A Convenient Colorimetric Bacteria Detection Method Utilizing Chitosan-Coated Magnetic Nanoparticles
Abstract
1. Introduction
2. Experimental Section
2.1. Materials
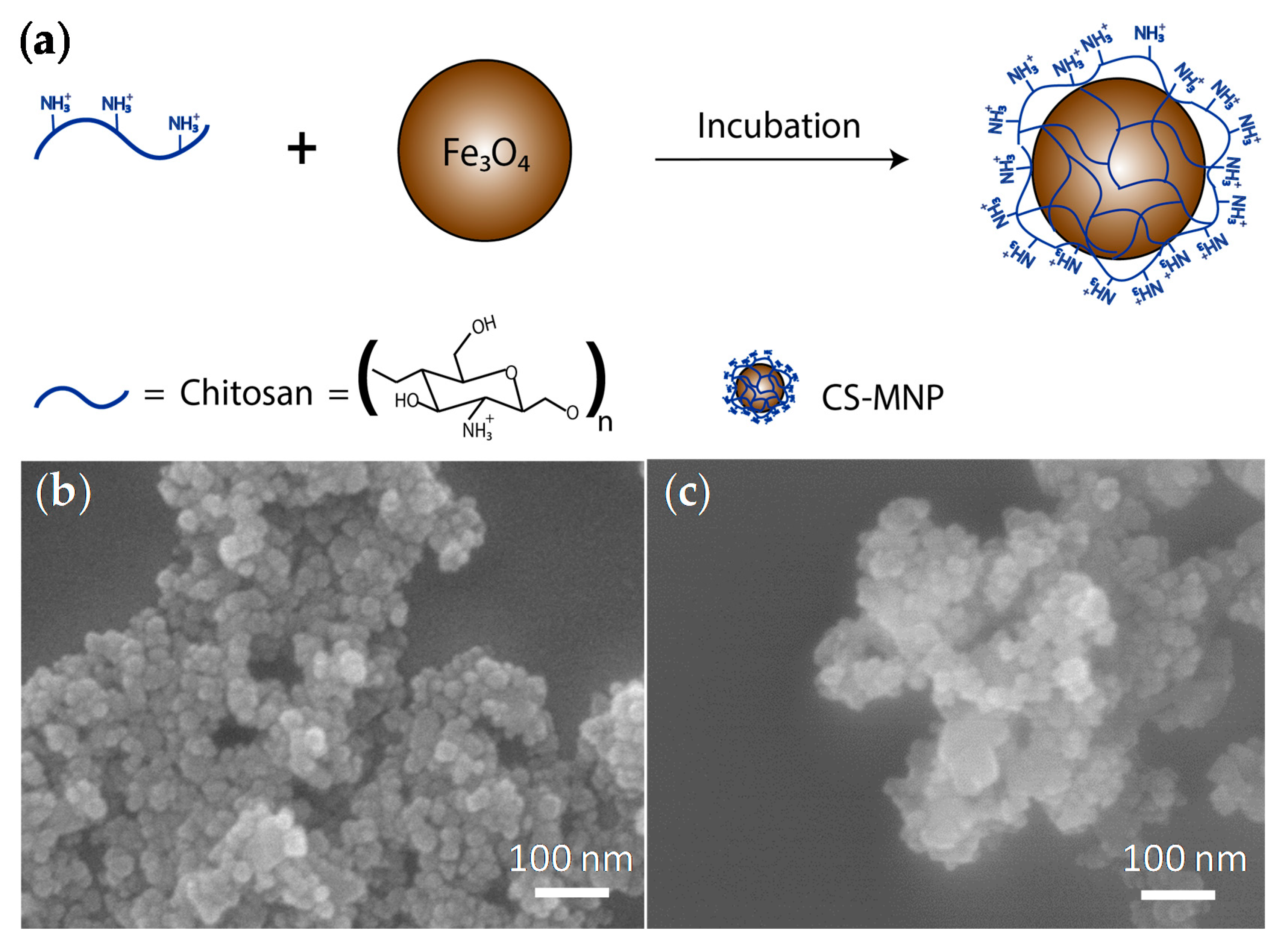
2.2. Synthesis of MNPs and CS-MNPs
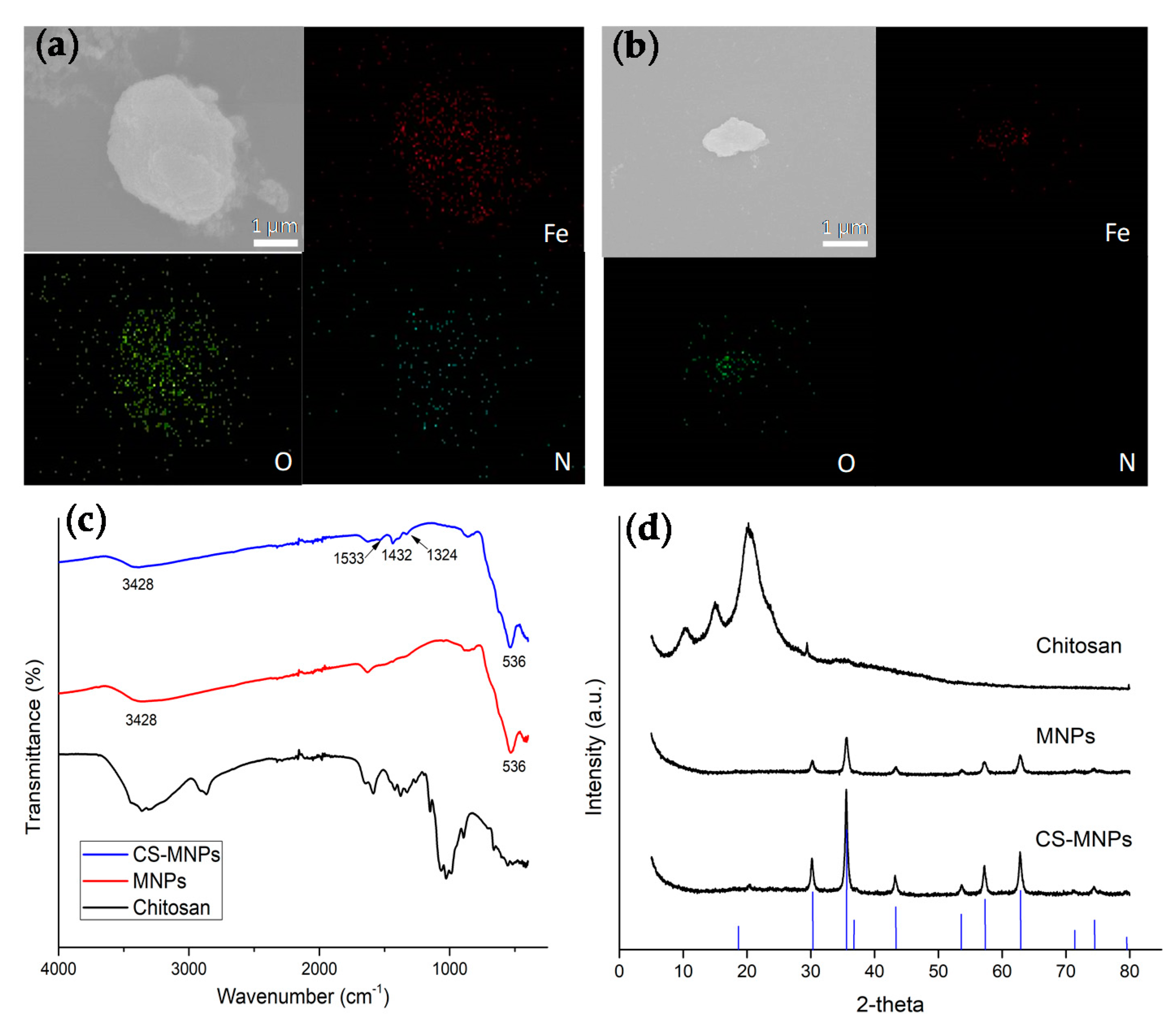
2.3. Characterization of MNPs, CS-MNPs, and Bacteria with Nanoparticles
2.4. Selected Bacteria and Culture Conditions
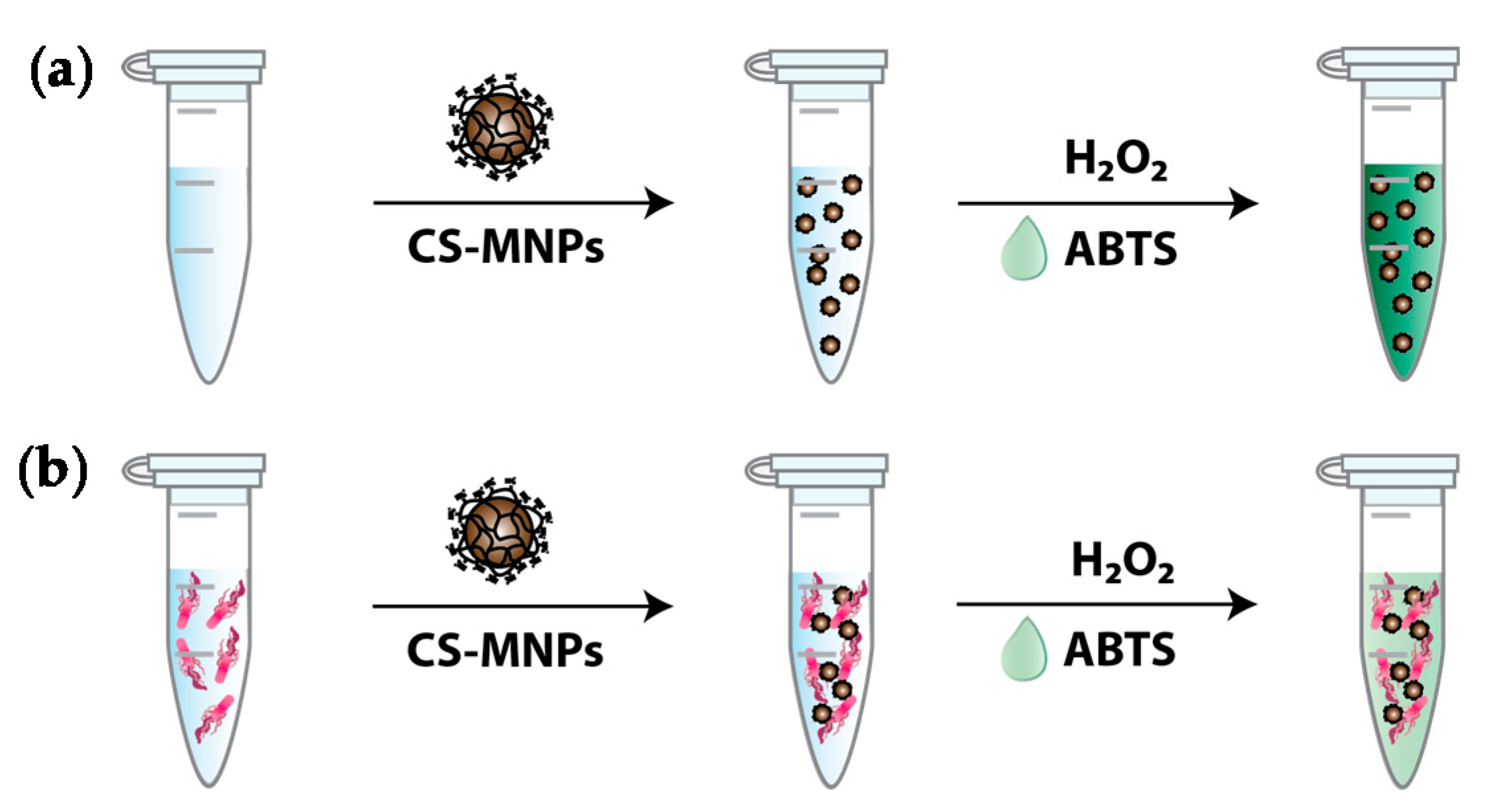
2.5. Colorimetric Bacteria Detection Using CS-MNPs
3. Results and Discussion
3.1. Preparation and Structural Characterization of MNPs and CS-MNPs
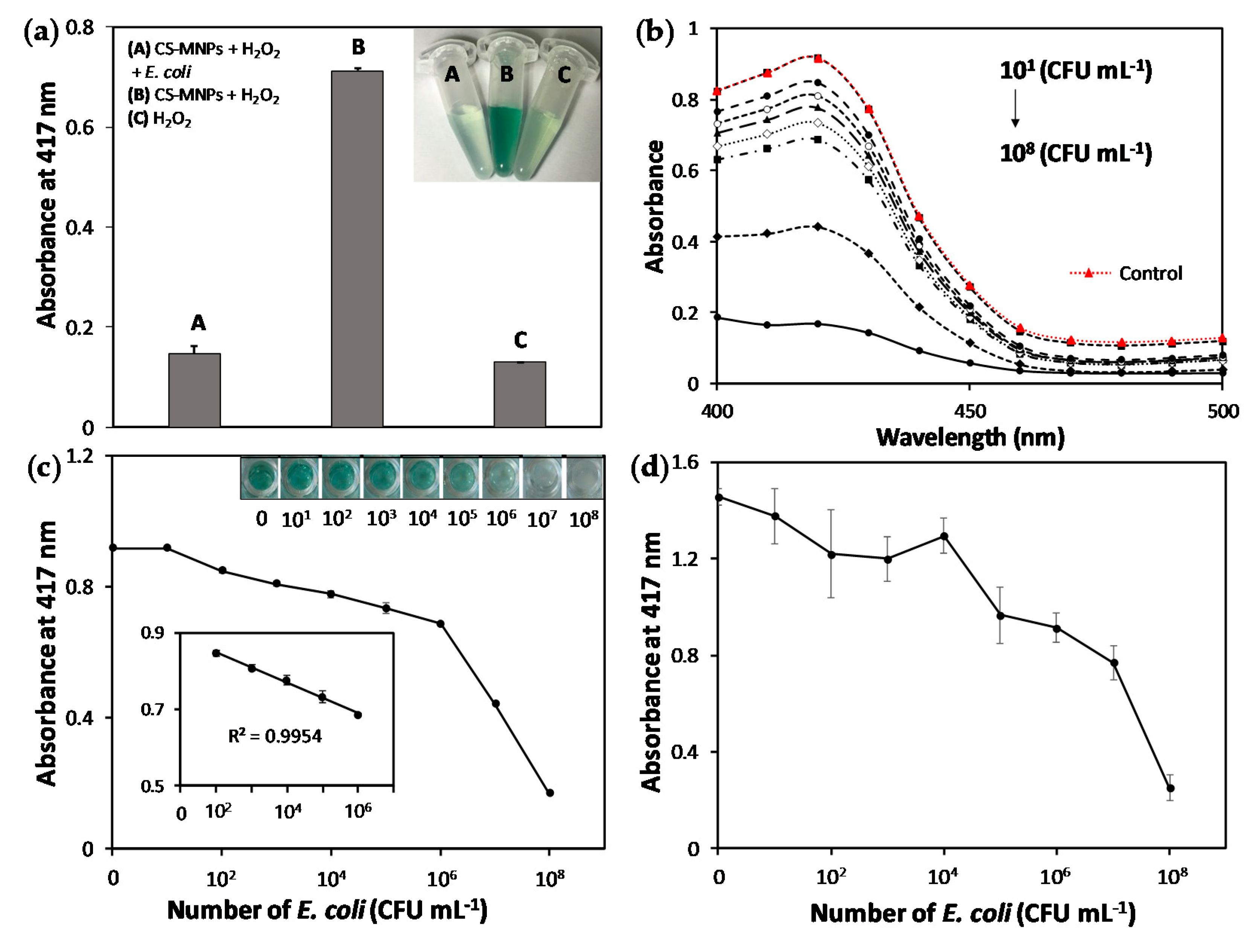
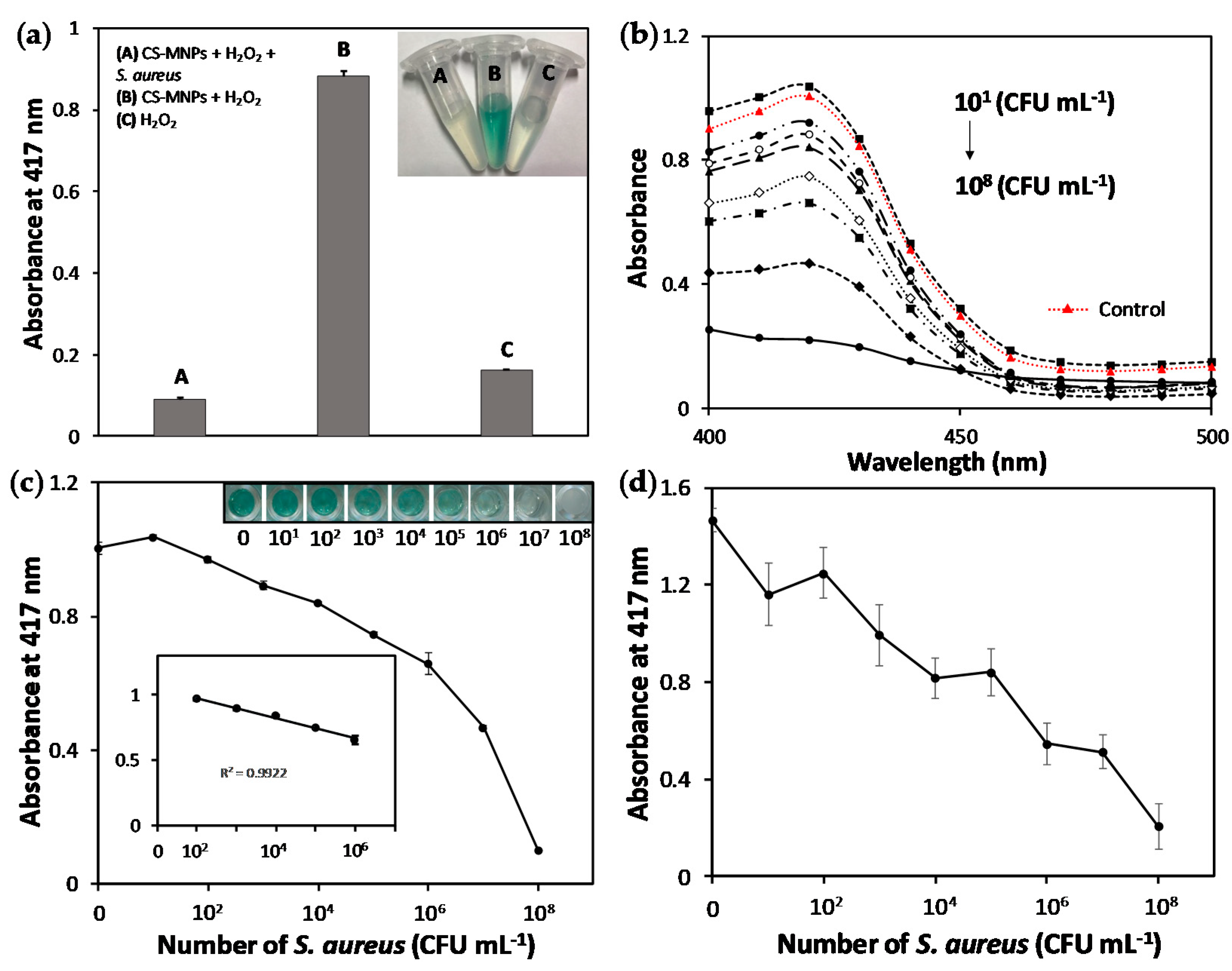
3.2. Rapid and Sensitive Colorimetric Bacteria Detection Using CS-MNPs
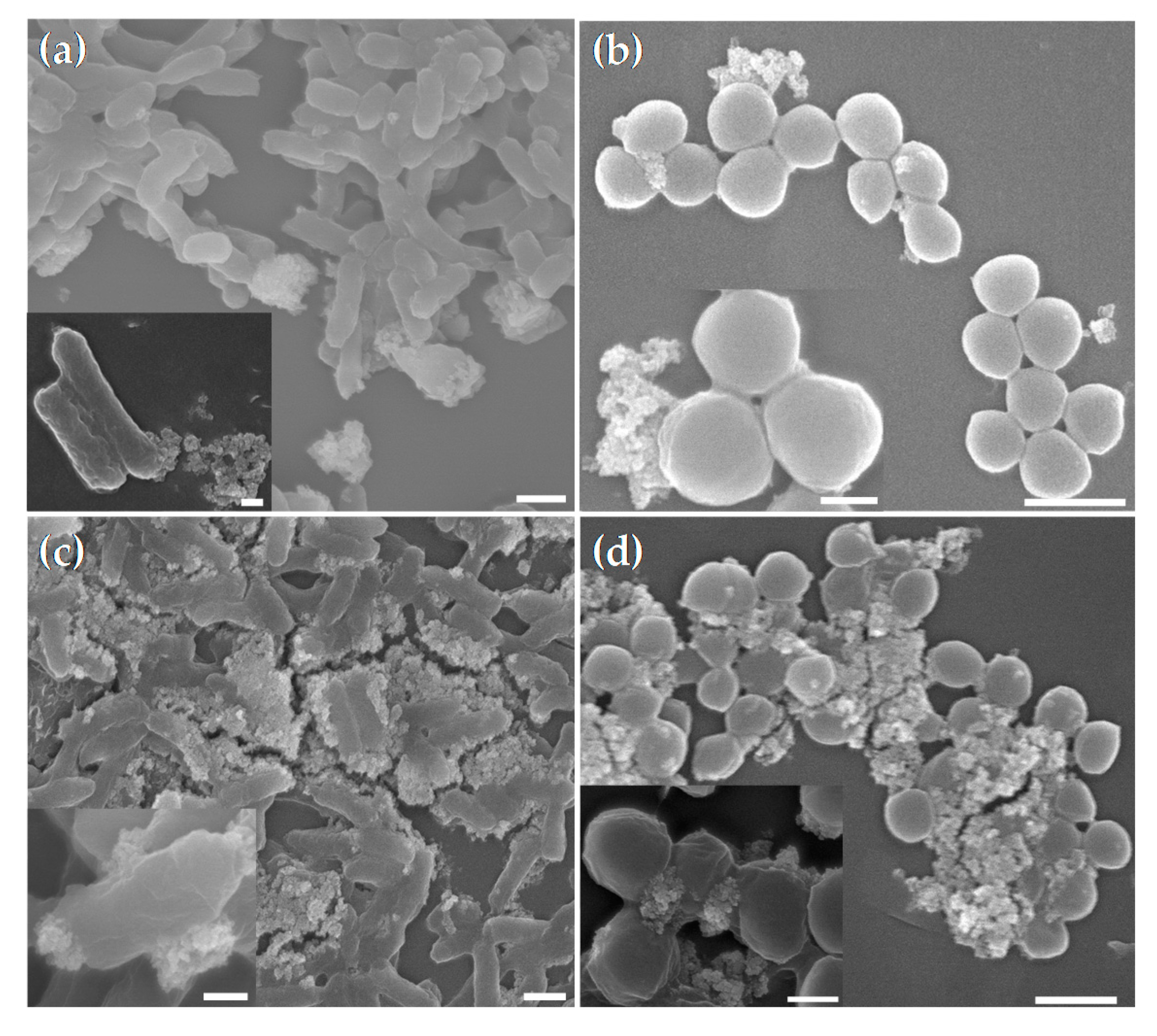
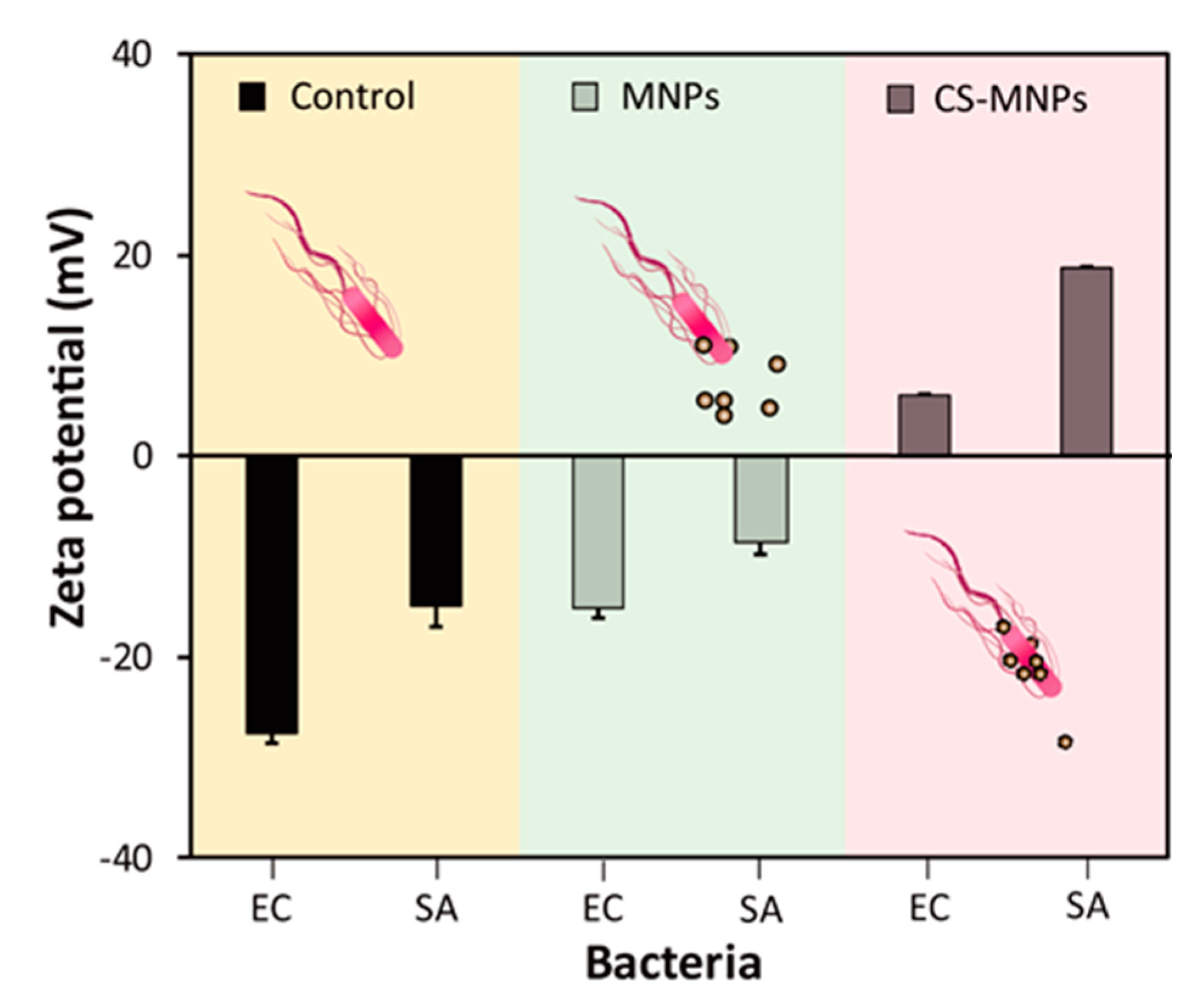
3.3. Demonstration of the Interaction between CS-MNPs and Bacteria
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [CrossRef] [PubMed]
- Twele, L.; Moyen, E.; Zhang, K.; Dalton, B.; Church, D.; Conly, J. Methicillin-resistant Staphylococcus aureus endocarditis and de novo development of daptomycin resistance during therapy. Can. J. Infect. Dis. Med. Microbiol. 2010, 21, 89–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lâm, T.-T.; Giese, B.; Chikkaballi, D.; Kühn, A.; Wolber, W.; Pané-Farré, J.; Schäfer, D.; Engelmann, S.; Fraunholz, M.; Sinha, B. Phagolysosomal integrity is generally maintained after Staphylococcus aureus invasion of nonprofessional phagocytes but is modulated by strain 6850. Infect. Immun. 2010, 78, 3392–3403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Global Burden of Foodborne Diseases; WHO: Geneva, Switzerland, 2015.
- Brooks, B.; Devenish, J.; Lutze-Wallace, C.; Milnes, D.; Robertson, R.; Berlie-Surujballi, G. Evaluation of a monoclonal antibody-based enzyme-linked immunosorbent assay for detection of Campylobacter fetus in bovine preputial washing and vaginal mucus samples. Vet. Microbiol. 2004, 103, 77–84. [Google Scholar] [CrossRef]
- Heo, J.; Hua, S.Z. An overview of recent strategies in pathogen sensing. Sensors 2009, 9, 4483–4502. [Google Scholar] [CrossRef]
- Zhu, L.; He, J.; Cao, X.; Huang, K.; Luo, Y.; Xu, W. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus Cereus in food. Sci. Rep. 2016, 6, 16092. [Google Scholar] [CrossRef]
- Alamer, S.; Eissa, S.; Chinnappan, R.; Herron, P.; Zourob, M. Rapid colorimetric lactoferrin-based sandwich immunoassay on cotton swabs for the detection of foodborne pathogenic bacteria. Talanta 2018, 185, 275–280. [Google Scholar] [CrossRef]
- Paniel, N.; Baudart, J. Colorimetric and electrochemical genosensors for the detection of Escherichia coli DNA without amplification in seawater. Talanta 2013, 115, 133–142. [Google Scholar] [CrossRef]
- Chen, J.; Alcaine, S.D.; Jackson, A.A.; Rotello, V.M.; Nugen, S.R. Development of engineered bacteriophages for Escherichia coli detection and high-throughput antibiotic resistance determination. ACS Sens. 2017, 2, 484–489. [Google Scholar] [CrossRef]
- Sun, J.; Huang, J.; Li, Y.; Lv, J.; Ding, X. A simple and rapid colorimetric bacteria detection method based on bacterial inhibition of glucose oxidase-catalyzed reaction. Talanta 2019, 197, 304–309. [Google Scholar] [CrossRef]
- Yuan, P.; Ding, X.; Yang, Y.Y.; Xu, Q.-H. Metal nanoparticles for diagnosis and therapy of bacterial infection. Adv. Healthc. Mater. 2018, 7, 1701392. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kakkar, S.; Kumar, R.; Bhalla, V. Rapid and sensitive colorimetric detection of pathogens based on silver-urease interaction. Chem. Commun. 2019, 55, 4765–4768. [Google Scholar] [CrossRef] [PubMed]
- Halas, N.J.; Lal, S.; Chang, W.-S.; Link, S.; Nordlander, P. Plasmons in strongly coupled metallic nanostructures. Chem. Rev. 2011, 111, 3913–3961. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Huang, X.; El-Sayed, I.H.; El-Sayed, M.A. Noble metals on the nanoscale: Optical and photothermal properties and some applications in imaging, sensing, biology, and medicine. Acc. Chem. Res. 2008, 41, 1578–1586. [Google Scholar] [CrossRef]
- Verma, M.S.; Rogowski, J.L.; Jones, L.; Gu, F.X. Colorimetric biosensing of pathogens using gold nanoparticles. Biotechnol. Adv. 2015, 33, 666–680. [Google Scholar] [CrossRef]
- Wu, S.; Duan, N.; Qiu, Y.; Li, J.; Wang, Z. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int. J. Food Microbiol. 2017, 261, 42–48. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577. [Google Scholar] [CrossRef]
- Jv, Y.; Li, B.; Cao, R. Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection. Chem. Commun. 2010, 46, 8017–8019. [Google Scholar] [CrossRef]
- Chen, J.; Andler, S.M.; Goddard, J.M.; Nugen, S.R.; Rotello, V.M. Integrating recognition elements with nanomaterials for bacteria sensing. Chem. Soc. Rev. 2017, 46, 1272–1283. [Google Scholar] [CrossRef]
- Kwon, D.; Lee, S.; Ahn, M.M.; Kang, I.S.; Park, K.-H.; Jeon, S. Colorimetric detection of pathogenic bacteria using platinum-coated magnetic nanoparticle clusters and magnetophoretic chromatography. Anal. Chim. Acta 2015, 883, 61–66. [Google Scholar] [CrossRef]
- Eltzov, E.; Marks, R.S. Colorimetric stack pad immunoassay for bacterial identification. Biosens. Bioelectron. 2017, 87, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Alamer, S.; Eissa, S.; Chinnappan, R.; Zourob, M. A rapid colorimetric immunoassay for the detection of pathogenic bacteria on poultry processing plants using cotton swabs and nanobeads. Mikrochim. Acta 2018, 185, 164. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhao, G.; Dou, W. Blue silica nanoparticle-based colorimetric immunoassay for detection of Salmonella pullorum. Anal. Methods 2015, 7, 8647–8654. [Google Scholar] [CrossRef]
- Park, J.Y.; Jeong, H.Y.; Kim, M.I.; Park, T.J. Colorimetric detection system for Salmonella typhimurium based on peroxidase-like activity of magnetic nanoparticles with DNA aptamers. J. Nanomater. 2015, 2015, 2. [Google Scholar] [CrossRef]
- Luo, K.; Jeong, K.B.; You, S.M.; Lee, D.H.; Jung, J.Y.; Kim, Y.R. Surface-engineered starch magnetic microparticles for highly effective separation of a broad range of bacteria. ACS Sustain. Chem. Eng. 2018, 6, 13524–13531. [Google Scholar] [CrossRef]
- Lu, A.H.; Salabas, E.E.L.; Schüth, F. Magnetic nanoparticles: Synthesis, protection, functionalization, and application. Angew. Chem. Int. Ed. 2007, 46, 1222–1244. [Google Scholar] [CrossRef]
- Safari, J.; Javadian, L. Chitosan decorated Fe3O4 nanoparticles as a magnetic catalyst in the synthesis of phenytoin derivatives. RSC Adv. 2014, 4, 48973–48979. [Google Scholar] [CrossRef]
- Juang, R.-S.; Shiau, R.-C. Metal removal from aqueous solutions using chitosan-enhanced membrane filtration. J. Membr. Sci 2000, 165, 159–167. [Google Scholar] [CrossRef]
- Konwar, A.; Chowdhury, D.; Dan, A. Chitosan based in situ and ex situ magnetic iron oxide nanoparticles for rapid endotoxin removal from protein solutions. Mater. Chem. Front. 2019, 3, 716–725. [Google Scholar] [CrossRef]
- Cheon, H.J.; Adhikari, M.D.; Chung, M.; Tran, T.D.; Kim, J.; Kim, M.I. Magnetic nanoparticles-embedded enzyme-inorganic hybrid nanoflowers with enhanced peroxidase-like activity and substrate channeling for glucose biosensing. Adv. Healthc. Mater. 2019, 8, 1801507. [Google Scholar] [CrossRef]
- Katas, H.; Hussain, Z.; Ling, T.C. Chitosan nanoparticles as a percutaneous drug delivery system for hydrocortisone. J. Nanomater. 2012, 2012, 45. [Google Scholar] [CrossRef]
- Rao, K.K.; Rao, K.M.; Kumar, P.N.; Chung, I.-D. Novel chitosan-based pH sensitive micro-networks for the controlled release of 5-fluorouracil. Iran. Polym. J. 2010, 19, 265–276. [Google Scholar]
- Vijayalekshmi, V.; Khastgir, D. Eco-friendly methanesulfonic acid and sodium salt of dodecylbenzene sulfonic acid doped cross-linked chitosan based green polymer electrolyte membranes for fuel cell applications. J. Membr. Sci. 2017, 523, 45–59. [Google Scholar] [CrossRef]
- Silva, V.; Andrade, P.; Silva, M.; Valladares, L.D.L.S.; Aguiar, J.A. Synthesis and characterization of Fe3O4 nanoparticles coated with fucan polysaccharides. J. Magn. Magn. Mater 2013, 343, 138–143. [Google Scholar] [CrossRef]
- Jia, X.; Ahmad, I.; Yang, R.; Wang, C. Versatile graphene-based photothermal nanocomposites for effectively capturing and killing bacteria, and for destroying bacterial biofilms. J. Mater. Chem. B 2017, 5, 2459–2467. [Google Scholar] [CrossRef]
- Zhang, W.; Shi, S.; Wang, Y.; Yu, S.; Zhu, W.; Zhang, X.; Zhang, D.; Yang, B.; Wang, X.; Wang, J. Versatile molybdenum disulfide based antibacterial composites for in vitro enhanced sterilization and in vivo focal infection therapy. Nanoscale 2016, 8, 11642–11648. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, T.N.; Tran, T.D.; Kim, M.I. A Convenient Colorimetric Bacteria Detection Method Utilizing Chitosan-Coated Magnetic Nanoparticles. Nanomaterials 2020, 10, 92. https://doi.org/10.3390/nano10010092
Le TN, Tran TD, Kim MI. A Convenient Colorimetric Bacteria Detection Method Utilizing Chitosan-Coated Magnetic Nanoparticles. Nanomaterials. 2020; 10(1):92. https://doi.org/10.3390/nano10010092
Chicago/Turabian StyleLe, Thao Nguyen, Tai Duc Tran, and Moon Il Kim. 2020. "A Convenient Colorimetric Bacteria Detection Method Utilizing Chitosan-Coated Magnetic Nanoparticles" Nanomaterials 10, no. 1: 92. https://doi.org/10.3390/nano10010092
APA StyleLe, T. N., Tran, T. D., & Kim, M. I. (2020). A Convenient Colorimetric Bacteria Detection Method Utilizing Chitosan-Coated Magnetic Nanoparticles. Nanomaterials, 10(1), 92. https://doi.org/10.3390/nano10010092





