The Fate of Lipid-Coated and Uncoated Fluorescent Nanodiamonds during Cell Division in Yeast
Abstract
1. Introduction
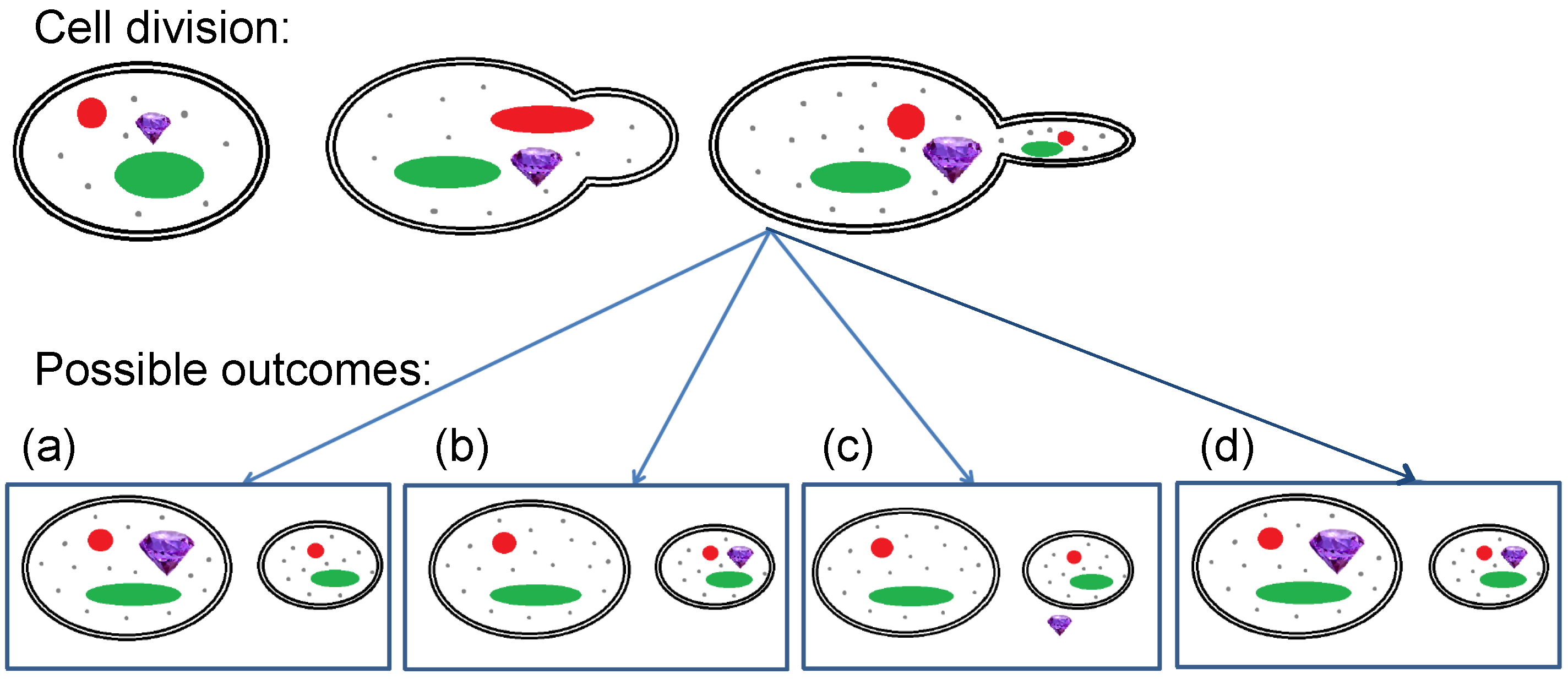
- FNDs could remain (preferentially) with the mother, for example, because they are regarded as harmful by the cell (see Figure 1a).
- FNDs might (preferentially) move into the daughter cells (see Figure 1b).
- FNDs might be excreted (see Figure 1c).
- FNDs might end up randomly distributed between both mother and daughter cells (see Figure 1d).
2. Materials and Methods
2.1. Diamond Starting Material
2.1.1. Bare Particles
2.1.2. Coated Particles
2.1.3. Particle Characterization
2.2. Fluorescence Nanodiamond Particles Uptake
2.3. Immobilizing Yeast Cells
2.4. Equipment
2.5. Tracking FND Movement during Cell Division
2.6. FND Quantification during Cell Division
2.7. Single Particle Tracking
2.8. Statistical Analysis
3. Results
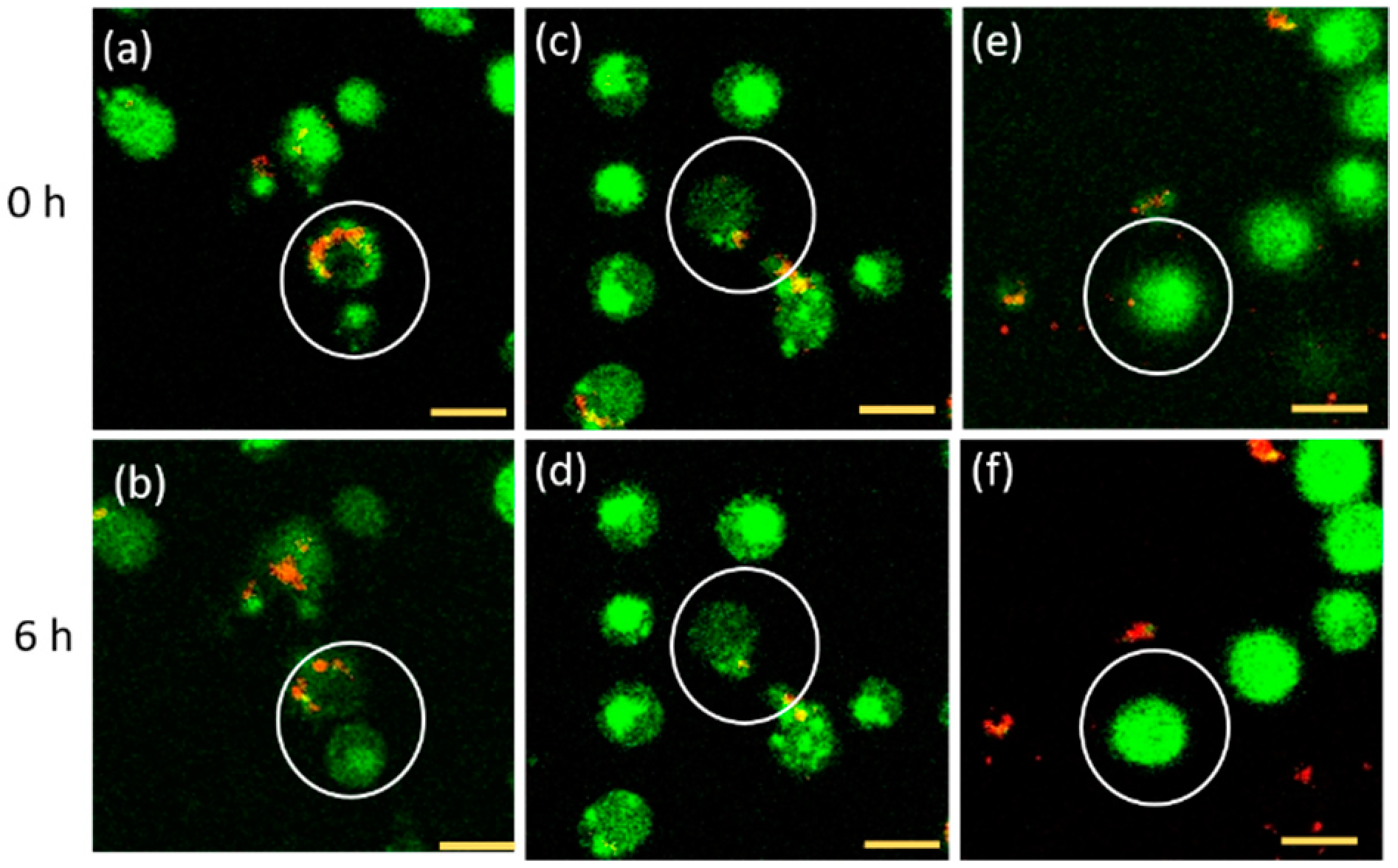
3.1. FNDs Movement during Cell Division
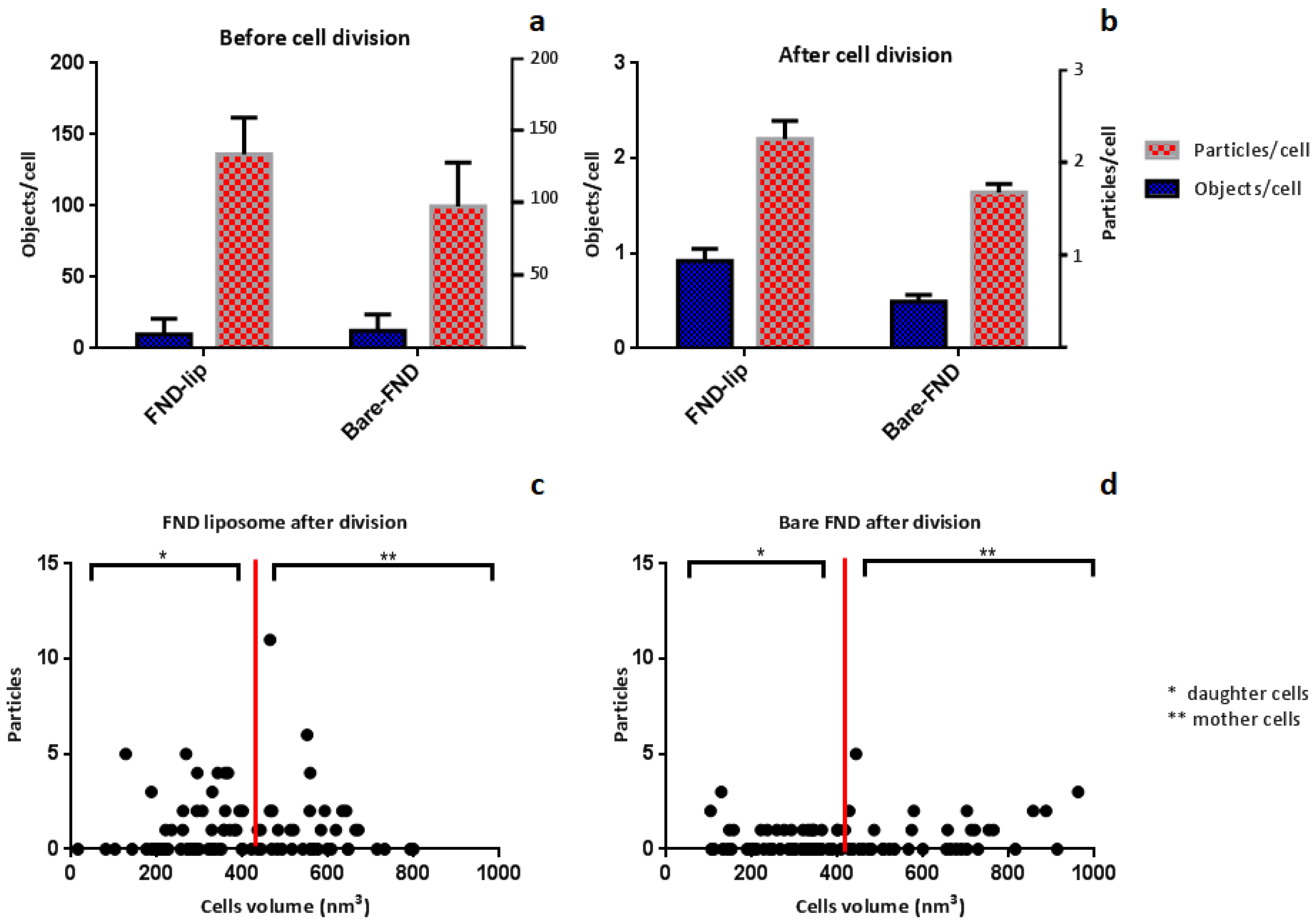
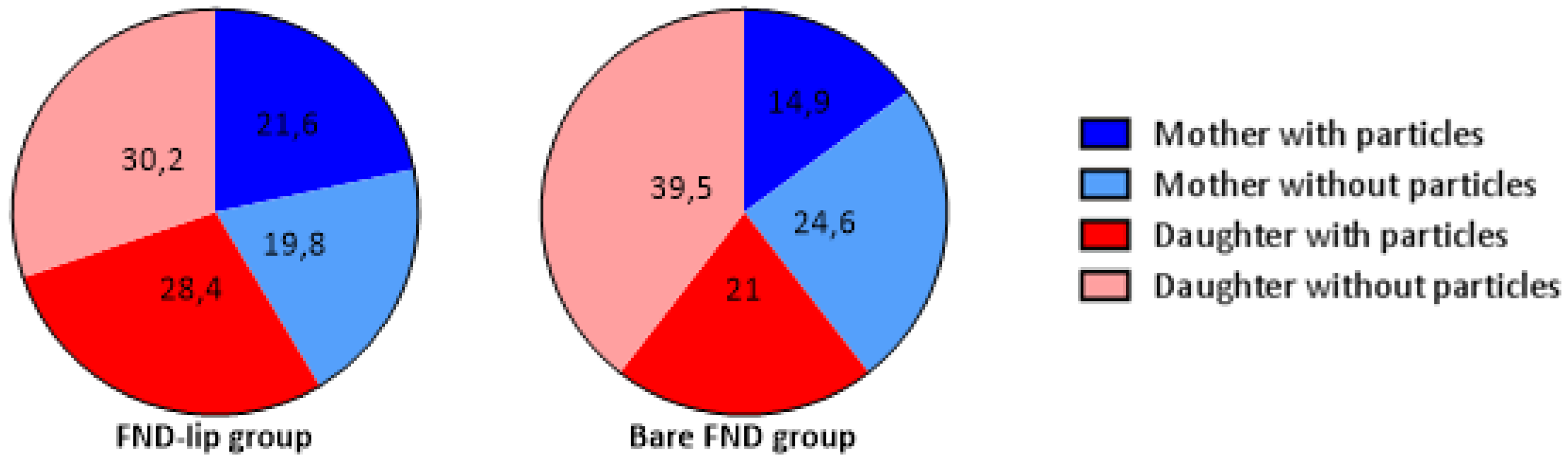
3.2. FND Quantification after Cell Division
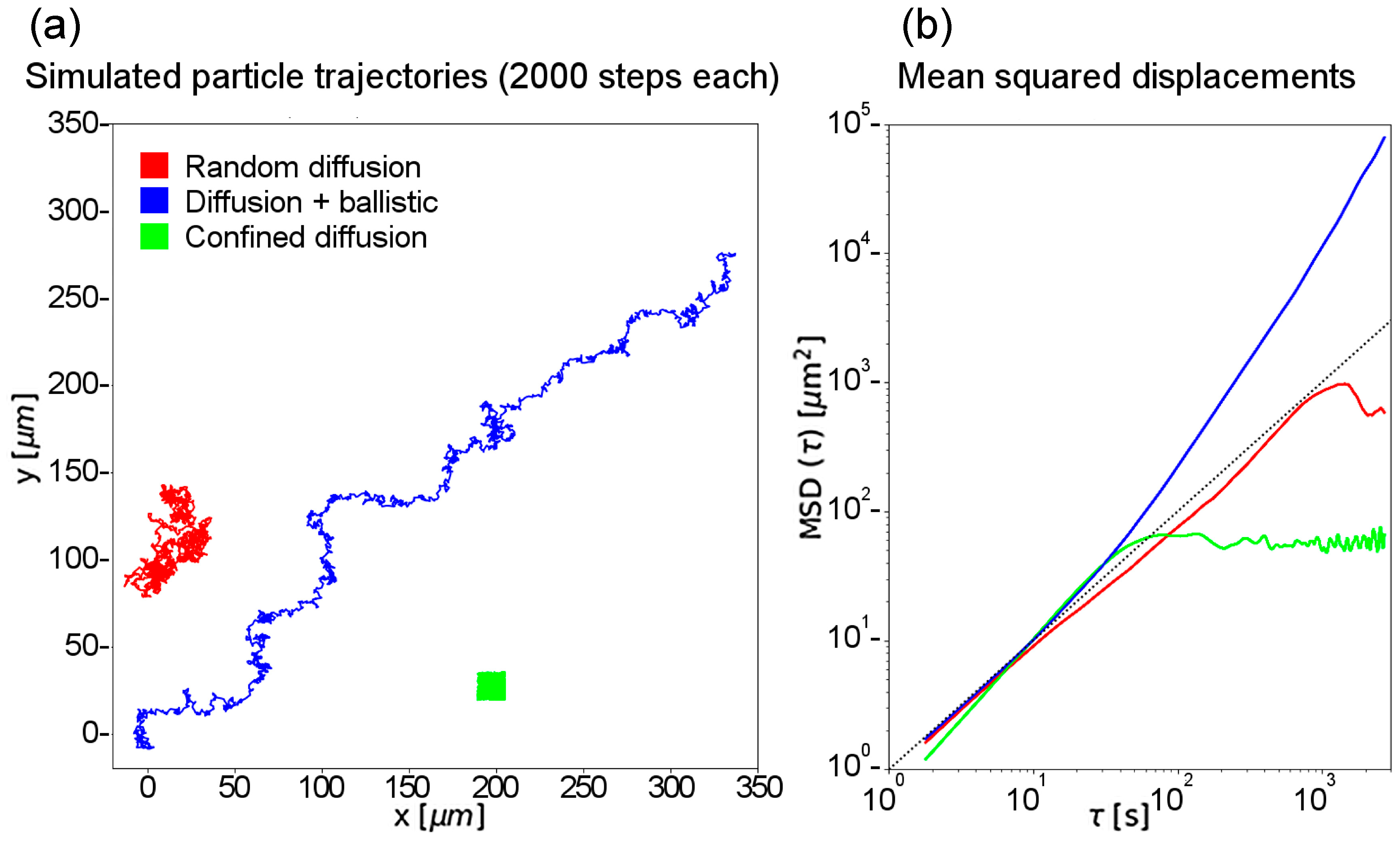
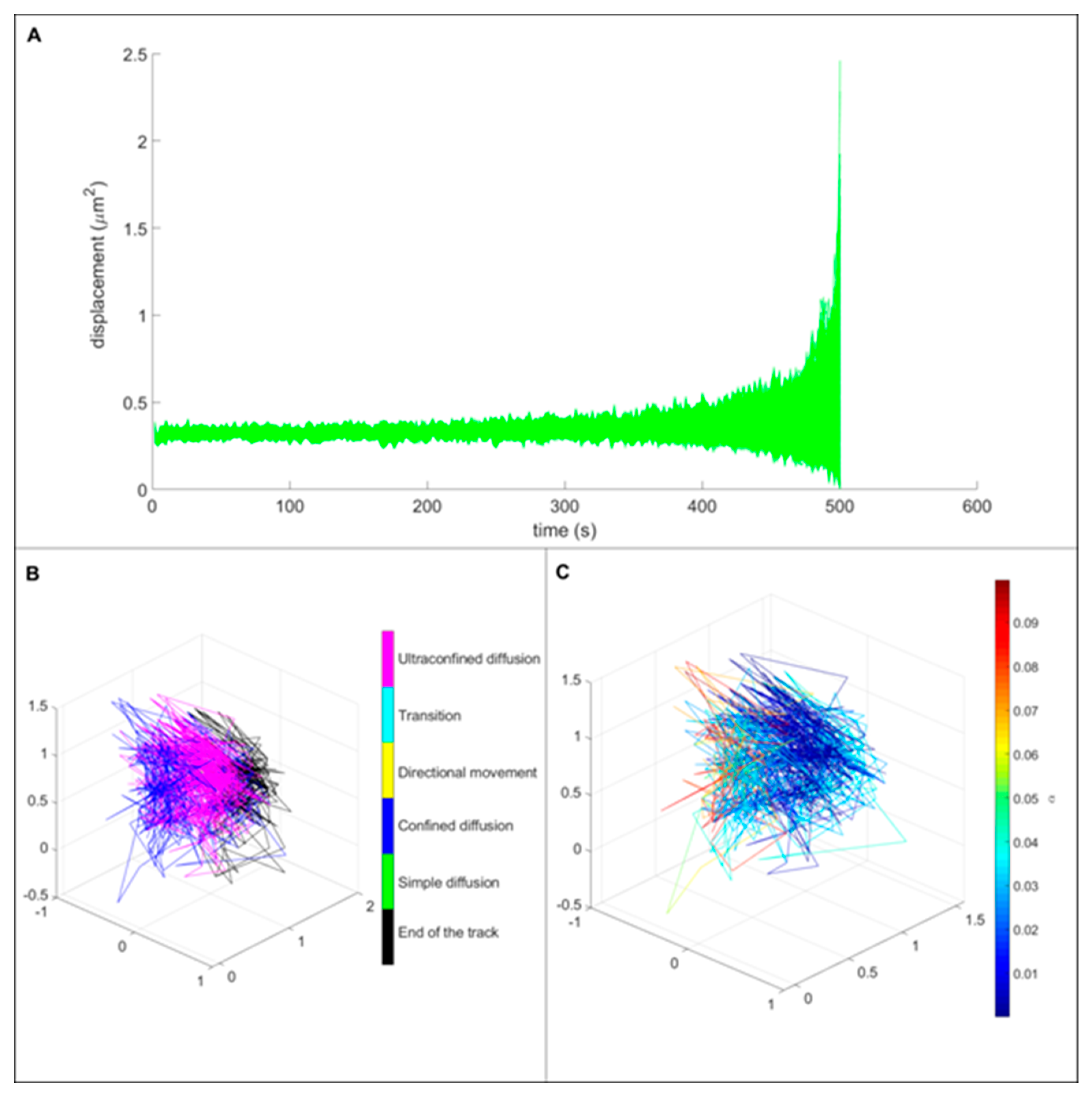
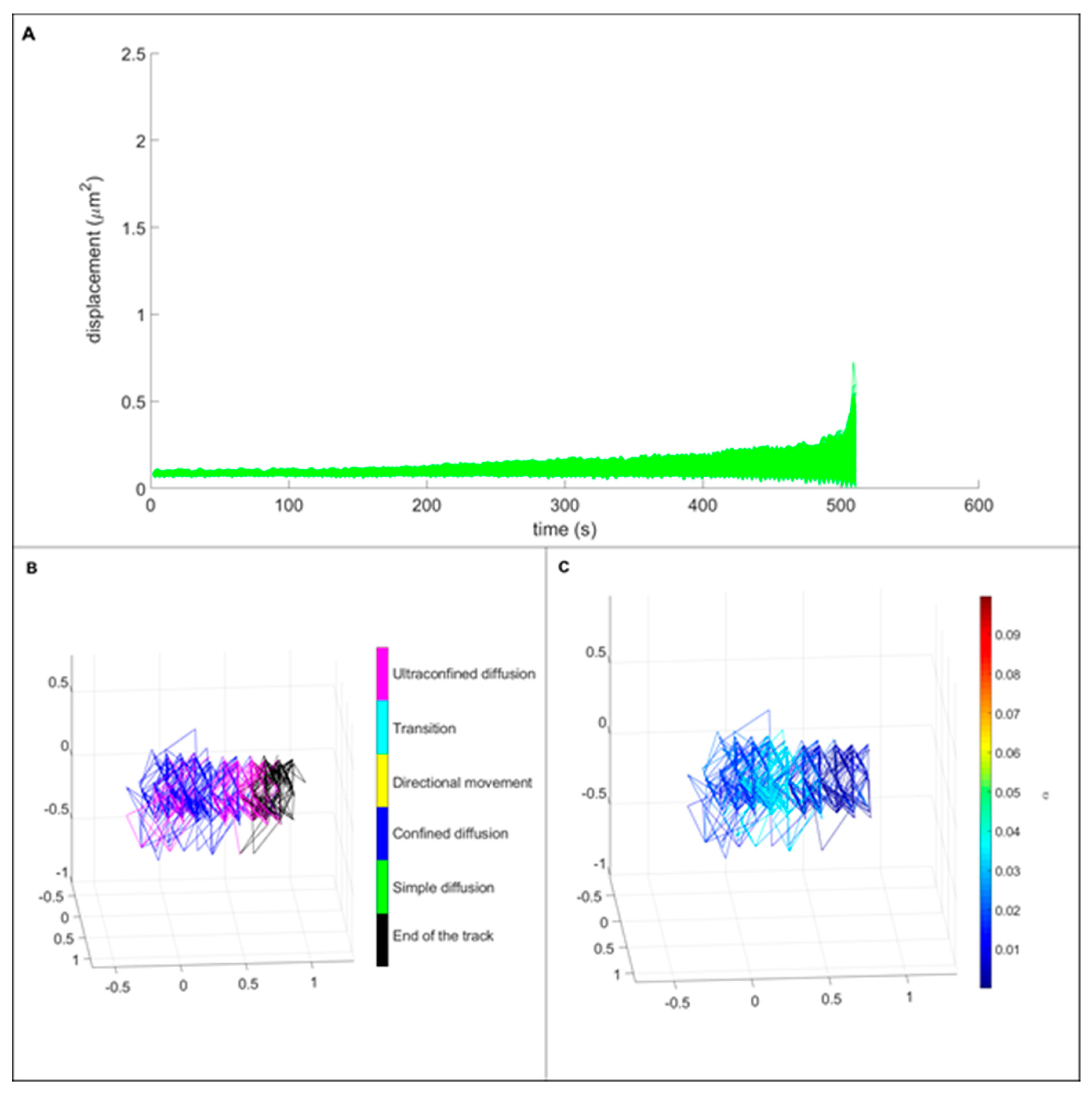
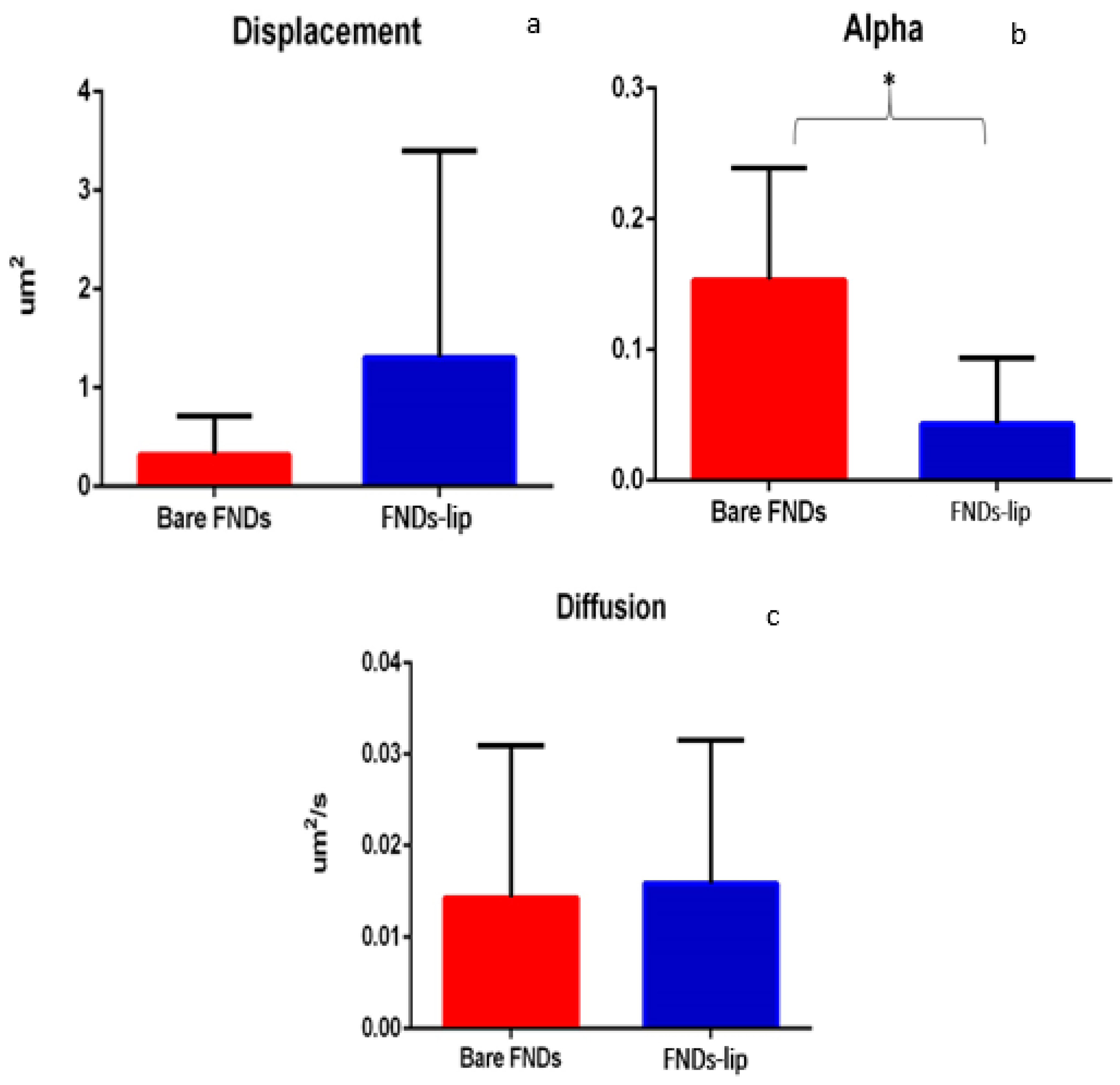
3.3. Single Particle Tracking
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Balasubramanian, G.; Chan, I.Y.; Kolesov, R.; Al-Hmoud, M.; Tisler, J.; Shin, C.; Kim, C.; Wojcik, A.; Hemmer, P.R.; Krueger, A.; et al. Nanoscale imaging magnetometry with diamond spins under ambient conditions. Nature 2008, 455, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Bradac, C.; Gaebel, T.; Naidoo, N.; Sellars, M.J.; Twamley, J.; Brown, L.J.; Barnard, A.S.; Plakhotnik, T.; Zvyagin, A.V.; Rabeau, J.R. Observation and control of blinking nitrogen-vacancy centres in discrete nanodiamonds. Nat. Nanotechnol. 2010, 5, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Dolde, F.; Fedder, H.; Doherty, M.W.; Nobauer, T.; Rempp, F.; Balasubramanian, G.; Wolf, T.; Reinhard, F.; Hollenberg, L.C.L.; Jelezko, F.; et al. Electric-field sensing using single diamond spins. Lett. Nat. Phys. 2011, 7, 459–463. [Google Scholar] [CrossRef]
- Laube, C.; Oeckinghaus, T.; Lehnert, J.; Griebel, J.; Knolle, W.; Denisenko, A.; Kahnt, A.; Meijer, J.; Wrachtrup, J.; Abel, B. Controlling the fluorescence properties of nitrogen vacancy centers in nanodiamonds. Nanoscale 2019, 11, 1770–1783. [Google Scholar] [CrossRef]
- Hickey, D.P.; Jones, K.S.; Elliman, R.G. Amorphization and graphitization of single-crystal diamond—A transmission electron microscopy study. Diam. Relat. Mater. 2009, 18, 1353–1359. [Google Scholar] [CrossRef]
- Bosia, F.; Argiolas, N.; Bazzan, M.; Olivero, P.; Picollo, F.; Sordini, A.; Vannoni, M.; Vittone, E. Modification of the structure of diamond with MeV ion implantation. Diam. Relat. Mater. 2011, 20, 774–778. [Google Scholar] [CrossRef]
- Dantelle, G.; Slablab, A.; Rondin, L.; Laine, F.; Carrel, F.; Bergonzo, P.; Perruchas, S.; Gacoin, T.; Treussart, F.; Roch, J.-F. Efficient production of NV colour centres in nanodiamonds using high-energy electron irradiation. J. Lumin. 2010, 130, 1655–1658. [Google Scholar] [CrossRef]
- Boudou, J.; Curmi, P.A.; Jelezko, F.; Wrachtrup, J.; Aubert, P.; Sennour, M.; Balasubramanian, G.; Reuter, R.; Thorel, A.; Gaffet, E. High yield fabrication of fluorescent nanodiamonds. Nanotechnology 2009, 20, 1–11. [Google Scholar] [CrossRef]
- Chipaux, M.; van der Laan, K.J.; Hemelaar, S.R.; Hasani, M.; Zheng, T.; Schirhagl, R. Nanodiamonds and their Applications in cells. Small 2018, 14, 1704263. [Google Scholar] [CrossRef]
- Mitura, K.A.; Włodarczyk, E. Fluorescent nanodiamonds in biomedical applications. J. AOAC Int. 2018, 101, 1297–1307. [Google Scholar] [CrossRef]
- Hemelaar, S.R.; Saspaanithy, B.; L’Hommelet, S.R.M.; Perona Martinez, F.P.; van der Laan, K.J.; Schirhagl, R. The response of HeLa cells to fluorescent nanodiamond uptake. Sensors 2018, 18, 355. [Google Scholar] [CrossRef] [PubMed]
- Vaijayanthimala, V.; Tzeng, Y.; Chang, H.; Li, C. The biocompatibility of fluorescent nanodiamonds and their mechanism of cellular uptake. Nanotechnology 2009, 20, 425103. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Miu, K.; Lung, P.; Zhang, S.; Zhao, S.; Chang, H.; Lin, G.; Li, Q. Rapid endosomal escape of prickly nanodiamonds: Implications for gene delivery. Sci. Rep. 2015, 5, 1161. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Vaijayanthimala, V.; Cheng, C.; Yeh, S.; Chang, C.; Li, C.; Chang, H. The Exocytosis of fluorescent nanodiamond and its use as a long-term cell tracker. Small 2011, 7, 3363–3370. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Wang, C.; Cheng, C.; Chao, J. Endocytic carboxylated nanodiamond for the labeling and tracking of cell division and differentiation in cancer and stem cells. Biomaterials 2009, 30, 4249–4259. [Google Scholar] [CrossRef]
- Hemelaar, S.R.; van der Laan, K.J.; Hinterding, S.R.; Koot, M.V.; Ellermann, E.; Perona-Martinez, F.P.; Roig, D.; Hommelet, S.; Novarina, D.; Takahashi, H.; et al. Generally applicable transformation protocols for fluorescent nanodiamond internalization into cells. Sci. Rep. 2017, 7, 5862. [Google Scholar] [CrossRef]
- van der Laan, K.J.; Naulleau, J.; Damle, V.G.; Sigaeva, A.; Jamot, N.; Perona-Martinez, F.P.; Chipaux, M.; Schirhagl, R. Towards using fluorescent nanodiamonds to study chronological ageing in Saccharomyces cerevisiae. Anal. Chem. 2018, 90, 13506–13513. [Google Scholar] [CrossRef]
- Morita, A.; Martinez, F.P.P.; Chipaux, M.; Jamot, N.; Hemelaar, S.R.; van der Laan, K.J.; Schirhagl, R. Cell uptake of lipid-coated diamond. Part. Part. Syst. Charact. 2019, 36, 1900116. [Google Scholar] [CrossRef]
- Neiman, A.M. Sporulation in budding yeast. Genetics 2011, 189, 737–765. [Google Scholar] [CrossRef]
- Baldi, S.; Bolognesi, A.; Meinema, A.C.; Barral, Y. Heat stress promotes longevity in budding yeast by relaxing the confinement of age-promoting factors in the mother cell. Elife 2017, 6, e28329. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Burtner, C.R.; Kennedy, B.K. Recent developments in yeast aging. PLoS Genet. 2007, 3, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Qiu, C.; Shen, Y.; Li, H.; Bao, X. Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 2017, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Walker, G.; Hill, A. Saccharomyces cerevisiae in the Production of Whisk(e)y. Beverages 2016, 2, 38. [Google Scholar] [CrossRef]
- Karas, B.J.; Jablanovic, J.; Irvine, E.; Sun, L.; Ma, L.; Weyman, P.D.; Gibson, D.G.; Glass, J.I.; Venter, J.C.; Hutchison, C.A., III; et al. Transferring whole genomes from bacteria to yeast spheroplasts using entire bacterial cells to reduce DNA shearing. Nat. Protoc. 2014, 9, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Hemelaar, S.R.; De Boer, P.; Chipaux, M.; Zuidema, W.; Hamoh, T.; Perona Martinez, F.; Nagl, A.; Hoogenboom, J.P.; Giepmans, B.N.G.; Schirhagl, R. Nanodiamonds as multi-purpose labels for microscopy. Sci. Rep. 2017, 7, 720. [Google Scholar] [CrossRef]
- Shenderova, O.A.; Shames, A.I.; Nunn, N.A.; Torelli, M.D.; Vlasov, I.; Zaitsev, A. Synthesis, properties, and applications of fluorescent diamond particles. J. Vac. Sci. Technol. B 2019, 37, 030802. [Google Scholar] [CrossRef]
- Fu, C.; Lee, H.; Chen, K.; Lim, T.; Wu, H.; Lin, P.; Wei, P.; Tsao, P.; Chang, H.; Fann, W. Characterization and application of single fluorescent nanodiamonds as cellular biomarkers. Proc. Natl. Acad. Sci. USA 2007, 104, 727–732. [Google Scholar] [CrossRef]
- Rondin, L.; Dantelle, G.; Slablab, A.; Grosshans, F.; Treussart, F.; Bergonzo, P.; Perruchas, S.; Gacoin, T.; Chaigneau, M.; Chang, H.-C.; et al. Surface-induced charge state conversion of nitrogen-vacancy defects in nanodiamonds. Phys. Rev. B 2010, 82, 115449. [Google Scholar] [CrossRef]
- Van der Laan, K.; Hasani, M.; Zheng, T.; Schirhagl, R. Nanodiamonds for In Vivo Applications. Small 2018, 1703838, e1703838. [Google Scholar] [CrossRef]
- Loretz, M.; Pezzagna, S.; Meijer, J.; Degen, C.L. Nanoscale nuclear magnetic resonance with a 1.9-nm-deep nitrogen-vacancy sensor. Appl. Phys. Lett. 2014, 104, 033102. [Google Scholar] [CrossRef]
- Zheng, T.; Perona Martínez, F.; Storm, I.M.; Rombouts, W.; Sprakel, J.; Schirhagl, R.; de Vries, R. Recombinant Protein Polymers for Colloidal Stabilization and Improvement of Cellular Uptake of Diamond Nanosensors. Anal. Chem. 2017, 89, 12812–12820. [Google Scholar] [CrossRef] [PubMed]
- Torrano, A.A.; Blechinger, J.; Osseforth, C.; Argyo, C.; Reller, A.; Bein, T.; Michaelis, J.; Brauchle, C. A fast analysis method to quantify nanoparticle uptake on a single cell level. Nanomedicine 2013, 8, 1815–1828. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, R. Extensions of DAMAS and Benefits and Limitations of Deconvolution in Beamforming. In Proceedings of the 11th AIAA/CEAS Aeroacoustics Conference, Monterey, CA, USA, 23–25 May 2005. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhu, X.; Wang, X.; Yuan, R.; Zheng, W.; Xu, M.; Ao, P. Two programmed replicative lifespans of Saccharomyces cerevisiae formed by the endogenous molecular-cellular network. J. Theor. Biol. 2014. [Google Scholar] [CrossRef]
- Velasco, I.; Tenreiro, S.; Calderon, I.L.; Andre, B. Saccharomyces cerevisiae Aqr1 Is an Internal-Membrane Transporter Involved in Excretion of Amino Acids. Eukaryot. Cell 2004, 3, 1492–1503. [Google Scholar] [CrossRef]
- Jorgensen, P.; Nishikawa, J.L.; Breitkreutz, B.; Tyers, M. Systematic identification of pathways that couple cell growth and division in yeast. Science 2002, 297, 1070850. [Google Scholar] [CrossRef]
- Andrade-Restrepo, M. Is aggregate-dependent yeast aging fortuitous? A model of damage segregation and aggregate dynamics. Biophys. J. 2017, 113, 2464–2476. [Google Scholar] [CrossRef][Green Version]
- Paoletti, C.; Quintin, S.; Matifas, A.; Charvin, G. Kinetics of formation and asymmetrical distribution of Hsp104-bound protein aggregates in yeast. Biophys. J. 2016, 110, 1605–1614. [Google Scholar] [CrossRef][Green Version]
- Liu, B.; Larsson, L.; Caballero, A.; Hao, X.; Oling, D.; Grantham, J.; Nystrom, T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell 2010, 140, 257–267. [Google Scholar] [CrossRef]
- Spokoini, R.; Moldavski, O.; Nahmias, Y.; England, J.L.; Schuldiner, M.; Kaganovich, D. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012, 2, 738–747. [Google Scholar] [CrossRef]
- Zhou, C.; Slaughter, B.D.; Unruh, J.R.; Guo, F.; Yu, Z.; Mickey, K.; Narkar, A.; Ross, R.T.; McClain, M.; Li, R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell 2014, 159, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-peña, J.M.; Diez-Muniz, S.; Bermejo, C.; Nombela, C.; Arroyo, J. Activation of the yeast cell wall integrity MAPK pathway by zymolyase depends on protease and glucanase activities and requires the mucin-like protein Hkr1 but not Msb2. FEBS Lett. 2013, 587, 3675–3680. [Google Scholar] [CrossRef]
- Lesage, G.; Bussey, H. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morita, A.; Hamoh, T.; Perona Martinez, F.P.; Chipaux, M.; Sigaeva, A.; Mignon, C.; van der Laan, K.J.; Hochstetter, A.; Schirhagl, R. The Fate of Lipid-Coated and Uncoated Fluorescent Nanodiamonds during Cell Division in Yeast. Nanomaterials 2020, 10, 516. https://doi.org/10.3390/nano10030516
Morita A, Hamoh T, Perona Martinez FP, Chipaux M, Sigaeva A, Mignon C, van der Laan KJ, Hochstetter A, Schirhagl R. The Fate of Lipid-Coated and Uncoated Fluorescent Nanodiamonds during Cell Division in Yeast. Nanomaterials. 2020; 10(3):516. https://doi.org/10.3390/nano10030516
Chicago/Turabian StyleMorita, Aryan, Thamir Hamoh, Felipe P. Perona Martinez, Mayeul Chipaux, Alina Sigaeva, Charles Mignon, Kiran J. van der Laan, Axel Hochstetter, and Romana Schirhagl. 2020. "The Fate of Lipid-Coated and Uncoated Fluorescent Nanodiamonds during Cell Division in Yeast" Nanomaterials 10, no. 3: 516. https://doi.org/10.3390/nano10030516
APA StyleMorita, A., Hamoh, T., Perona Martinez, F. P., Chipaux, M., Sigaeva, A., Mignon, C., van der Laan, K. J., Hochstetter, A., & Schirhagl, R. (2020). The Fate of Lipid-Coated and Uncoated Fluorescent Nanodiamonds during Cell Division in Yeast. Nanomaterials, 10(3), 516. https://doi.org/10.3390/nano10030516






