Adhesion of Escherichia Coli to Nanostructured Surfaces and the Role of Type 1 Fimbriae
Abstract
1. Introduction
2. Materials and Methods
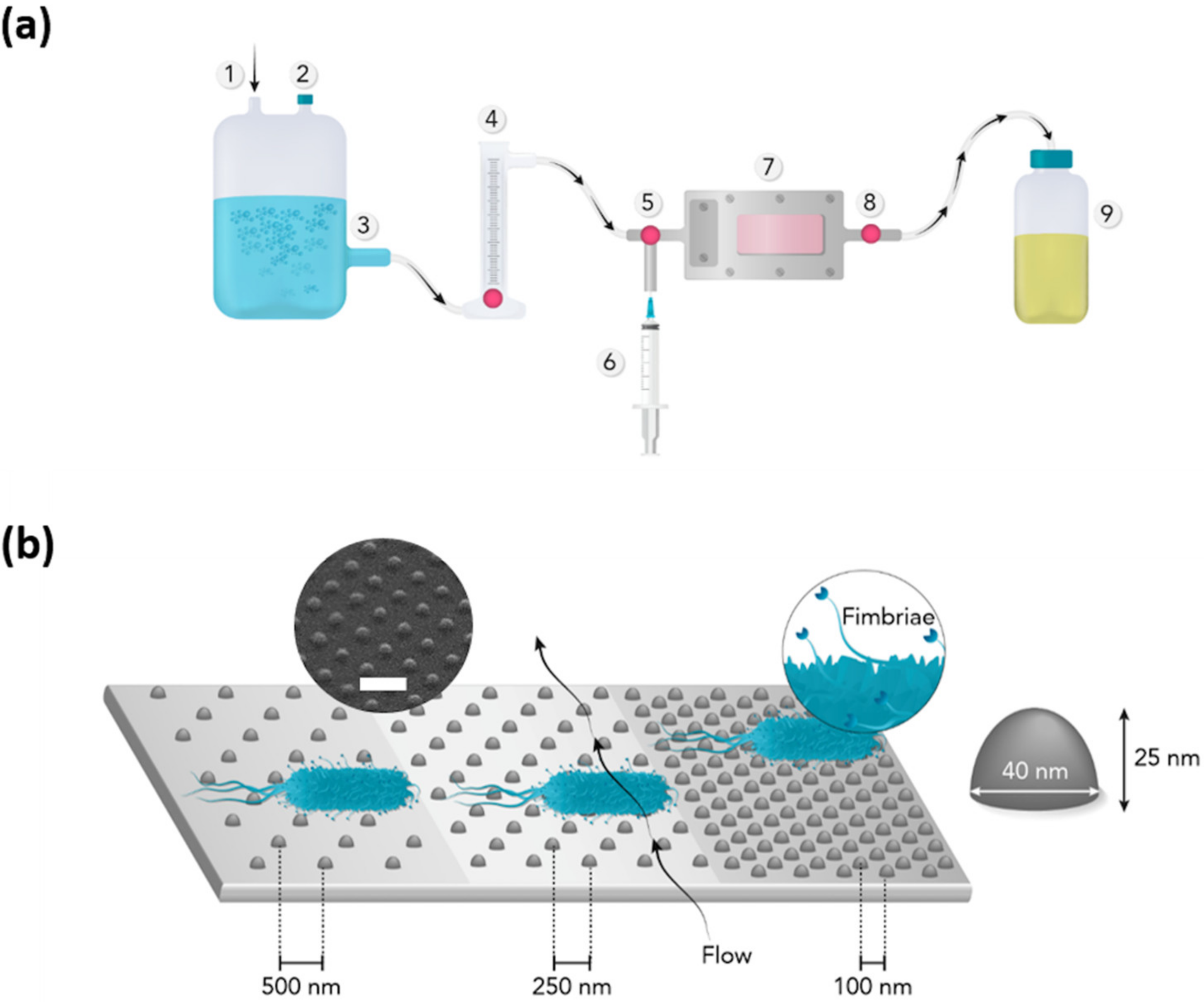
2.1. Preparation of Nanostructured Surfaces
2.2. Surface Characterization
2.3. Bacterial Preparation and Growth
2.4. Bacterial Adhesion
2.5. Fluorescence Microscopy
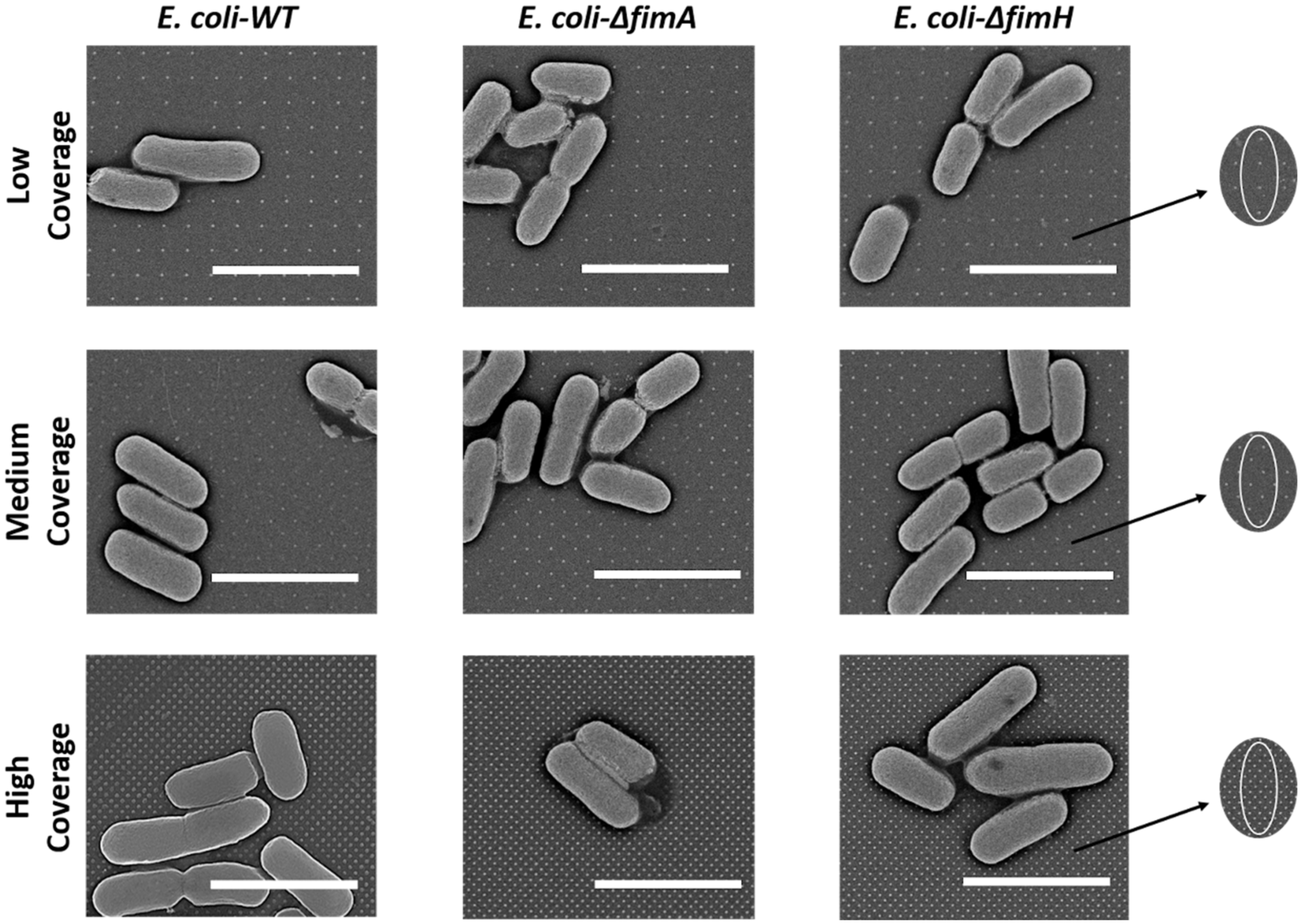
2.6. Scanning Electron Microscopy
2.7. ImageJ Analysis
2.8. Statistical Analysis
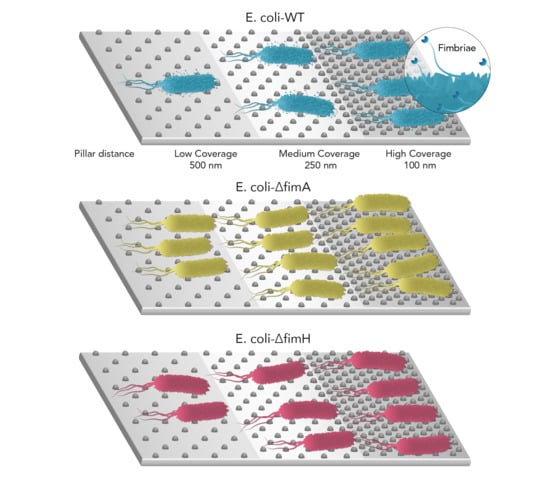
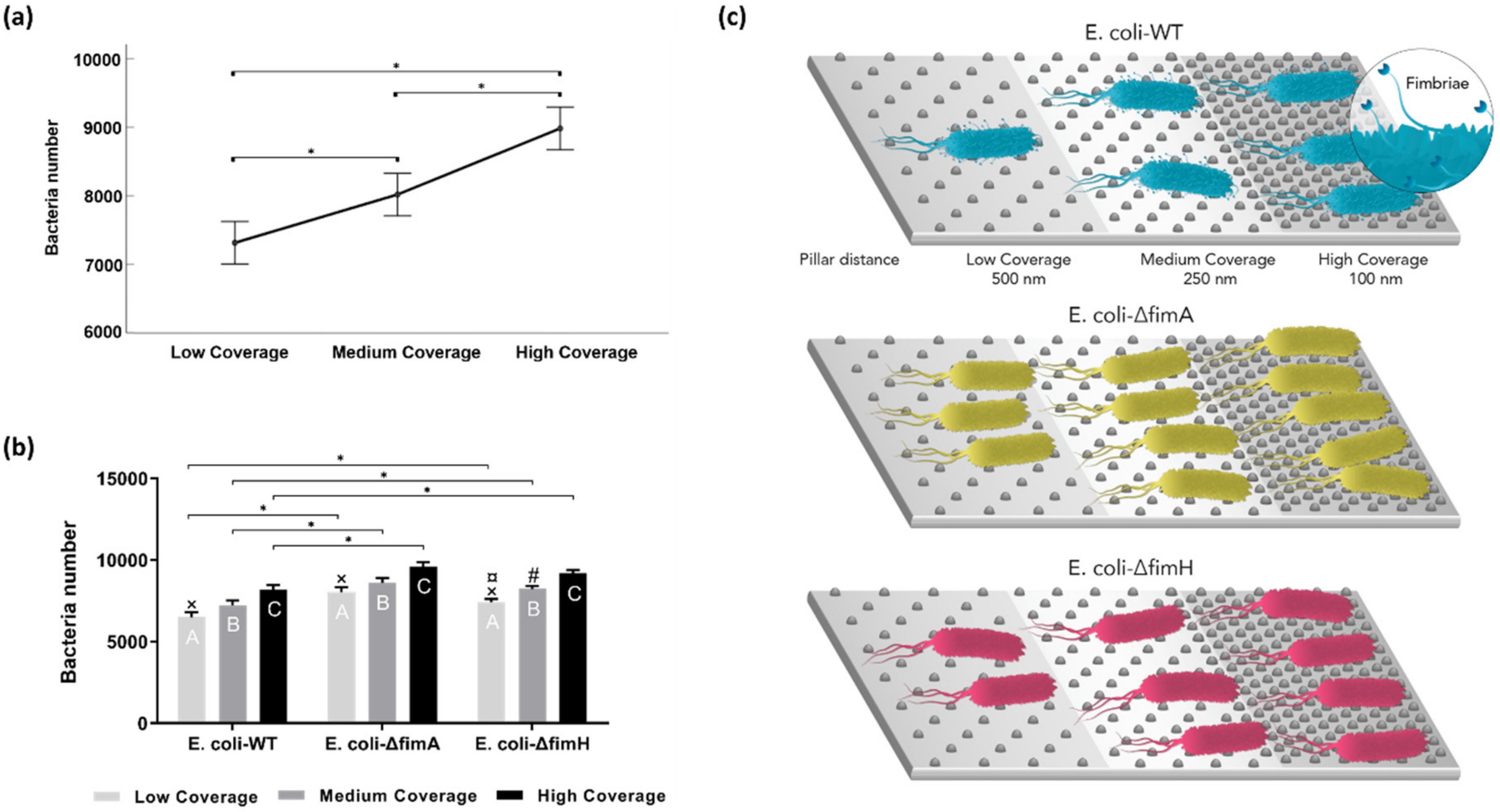
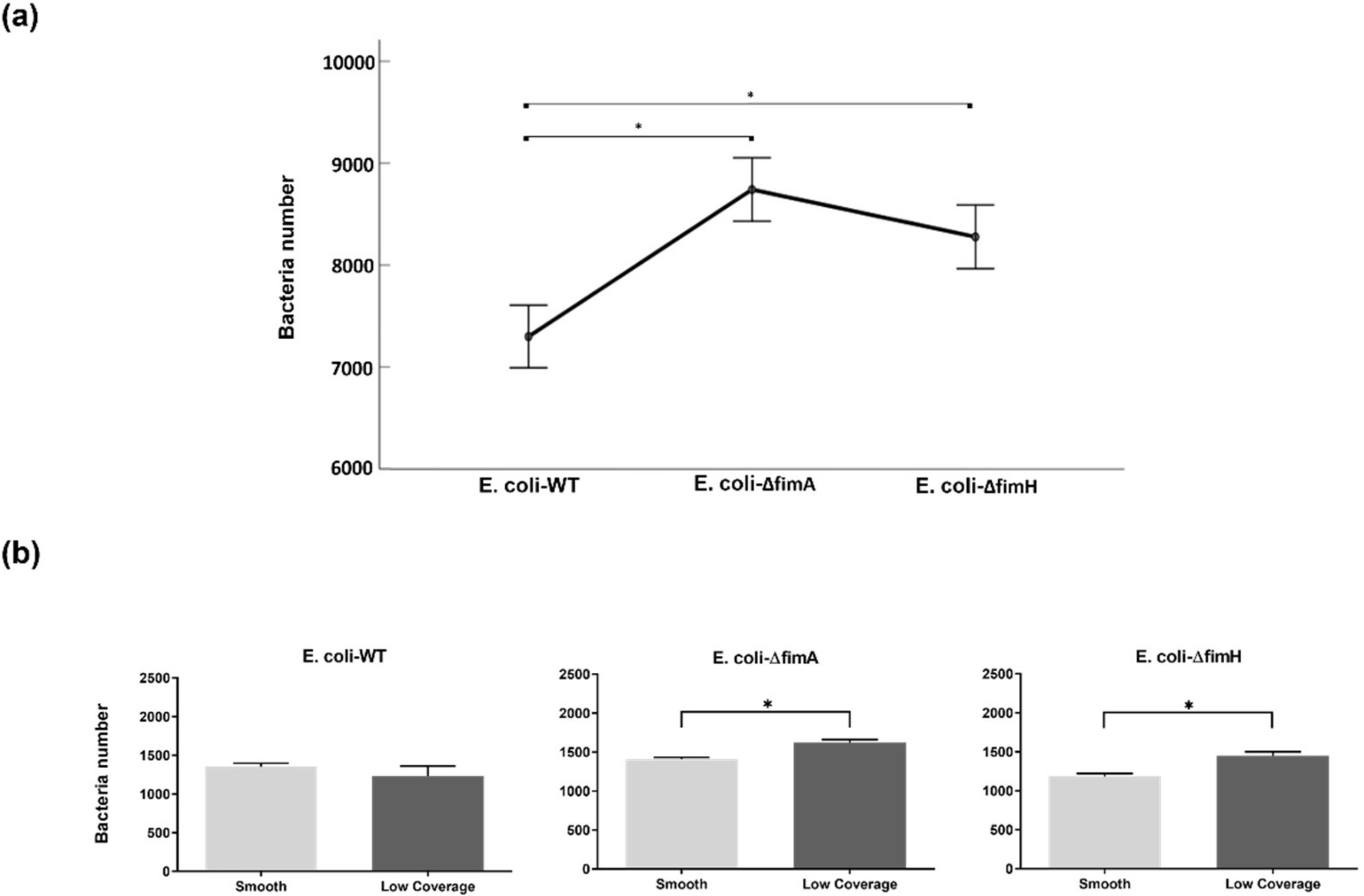
3. Results
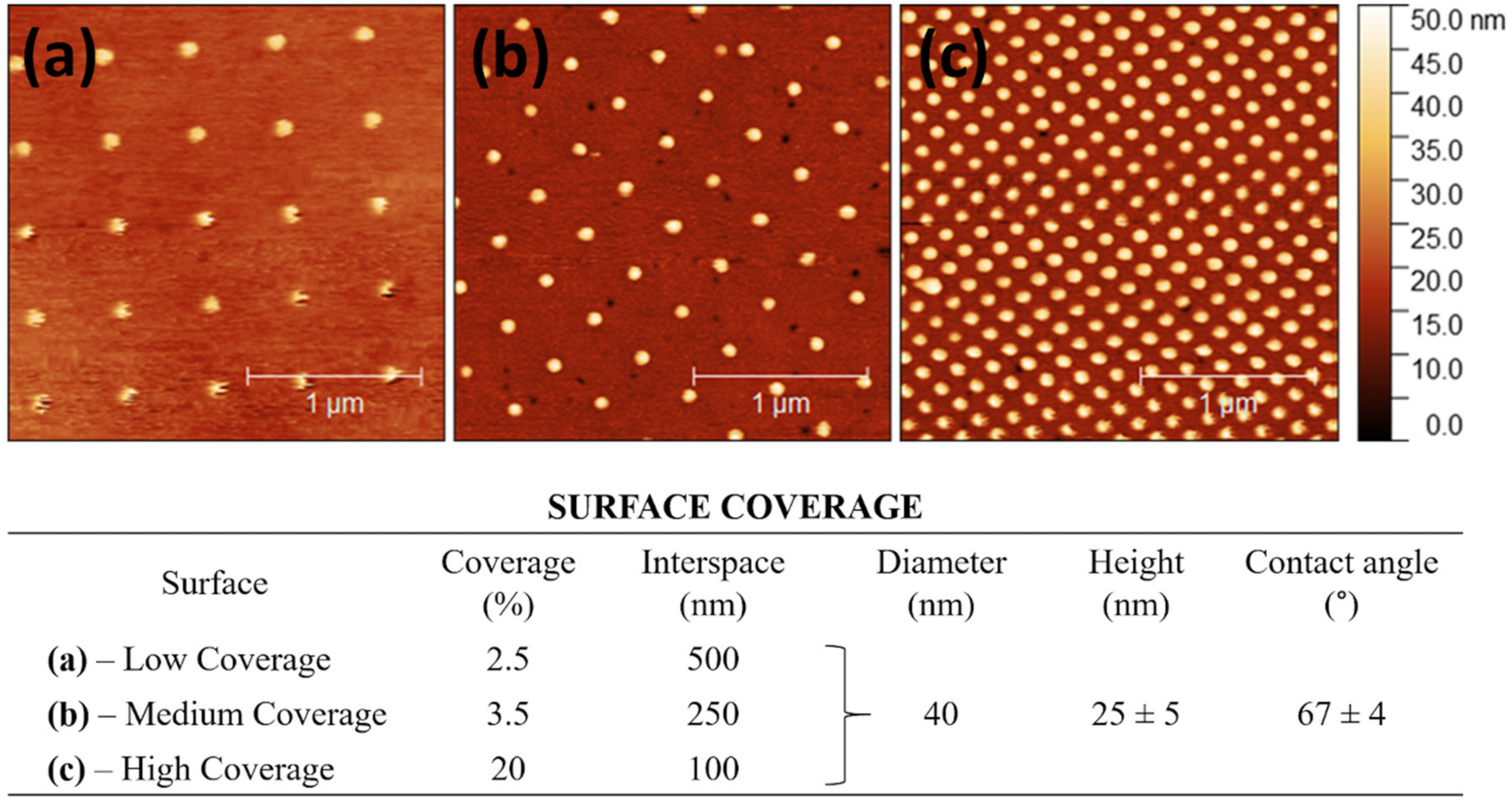
3.1. Surface Characteristics
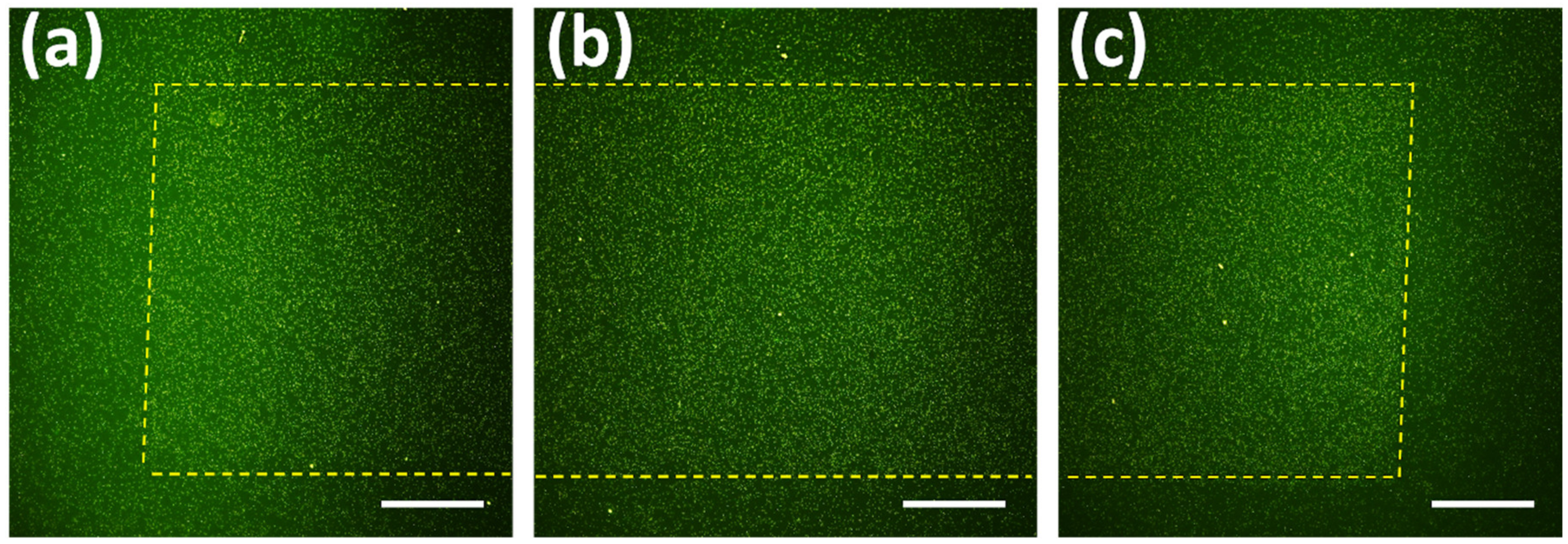
3.2. Bacterial Adhesion
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian. Pac. J. Trop. Bio. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Inweregbu, K.; Dave, J.; Pittard, A. Nosocomial infections. Cont. Care Pain 2005, 5, 14–17. [Google Scholar] [CrossRef]
- Darouiche, R.O. Device-associated infections: A macroproblem that starts with microadherence. Clin. Infect. Dis. 2001, 33, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Afle, F.C.D.; Agbankpe, A.J.; Johnson, R.C.; Houngbegnon, O.; Houssou, S.C.; Bankole, H.S. Healthcare-associated infections: Bacteriological characterization of the hospital surfaces in the University Hospital of Abomey-Calavi/so-ava in South Benin (West Africa). BMC. Infect. Dis. 2019, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, R.; Murt, A. Epidemiology of urological infections: A global burden. World J. of Uro. 2020. [Google Scholar] [CrossRef]
- Friedman, C.; Newsom, W. IFIC Basic Concepts of Infection Control; International Federation of Infection Control: Portadown, UK, 2011; p. 259. [Google Scholar]
- Campbell, N.A.; Reece, J.B. Biology: International Edition, 6th ed.; Pearson Education, Inc.: London, UK, 2002; pp. 595–615, 6th ed. [Google Scholar]
- Salyers, A.A.; Whitt, D.D.; Whitt, D.D. Bacterial Pathogenesis: A Molecular Approach, 1st ed.; ASM press: Washington, DC, USA, 1994; p. 190, 1st ed. [Google Scholar]
- Stecher, B.; Hardt, W.D. The role of microbiota in infectious disease. Trends Microbiol. 2008, 16, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Moseley, S.L.; Roberts, P.L.; Stamm, W.E. Aerobactin and other virulence factor genes among strains of Escherichia coli causing urosepsis: Association with patient characteristics. Infect. Immun. 1988, 56, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.J.; Mulvey, M.A.; Schilling, J.D.; Pinkner, J.S.; Hultgren, S.J. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO. J. 2000, 19, 2803–2812. [Google Scholar] [CrossRef]
- Klemm, P.; Krogfelt, K.A.; Hedegaard, L.; Christiansen, G. The major subunit of Escherichia coli type 1 fimbriae is not required for D-mannose-specific adhesion. Mol. Microbiol. 1990, 4, 553–559. [Google Scholar] [CrossRef]
- Russell, P.W.; Orndorff, P.E. Lesions in two Escherichia coli type 1 pilus genes alter pilus number and length without affecting receptor binding. J. Bacteriol. 1992, 174, 5923–5935. [Google Scholar] [CrossRef]
- Jones, C.H.; Pinkner, J.S.; Roth, R.; Heuser, J.; Nicholes, A.V.; Abraham, S.N.; Hultgren, S.J. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 1995, 92, 2081–2085. [Google Scholar] [CrossRef] [PubMed]
- Blumer, C.; Kleefeld, A.; Lehnen, D.; Heintz, M.; Dobrindt, U.; Nagy, G.; Michaelis, K.; Emödy, L.; Polen, T.; Rachel, R.; et al. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology (Reading) 2005, 151, 3287–3298. [Google Scholar] [CrossRef] [PubMed]
- Reisner, A.; Maierl, M.; Jorger, M.; Krause, R.; Berger, D.; Haid, A.; Tesic, D.; Zechner, E.L. Type 1 fimbriae contribute to catheter-associated urinary tract infections caused by Escherichia coli. J. Bacteriol. 2014, 196, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms and device-associated infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Valen, H.; Scheie, A.A. Biofilms and their properties. Eur. J. Oral Sci. 2018, 126, 13–18. [Google Scholar] [CrossRef]
- Kalivoda, E.J.; Stella, N.A.; O’Dee, D.M.; Nau, G.J.; Shanks, R.M. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl. Environ. Microbiol. 2008, 74, 3461–3470. [Google Scholar] [CrossRef]
- Stahlhut, S.G.; Struve, C.; Krogfelt, K.A.; Reisner, A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol. Med. Microbiol. 2012, 65, 350–359. [Google Scholar] [CrossRef]
- Platt, R.; Polk, B.F.; Murdock, B.; Rosner, B. Mortality associated with nosocomial urinary-tract infection. N. Engl. J. Med. 1982, 307, 637–642. [Google Scholar] [CrossRef]
- Lee, D.S.; Lee, S.J.; Choe, H.S. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. Biomed. Res. Int. 2018, 2018, 7656752. [Google Scholar] [CrossRef]
- Alanazi, M.Q.; Alqahtani, F.Y.; Aleanizy, F.S. An evaluation of E. coli in urinary tract infection in emergency department at KAMC in Riyadh, Saudi Arabia: Retrospective study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 3. [Google Scholar] [CrossRef]
- Garofalo, C.K.; Hooton, T.M.; Martin, S.M.; Stamm, W.E.; Palermo, J.J.; Gordon, J.I.; Hultgren, S.J. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun. 2007, 75, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Marrs, C.F.; Zhang, L.; Foxman, B. Escherichia coli mediated urinary tract infections: Are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol. Lett. 2005, 252, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Connell, I.; Agace, W.; Klemm, P.; Schembri, M.; Marild, S.; Svanborg, C. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc. Natl. Acad. Sci. USA 1996, 93, 9827–9832. [Google Scholar] [CrossRef] [PubMed]
- Mozetic, M. Surface Modification to Improve Properties of Materials. Materials (Basel) 2019, 12, 441. [Google Scholar] [CrossRef]
- Rahmati, M.; Silva, E.A.; Reseland, J.E.; C, A.H.; Haugen, H.J. Biological responses to physicochemical properties of biomaterial surface. Chem. Soc. Rev. 2020, 49, 5178–5224. [Google Scholar] [CrossRef]
- Tripathy, A.; Sen, P.; Su, B.; Briscoe, W.H. Natural and bioinspired nanostructured bactericidal surfaces. Adv. Colloid Interface Sci. 2017, 248, 85–104. [Google Scholar] [CrossRef]
- Øilo, M.; Bakken, V. Biofilm and Dental Biomaterials. Materials 2015, 8, 2887–2900. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef]
- Xing, R.; Lyngstadaas, S.P.; Ellingsen, J.E.; Taxt-Lamolle, S.; Haugen, H.J. The influence of surface nanoroughness, texture and chemistry of TiZr implant abutment on oral biofilm accumulation. Clin. Oral Implants Res. 2015, 26, 649–656. [Google Scholar] [CrossRef]
- Haugen, H.; Lyngstadaas, S. Antibacterial effects of titanium dioxide in wounds. In Wound Healing Biomaterials-Volume 2: Functional Biomaterials; Ågren, M.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; p. 439. [Google Scholar]
- Dhull, N.; Nidhi; Gupta, V.; Tomar, M. Antimicrobial properties of metallic nanoparticles: A qualitative analysis. Mater. Today: Proc. 2019, 17, 155–160. [Google Scholar] [CrossRef]
- Smirnov, N.A.; Kudryashov, S.I.; Nastulyavichus, A.A.; Rudenko, A.A.; Saraeva, I.N.; Tolordava, E.R.; Gonchukov, S.A.; Romanova, Y.M.; Ionin, A.A.; Zayarny, D.A. Antibacterial properties of silicon nanoparticles. Laser Phys. Lett. 2018, 15, 105602. [Google Scholar] [CrossRef]
- Asefa, T.; Tao, Z. Biocompatibility of mesoporous silica nanoparticles. Chem. Res. Toxicol. 2012, 25, 2265–2284. [Google Scholar] [CrossRef] [PubMed]
- Sviridov, A.P.; Osminkina, L.A.; Kharin, A.Y.; Gongalsky, M.B.; Kargina, J.V.; Kudryavtsev, A.A.; Bezsudnova, Y.I.; Perova, T.S.; Geloen, A.; Lysenko, V.; et al. Cytotoxicity control of silicon nanoparticles by biopolymer coating and ultrasound irradiation for cancer theranostic applications. Nanotechnology 2017, 28, 105102. [Google Scholar] [CrossRef] [PubMed]
- Tamarov, K.P.; Osminkina, L.A.; Zinovyev, S.V.; Maximova, K.A.; Kargina, J.V.; Gongalsky, M.B.; Ryabchikov, Y.; Al-Kattan, A.; Sviridov, A.P.; Sentis, M.; et al. Radio frequency radiation-induced hyperthermia using Si nanoparticle-based sensitizers for mild cancer therapy. Sci. Rep. 2014, 4, 7034. [Google Scholar] [CrossRef]
- Charitidis, C.A.; Georgiou, P.; Koklioti, M.A.; Trompeta, A.-F.; Markakis, V. Manufacturing nanomaterials: From research to industry. Manufacturing Rev. 2014, 1, 11. [Google Scholar] [CrossRef]
- Altissimo, M. E-beam lithography for micro-nanofabrication. Biomicrofluidics 2010, 4, 026503. [Google Scholar] [CrossRef]
- Stormonth-Darling, J.M.; Gadegaard, N. Injection Moulding Difficult Nanopatterns with Hybrid Polymer Inlays. Macromol. Mater. Eng. 2012, 297, 1075–1080. [Google Scholar] [CrossRef]
- Chao, Y.; Zhang, T. Probing Roles of Lipopolysaccharide, Type 1 Fimbria, and Colanic Acid in the Attachment of Escherichia coli Strains on Inert Surfaces. Langmuir 2011, 27, 11545–11553. [Google Scholar] [CrossRef]
- Lönn-Stensrud, J.; Benneche, T.; Scheie, A.A. Furanones and Thiophenones in Control of Staphylococcus epidermidis Biofilm Infections? In Science and Technology Against Microbial Pathogens; World Scientific, Toh Tunk Link: Singapore, 2011; pp. 155–159. [Google Scholar] [CrossRef]
- Hulander, M.; Valen-Rukke, H.; Sundell, G.; Andersson, M. Influence of Fibrinogen on Staphylococcus epidermidis Adhesion Can Be Reversed by Tuning Surface Nanotopography. ACS Biomater. Sci. Eng. 2019, 5, 4323–4330. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Buchner, A.; Lang, A.G. Statistical power analyses using G*Power 3.1: Tests for correlation and regression analyses. Behav. Res. Methods 2009, 41, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Stormonth-Darling, J.M.; Pedersen, R.H.; How, C.; Gadegaard, N. Injection moulding of ultra high aspect ratio nanostructures using coated polymer tooling. J. Micromech. Microeng. 2014, 24, 12. [Google Scholar] [CrossRef]
- Darling, J.M.; Saeed, A.; Reynolds, P.M.; Gadegaard, N. Injection Molding Micro- and Nanostructures in Thermoplastic Elastomers. Macromol. Mater. Eng. 2016, 301, 964–971. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Cho, J.; Shivapooja, P.; Ista, L.K.; Lopez, G.P. Nanopatterned smart polymer surfaces for controlled attachment, killing, and release of bacteria. ACS Appl. Mater. Interfaces 2013, 5, 9295–9304. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.T.; Ercan, B.; Sun, L.L.; Ziemer, K.S.; Webster, T.J. Understanding the Role of Polymer Surface Nanoscale Topography on Inhibiting Bacteria Adhesion and Growth. ACS Biomater Sci. Eng. 2016, 2, 122–130. [Google Scholar] [CrossRef]
- Linklater, D.P.; Baulin, V.A.; Juodkazis, S.; Crawford, R.J.; Stoodley, P.; Ivanova, E.P. Mechano-bactericidal actions of nanostructured surfaces. Nat. Rev. Microbiol 2020, 1–15, Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Melican, K.; Sandoval, R.M.; Kader, A.; Josefsson, L.; Tanner, G.A.; Molitoris, B.A.; Richter-Dahlfors, A. Uropathogenic Escherichia coli P and Type 1 fimbriae act in synergy in a living host to facilitate renal colonization leading to nephron obstruction. PLoS. Pathog. 2011, 7, e1001298. [Google Scholar] [CrossRef]
- Klemm, P. Fimbrial adhesions of Escherichia coli. Rev. Infect. Dis. 1985, 7, 321–340. [Google Scholar] [CrossRef]
- Otto, K.; Elwing, H.; Hermansson, M. The role of type 1 fimbriae in adhesion of Escherichia coli to hydrophilic and hydrophobic surfaces. Colloids Surf. B-Biointerfaces 1999, 15, 99–111. [Google Scholar] [CrossRef]
- Widyaratih, D.S.; Hagedoorn, P.L.; Otten, L.G.; Ganjian, M.; Tumer, N.; Apachitei, I.; Hagen, C.W.; Fratila-Apachitei, L.E.; Zadpoor, A.A. Towards osteogenic and bactericidal nanopatterns? Nanotechnology 2019, 30, 20LT01. [Google Scholar] [CrossRef]
- Bandara, C.D.; Singh, S.; Afara, I.O.; Wolff, A.; Tesfamichael, T.; Ostrikov, K.; Oloyede, A. Bactericidal Effects of Natural Nanotopography of Dragonfly Wing on Escherichia coli. ACS Appl. Mater. Interfaces 2017, 9, 6746–6760. [Google Scholar] [CrossRef] [PubMed]
- Hasan, J.; Raj, S.; Yadav, L.; Chatterjee, K. Engineering a nanostructured "super surface" with superhydrophobic and superkilling properties. RSC Adv. 2015, 5, 44953–44959. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.N.; Liang, E.I.; Rodriguez, L.A.; Vollereaux, N.; Yee, A.F. Nanopatterned polymer surfaces with bactericidal properties. Biointerphases 2015, 10, 021010. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kallas, P.; Haugen, H.J.; Gadegaard, N.; Stormonth-Darling, J.; Hulander, M.; Andersson, M.; Valen, H. Adhesion of Escherichia Coli to Nanostructured Surfaces and the Role of Type 1 Fimbriae. Nanomaterials 2020, 10, 2247. https://doi.org/10.3390/nano10112247
Kallas P, Haugen HJ, Gadegaard N, Stormonth-Darling J, Hulander M, Andersson M, Valen H. Adhesion of Escherichia Coli to Nanostructured Surfaces and the Role of Type 1 Fimbriae. Nanomaterials. 2020; 10(11):2247. https://doi.org/10.3390/nano10112247
Chicago/Turabian StyleKallas, Pawel, Håvard J Haugen, Nikolaj Gadegaard, John Stormonth-Darling, Mats Hulander, Martin Andersson, and Håkon Valen. 2020. "Adhesion of Escherichia Coli to Nanostructured Surfaces and the Role of Type 1 Fimbriae" Nanomaterials 10, no. 11: 2247. https://doi.org/10.3390/nano10112247
APA StyleKallas, P., Haugen, H. J., Gadegaard, N., Stormonth-Darling, J., Hulander, M., Andersson, M., & Valen, H. (2020). Adhesion of Escherichia Coli to Nanostructured Surfaces and the Role of Type 1 Fimbriae. Nanomaterials, 10(11), 2247. https://doi.org/10.3390/nano10112247