 2011, 17, 713–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraley, S.I.; Feng, Y.; Krishnamurthy, R.; Kim, D.H.; Celedon, A.; Longmore, G.D.; Wirtz, D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010, 12, 598–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thery, M.; Racine, V.; Piel, M.; Pepin, A.; Dimitrov, A.; Chen, Y.; Sibarita, J.B.; Bornens, M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA 2006, 103, 19771–19776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Cui, C.; Ibrahim, M.M.; Han, B.; Li, Q.; Pacifici, M.; Lawrence, J.T.R.; Han, L.; Han, L.-H. Regulating Mechanotransduction in Three Dimensions using Sub-Cellular Scale, Crosslinkable Fibers of Controlled Diameter, Stiffness, and Alignment. Adv. Funct. Mater. 2019, 29, 1808967. [Google Scholar] [CrossRef]
- Isermann, P.; Lammerding, J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 2013, 23, R1113–R1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Kim, T.; Johnson, R.L.; Lim, D.S. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 2015, 11, 270–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Forward Primer (5′-3′) | Reverse Primer (5′-3′) | |
|---|---|---|
| ALP | TCAGAAGCTAACACCAACG | TTGTACGTCTTGGAGAGGGC |
| OPN | TCACCTGTGCCATACCAGTTAA | TGAGATGGGTCAGGGTTTAGC |
| RUNX2 | GGAGTGGACGAGGCAAGAGTTT | AGCTTCTGTCTGTGCCTTCTGG |
| OSX | GCAAAGCAGGCACAAAGAAG | CAGGTGAAAGGAGCCCATTAG |
| BSP | AGGCTGAGAATACCACACTTTC | GGATTGCAGCTAACCCTGTAT |
| 18s | TTCGGAACTGAGGCCATGAT | CGAACCTCCGACTTCGTTC |
| GAPDH | TCAGCAATGCATCCTGCAC | TCTGGGTGGCAGTGATGGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, P.; Vaquette, C.; Abdal-hay, A.; Ivanovski, S. The Mechanosensing and Global DNA Methylation of Human Osteoblasts on MEW Fibers. Nanomaterials 2021, 11, 2943. https://doi.org/10.3390/nano11112943
Han P, Vaquette C, Abdal-hay A, Ivanovski S. The Mechanosensing and Global DNA Methylation of Human Osteoblasts on MEW Fibers. Nanomaterials. 2021; 11(11):2943. https://doi.org/10.3390/nano11112943
Chicago/Turabian StyleHan, Pingping, Cedryck Vaquette, Abdalla Abdal-hay, and Sašo Ivanovski. 2021. "The Mechanosensing and Global DNA Methylation of Human Osteoblasts on MEW Fibers" Nanomaterials 11, no. 11: 2943. https://doi.org/10.3390/nano11112943
APA StyleHan, P., Vaquette, C., Abdal-hay, A., & Ivanovski, S. (2021). The Mechanosensing and Global DNA Methylation of Human Osteoblasts on MEW Fibers. Nanomaterials, 11(11), 2943. https://doi.org/10.3390/nano11112943









