Nanoporous Microsponge Particles (NMP) of Polysaccharides as Universal Carriers for Biomolecules Delivery
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis and Characterization of the Cross-Linking Solutions
2.3. Synthesis and Characterization of NMP
2.4. Preparation of Polysaccharide Fluorescently Labeled with Rhodamine B Isothiocyanate
2.5. Degradation Kinetics of NMP
2.6. Protein Adsorption and Release in Particles
2.7. Cell Assays for Cito-Toxicity and Adhesion/Internalization of NMPs
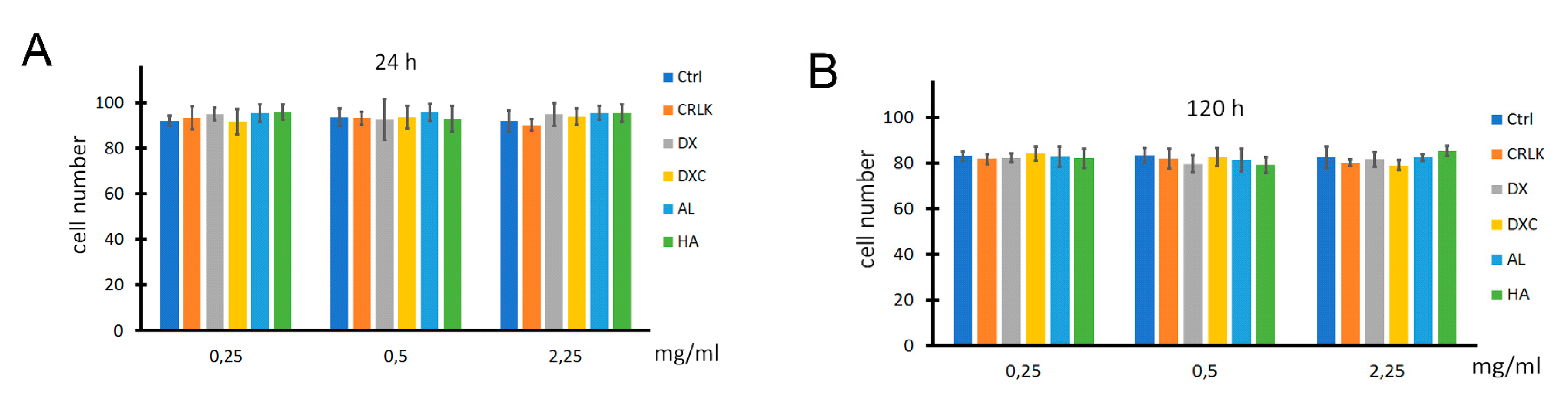
2.7.1. Cell Viability
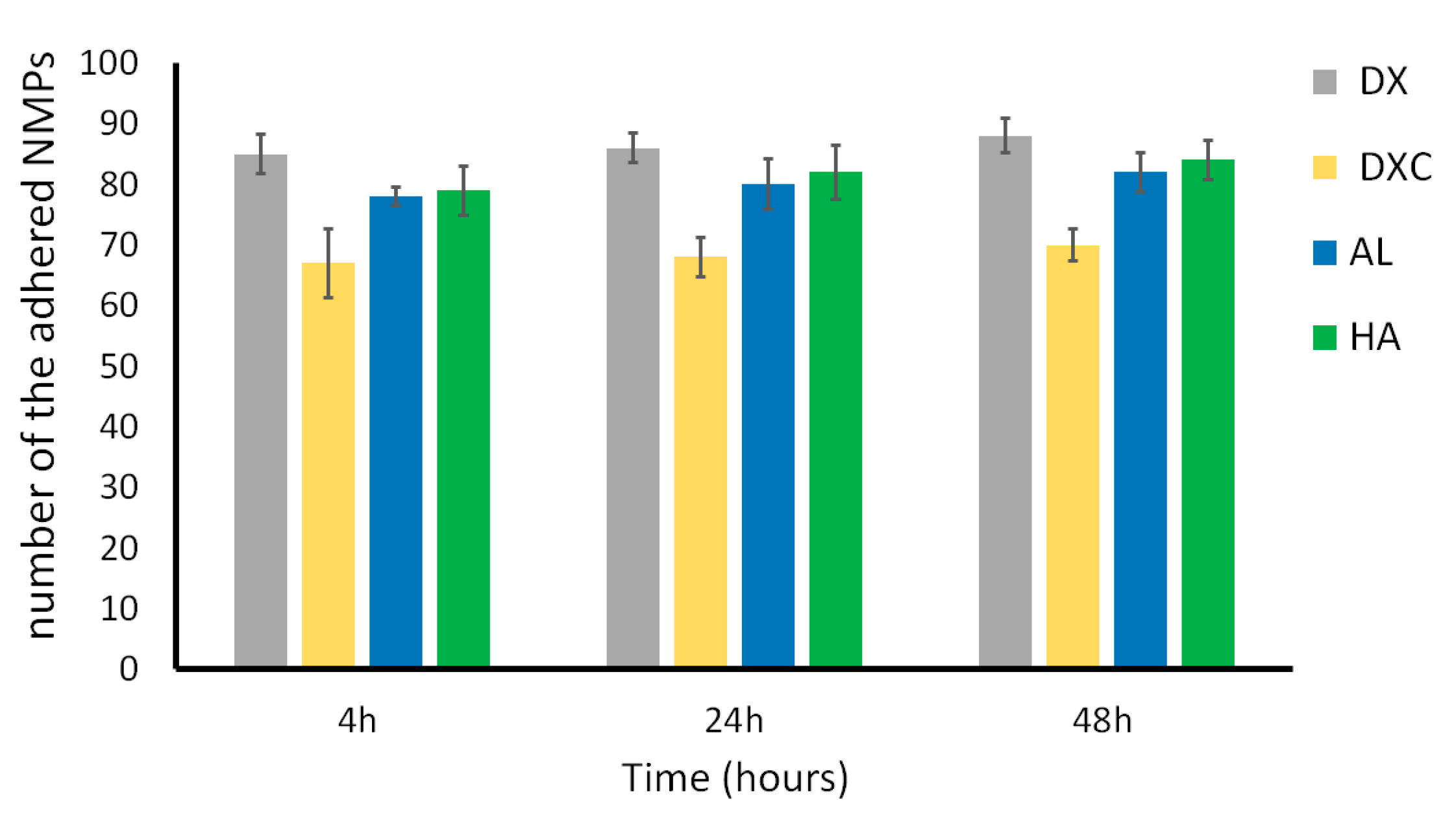
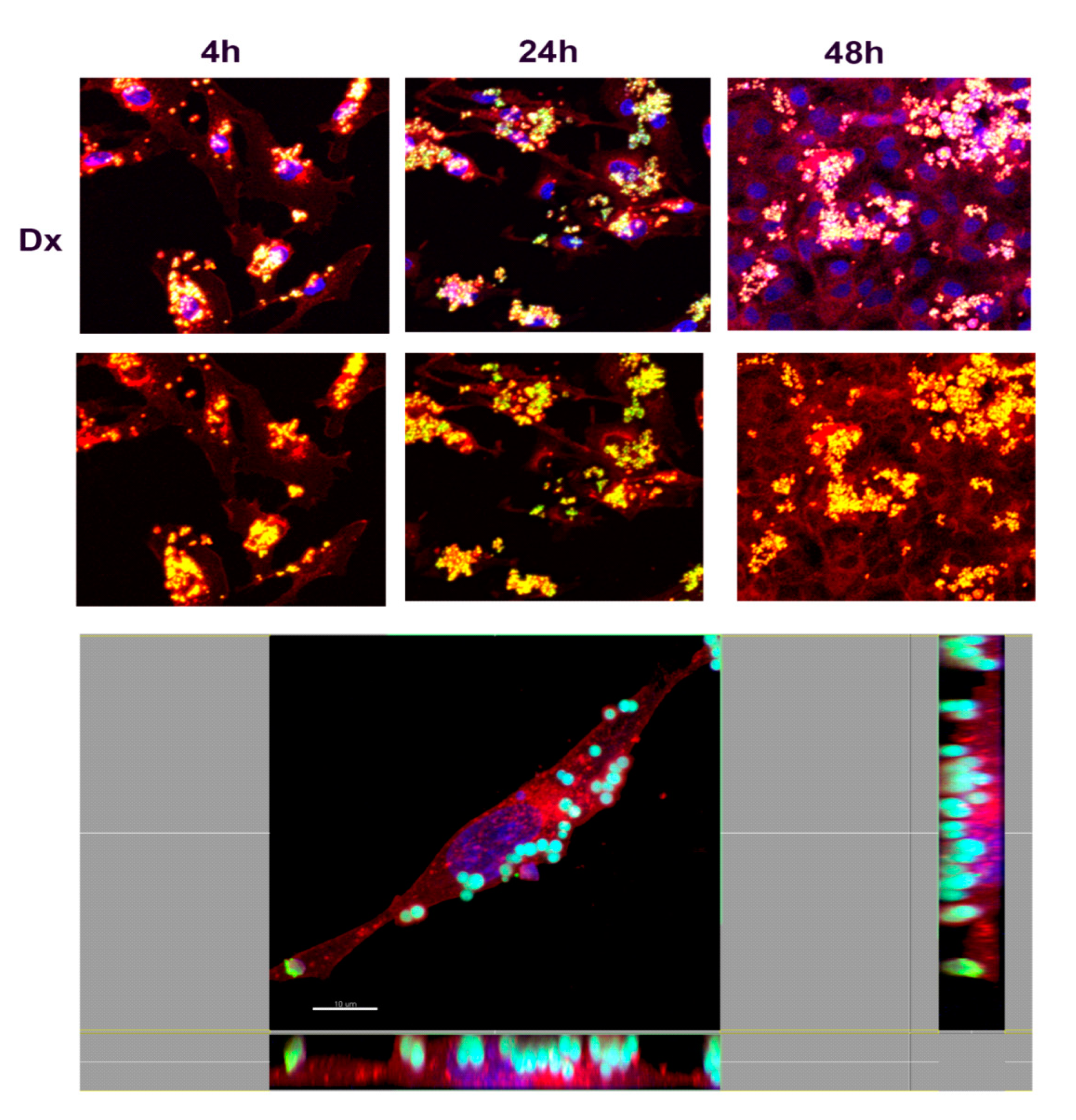
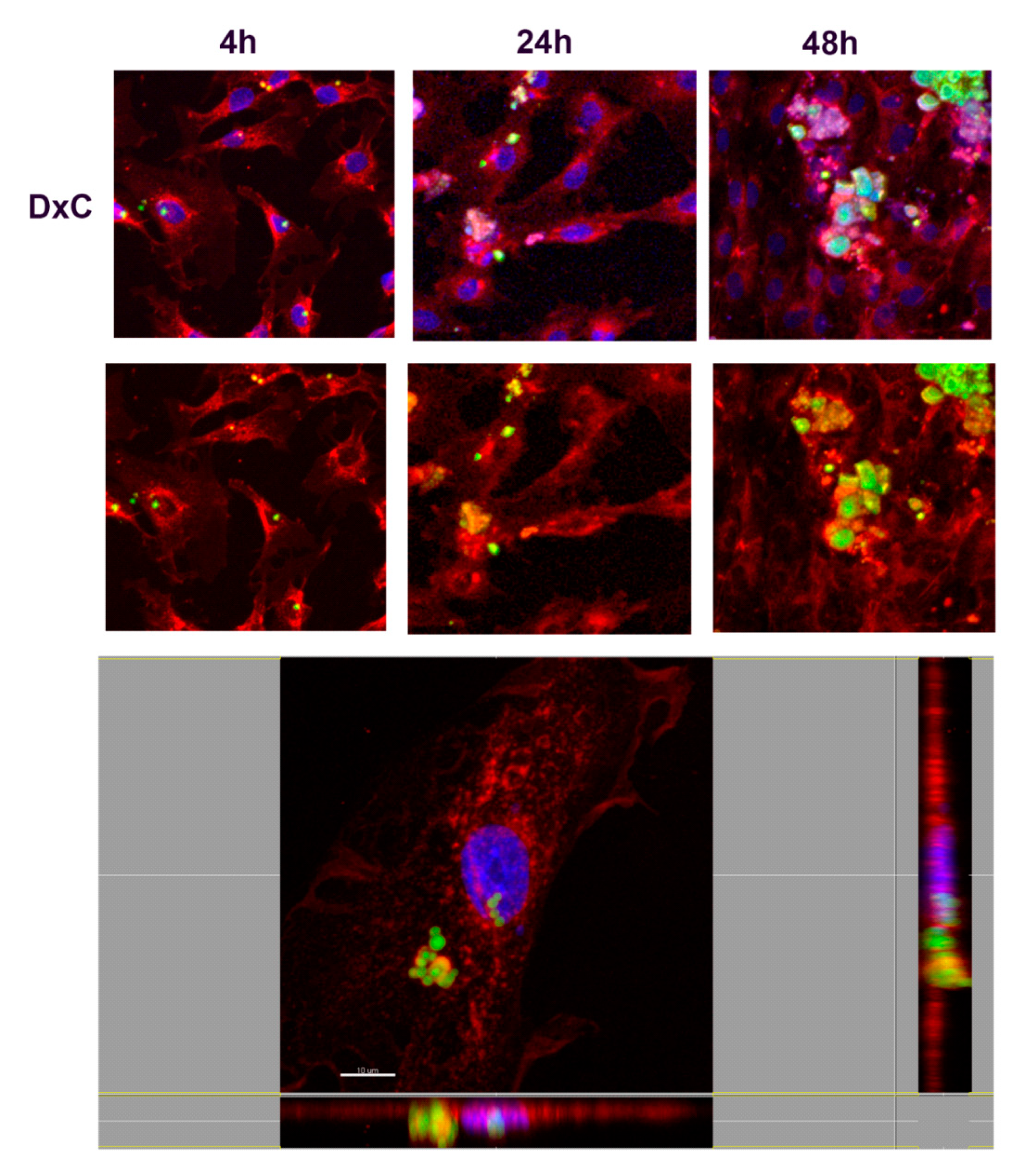
2.7.2. Study of Cell Adhesion/Internalization of NMPs by Confocal Microscopy
3. Results and Discussion
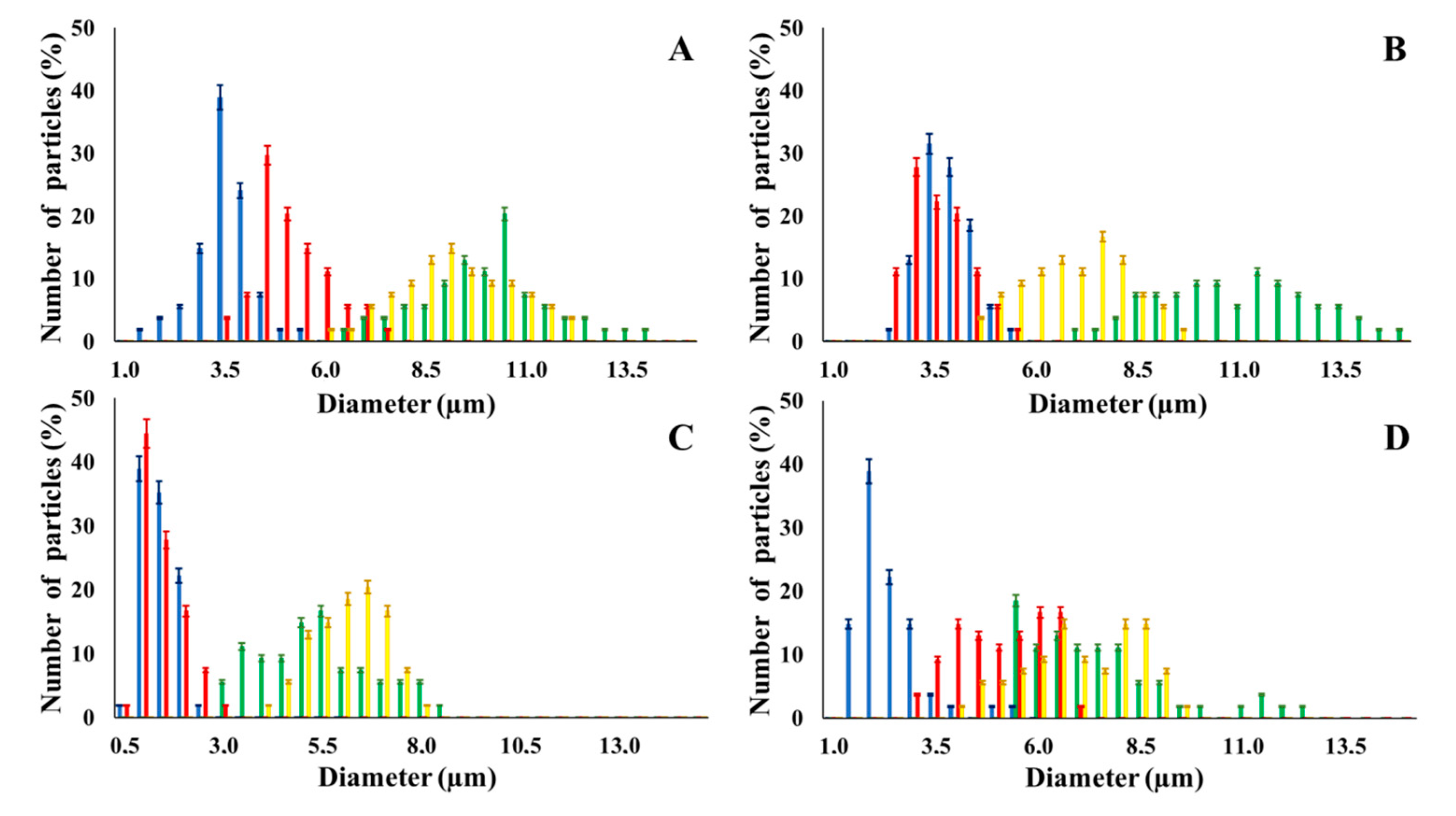
3.1. Two-Ways Parametric Study on NMP Synthesis vs. (i) Polymer and (ii) Cross- Linker Solvent
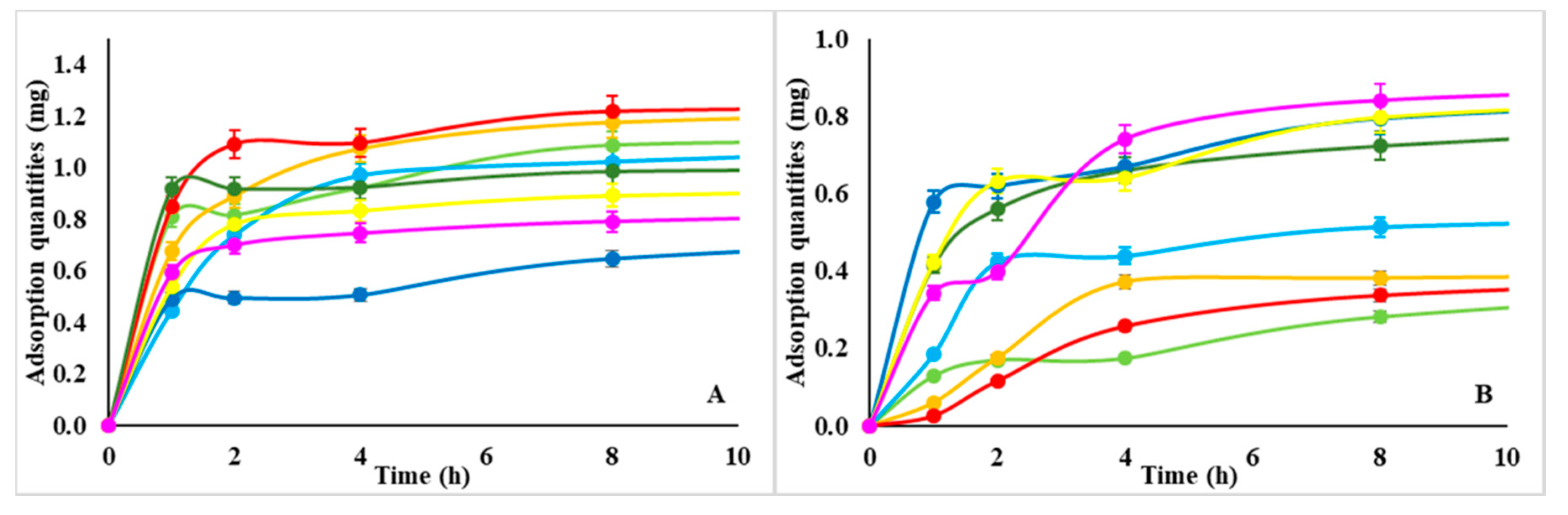
3.2. NMP as Platform for Sequestration and Release of Proteins.
- Proteins uptake of d-NMP and w-NMP varies between 0.8–1.2 and 0.4–0.8 mg protein/mg polysaccharide, respectively. Lysozyme is preferentially taken up by d-NMP, whereas serum albumin is equally adsorbed by both d-NMP and w-NMP. This indicates that the composite nanostructured template of d-NMP interacts more efficiently with different type of proteins.
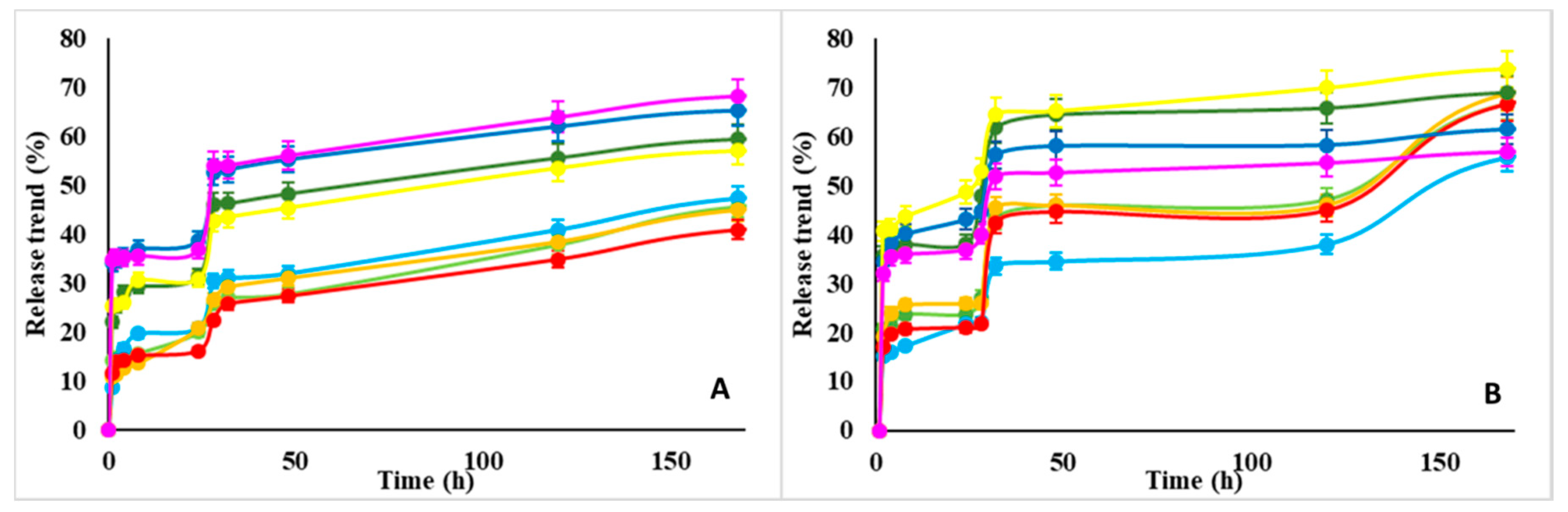
- On the other end, the efficiency of w-NMP (70–80%) in releasing the adsorbed proteins is higher than d-NMP efficiency (40–70%).
- The adsorption is nearly completed after 4 h while the release typically occurs on a scale of days.
Dependence of Adsorption/Release Profiles vs. pH and Protein Distribution within the NMP
- The absolute adsorption/release performance depends on the specific NMP polysaccharide (i.e., size, porosity etc., as discussed in Section 3.1);
- The adsorption and release rates tend to increase with the pH value (also Figure 6), although this effect is more marked on lysozyme than BSA.
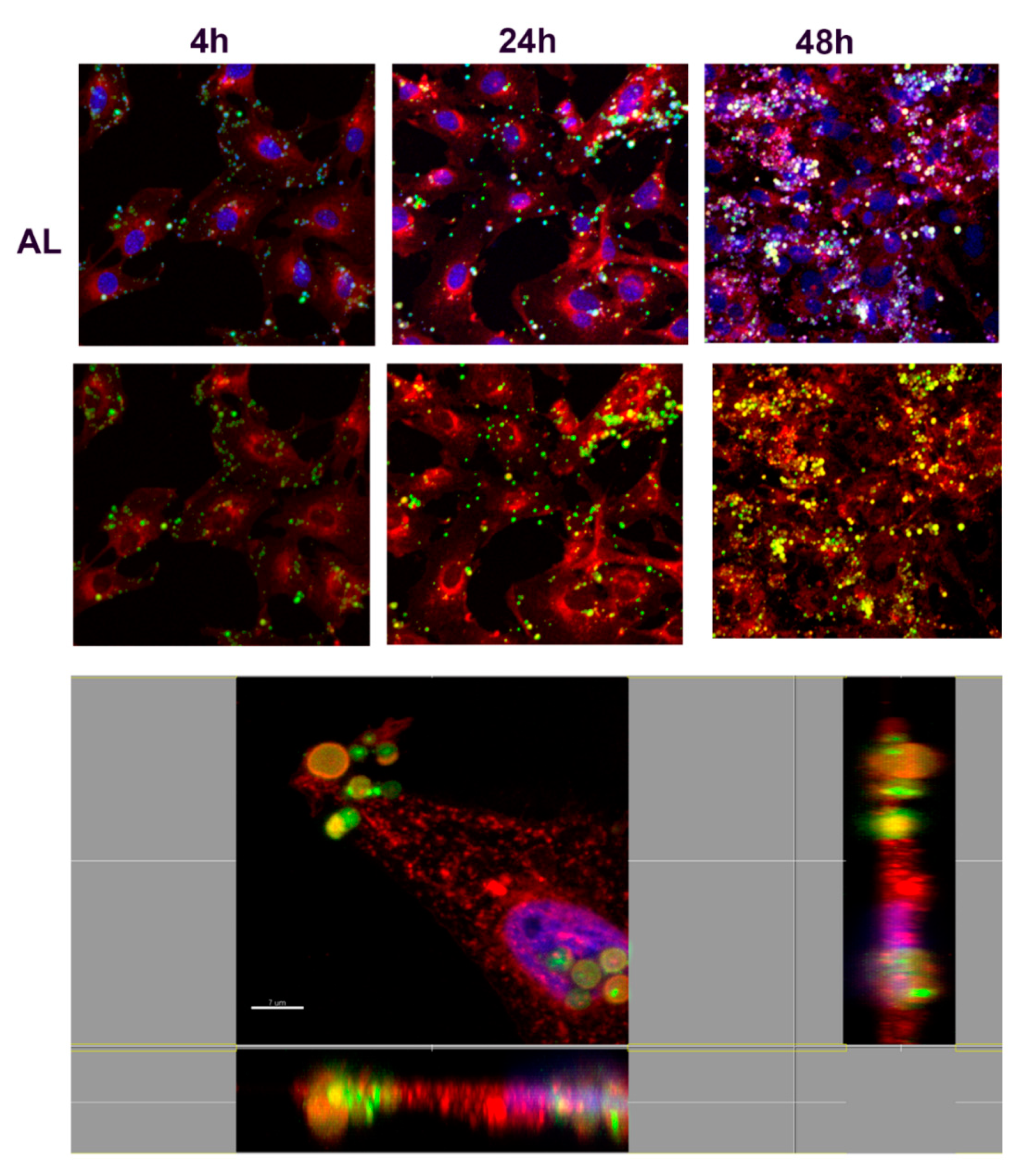
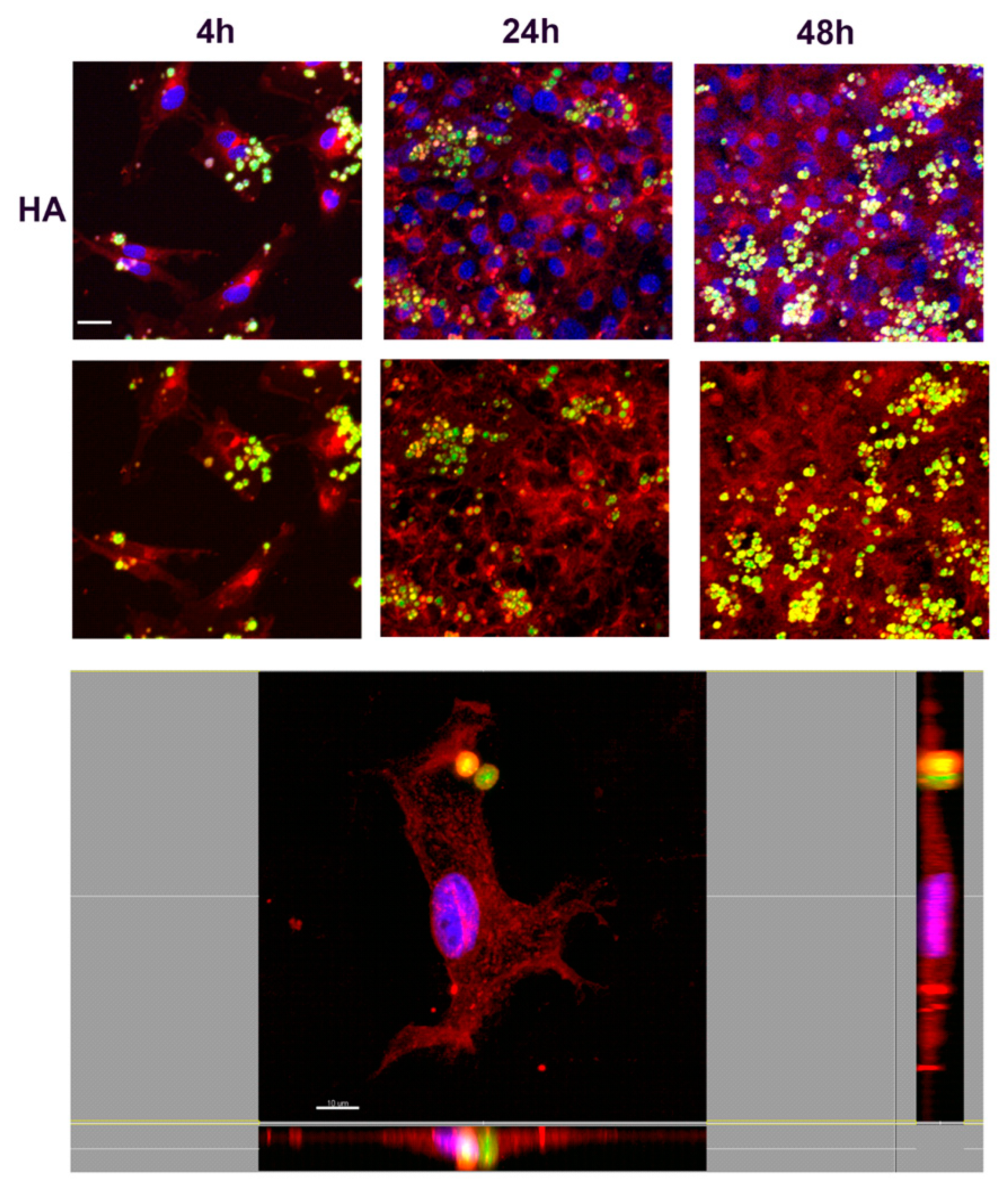
3.3. The Interaction of NMP with Fibroblast T3T Cells: Assessing Toxicity and Adhesion/Internalization by Cells
3.3.1. Toxicity Assays
3.3.2. Study of Cell Interaction and Uptake of NMPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shende, P.; Kasture, P.; Gaud, R. Nanoflowers: The future trend of nanotechnology for multi-applications. Artif. Cells Nanomed. Biotechnol. 2018, 46, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Pankaj, S. Formulation and Evaluation of Gel-Loaded Microsponges of Roxithromycin for Topical Drug Delivery. IOSR J. Pharm. 2019, 9, 14–22. [Google Scholar]
- Ruan, W.; Zheng, M.; An, Y.; Liu, Y.; Lovejoy, D.B.; Hao, M.; Zou, Y.; Lee, A.; Yang, S.; Lu, Y.; et al. DNA nanoclew templated spherical nucleic acids for siRNA delivery. Chem. Commun. 2018, 54, 3609–3612. [Google Scholar] [CrossRef]
- Ziegler, J.; Blaikie, A.; Fathalizadeh, A.; Miller, D.J.; Yasin, F.S.; Williams, K.; Mohrhardt, J.; McMorran, B.J.; Zettl, A.; Aleman, B.J. Single-Photon Emitters in Boron Nitride Nanococoons. Nano Lett. 2018, 18, 2683–2688. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Wang, Y.; Yip, X.; Glynn, F.; Shepherd, R.K.; Caruso, F. Nanoporous peptide particles for encapsulating and releasing neurotrophic factors in an animal model of neurodegeneration. Adv. Mater. 2012, 24, 3362–3366. [Google Scholar] [CrossRef]
- Reytblat, I.; Lipovsky, A.; Gedanken, A. DNA Microspheres Coated with Bioavailable Polymer as an Efficient Gene Expression Agent in Yeasts. J. Nanomater. 2016, 2016, 1–8. [Google Scholar] [CrossRef][Green Version]
- Gallo, L.; Bucalá, V.; Rigo, M.R. Formulation and Characterization of Polysaccharide Microparticles for Pulmonary Delivery of Sodium Cromoglycate. AAPS PharmSciTech 2016, 18, 1634–1645. [Google Scholar] [CrossRef] [PubMed]
- Keith, J.; Jefferson, K.; Shulga, O.V. Nanoporous Gold for Enzyme Immobilization. Enzym. Stab. Immobil. 2016, 1504, 37–60. [Google Scholar]
- Li, D.; Kaner, R.B.; Ho, M. MATERIALS SCIENCE: Graphene-Based Materials. Science 2008, 320, 1170–1171. [Google Scholar] [CrossRef]
- Kim, E.; Zwi-Dantsis, L.; Reznikov, N.; Hansel, C.S.; Agarwal, S.; Stevens, M.M. One-Pot Synthesis of Multiple Protein-Encapsulated DNA Flowers and Their Application in Intracellular Protein Delivery. Adv. Mater. 2017, 29, 1701086. [Google Scholar] [CrossRef]
- Hou, Y. Solvothermal Reduction Synthesis and Characterization of Superparamagnetic Magnetite Nanoparticles. J. Mater. Chem. 2013, 13, 1983–1987. [Google Scholar] [CrossRef]
- Xie, Y.; Kocaefe, D.; Chen, C.; Kocaefe, Y. Review of Research on Template Methods in Preparation of Nanomaterials. J. Nanomater. 2016, 2016, 1–10. [Google Scholar] [CrossRef]
- Cavalieri, F. Hyaluronic acid micro-sponges and method for the production thereof. Patent PCT/IB2016/052792, 17 November 2016. [Google Scholar]
- Palmieri, G.; Rinaldi, A.; Campagnolo, L.; Tortora, M.; Caso, M.F.; Mattei, M.; Notargiacomo, A.; Rosato, N.; Bottini, M.; Cavalieri, F. Hyaluronic Acid Nanoporous Microparticles with Long In Vivo Joint Residence Time and Sustained Release. Part. Part. Syst. Charact. 2017, 34, 1600411. [Google Scholar] [CrossRef]
- Au, J.L.-S.; Yeung, B.Z.; Lu, Z.; Wientjes, M.G.; Wientjes, M.G. Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities. Adv. Drug Deliv. Rev. 2016, 97, 280–301. [Google Scholar] [CrossRef] [PubMed]
- Raveendran, S.; Rochani, A.K.; Maekawa, T.; Kumar, D.S. Smart Carriers and Nanohealers: A Nanomedical Insight on Natural Polymers. Materials 2017, 10, 929. [Google Scholar] [CrossRef] [PubMed]
- Janarthanan, P.; Kumar, A.; Owh, C.; Jun Loh, X. Biodegradable Polysaccharides For Controlled Drug Delivery. ChemPlusChem 2016, 81, 504–514. [Google Scholar]
- Myrick, J.M.; Vendra, V.K.; Krishnan, S. Self-assembled polysaccharide nanostructures for controlled-release applications. Nanotechnol. Rev. 2014, 3. [Google Scholar] [CrossRef]
- Nur, M.; Vasiljevic, T. Can natural polymers assist in delivering insulin orally? Int. J. Boil. Macromol. 2017, 103, 889–901. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Natl. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef]
- Oh, N.; Park, J. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed. 2014, 9 (Suppl. 1), 51–63. [Google Scholar]
- Ispanixtlahuatl-Meráz, O.; Schins, R.P.F.; Chirino, Y.I. Cell type specific cytoskeleton disruption induced by engineered nanoparticles. Environ. Sci. Nano 2018, 5, 228–245. [Google Scholar] [CrossRef]


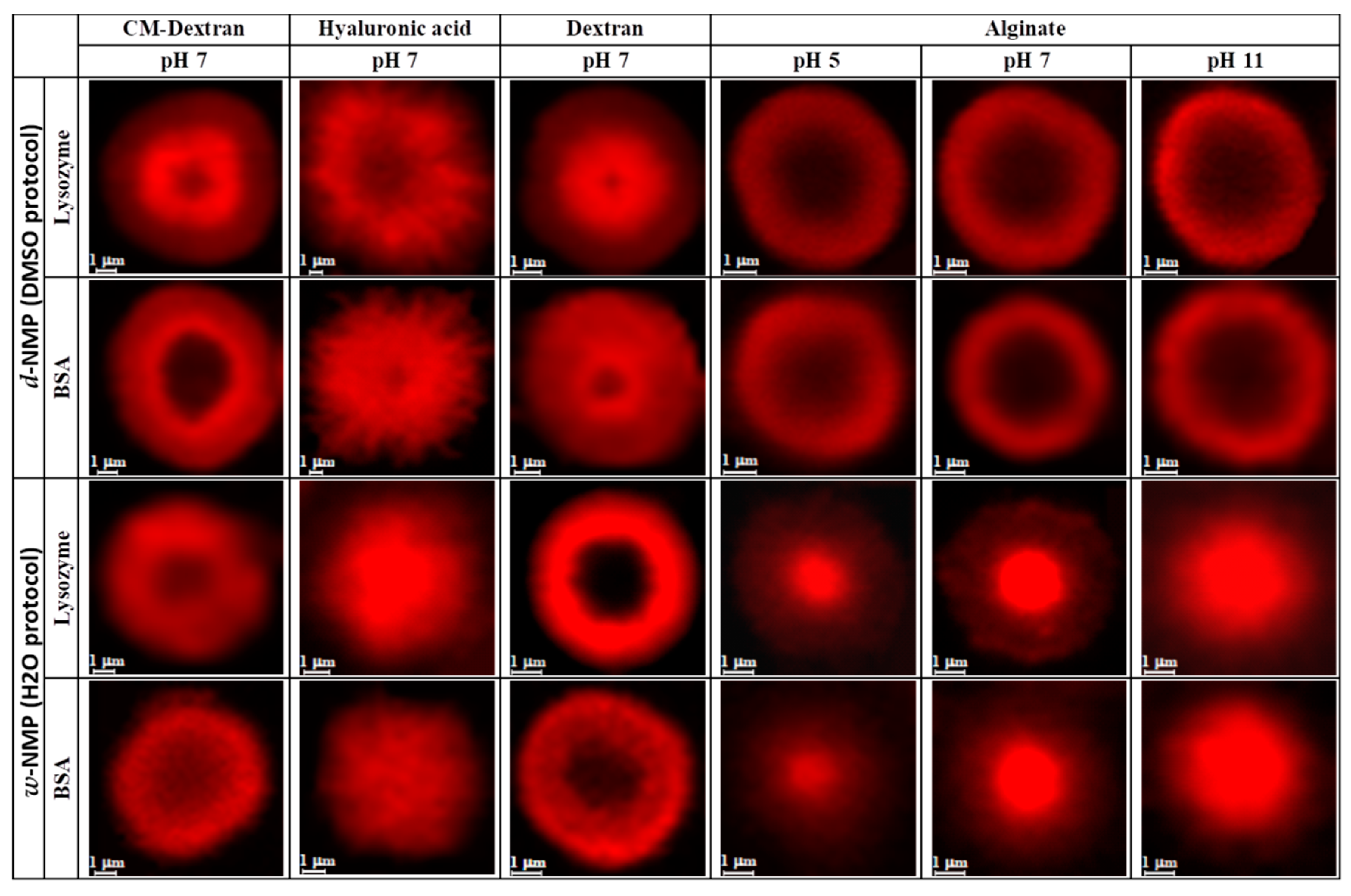
| Polysaccharide | Structure | DMSO-Protocol | H2O-Protocol | ||
|---|---|---|---|---|---|
| Confocal | SEM | Confocal | SEM | ||
| CM-Dextran | 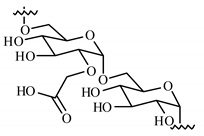 |  |  |  | 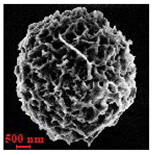 |
| Hyaluronic acid | 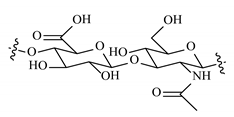 |  |  |  |  |
| Alginate | 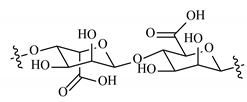 |  |  |  |  |
| Dextran |  |  |  |  |  |
| Polysaccharide | Solvent for CL | Average Yield (%) | NMP Diameter | NMP Pores Diameter | |||
|---|---|---|---|---|---|---|---|
| Dry (SEM) | Swollen | Swelling Ratio | |||||
| CM-Dextran | DMSO | → | 97 | 3.5 ± 0.8 µm | 9.8 ± 1.7 µm | 2.8 | 0.10 ± 0.04 µm |
| H2O | 94 | 5.0 ± 0.1 µm | 9.6 ± 1.8 µm | 1.9 | 0.13 ± 0.05 µm | ||
| Hyaluronic Acid | DMSO | 98 | 4.0 ± 0.8 µm | 11.2 ± 1.8 µm | 2.8 | 0.16 ± 0.06 µm | |
| H2O | 98 | 3.4 ± 0.7 µm | 7.3 ± 1.4 µm | 2.1 | 0.14 ± 0.05 µm | ||
| Alginate | DMSO | 98 | 1.4 ± 0.4 µm | 5.2 ± 1.6 µm | 3.5 | 0.06 ± 0.02 µm | |
| H2O | 98 | 1.2 ± 0.5 µm | 6.3 ± 1.0 µm | 5.3 | 0.08 ± 0.03 µm | ||
| Dextran | DMSO | 96 | 2.4 ± 0.8 µm | 7.4 ± 1.8 µm | 3.0 | 0.11 ± 0.04 µm | |
| H2O | 96 | 4.8 ± 1.0 µm | 7.5 ± 1.4 µm | 1.6 | 0.09 ± 0.04 µm | ||
| Polysaccharide | Adsorbed Lysozyme Quantity (mg) after 24 h | Adsorbed BSA Quantity (mg) after 24 h | ||
|---|---|---|---|---|
| d-NMP | w-NMP | d-NMP | w-NMP | |
| CM-Dextran | 1.099 | 0.441 | 0.999 | 0.850 |
| Hyaluronic acid | 1.155 | 0.553 | 0.814 | 0.894 |
| Alginate | 1.252 | 0.402 | 0.944 | 0.886 |
| Dextran | 1.225 | 0.432 | 0.868 | 0.913 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caso, M.F.; Carotenuto, F.; Di Nardo, P.; Migliore, A.; Aguilera, A.; Lopez, C.M.; Venanzi, M.; Cavalieri, F.; Rinaldi, A. Nanoporous Microsponge Particles (NMP) of Polysaccharides as Universal Carriers for Biomolecules Delivery. Nanomaterials 2020, 10, 1075. https://doi.org/10.3390/nano10061075
Caso MF, Carotenuto F, Di Nardo P, Migliore A, Aguilera A, Lopez CM, Venanzi M, Cavalieri F, Rinaldi A. Nanoporous Microsponge Particles (NMP) of Polysaccharides as Universal Carriers for Biomolecules Delivery. Nanomaterials. 2020; 10(6):1075. https://doi.org/10.3390/nano10061075
Chicago/Turabian StyleCaso, Maria Federica, Felicia Carotenuto, Paolo Di Nardo, Alberto Migliore, Ana Aguilera, Cruz Matilde Lopez, Mariano Venanzi, Francesca Cavalieri, and Antonio Rinaldi. 2020. "Nanoporous Microsponge Particles (NMP) of Polysaccharides as Universal Carriers for Biomolecules Delivery" Nanomaterials 10, no. 6: 1075. https://doi.org/10.3390/nano10061075
APA StyleCaso, M. F., Carotenuto, F., Di Nardo, P., Migliore, A., Aguilera, A., Lopez, C. M., Venanzi, M., Cavalieri, F., & Rinaldi, A. (2020). Nanoporous Microsponge Particles (NMP) of Polysaccharides as Universal Carriers for Biomolecules Delivery. Nanomaterials, 10(6), 1075. https://doi.org/10.3390/nano10061075







