Genetically Engineered Filamentous Bacteriophages Displaying TGF-β1 Promote Angiogenesis in 3D Microenvironments
Abstract
1. Introduction
2. Materials and Methods
2.1. Amplification and Purification of Phages
2.2. Enzyme-Linked Immunosorbent Assay (ELISA)
2.3. Fluorescence Assay
2.4. Cell Culture
2.5. Biocompatibility Test
2.6. Tube Formation Assay
2.7. Lap-on-a-Chip Migration Assay
2.8. RT-qPCR
2.9. Statistical Analysis
3. Results
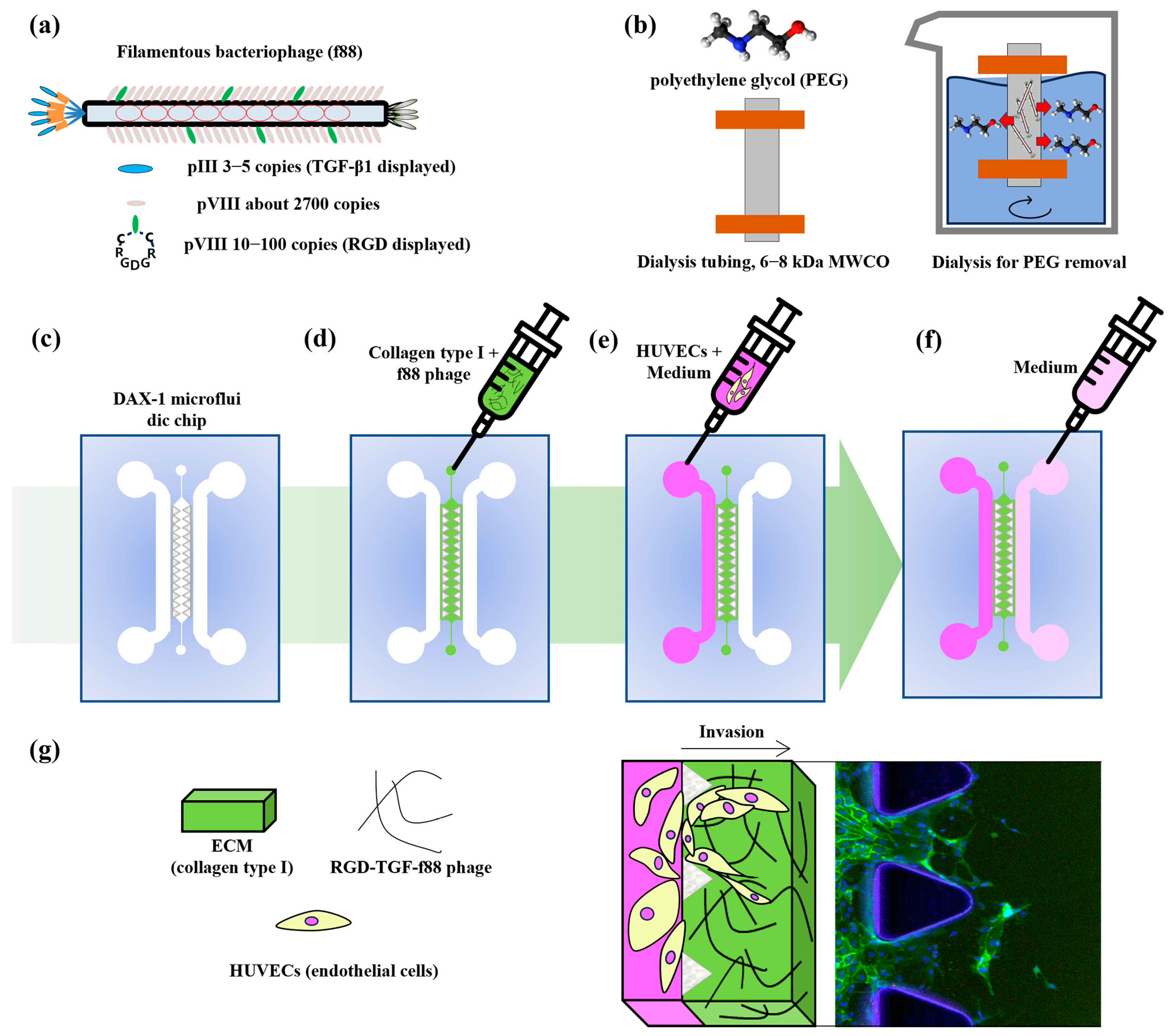
3.1. ELISA Results
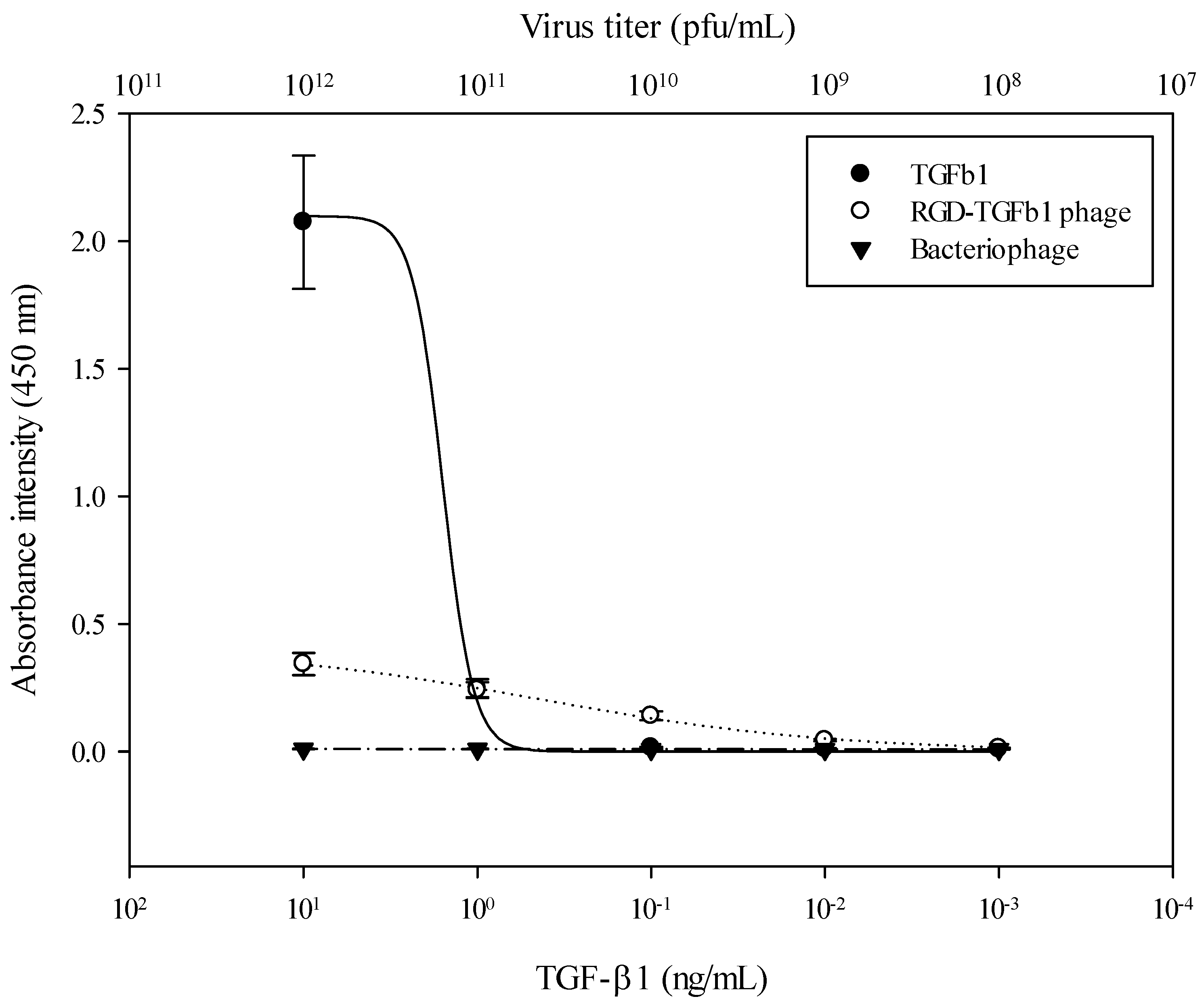
3.2. Fluorescence Results
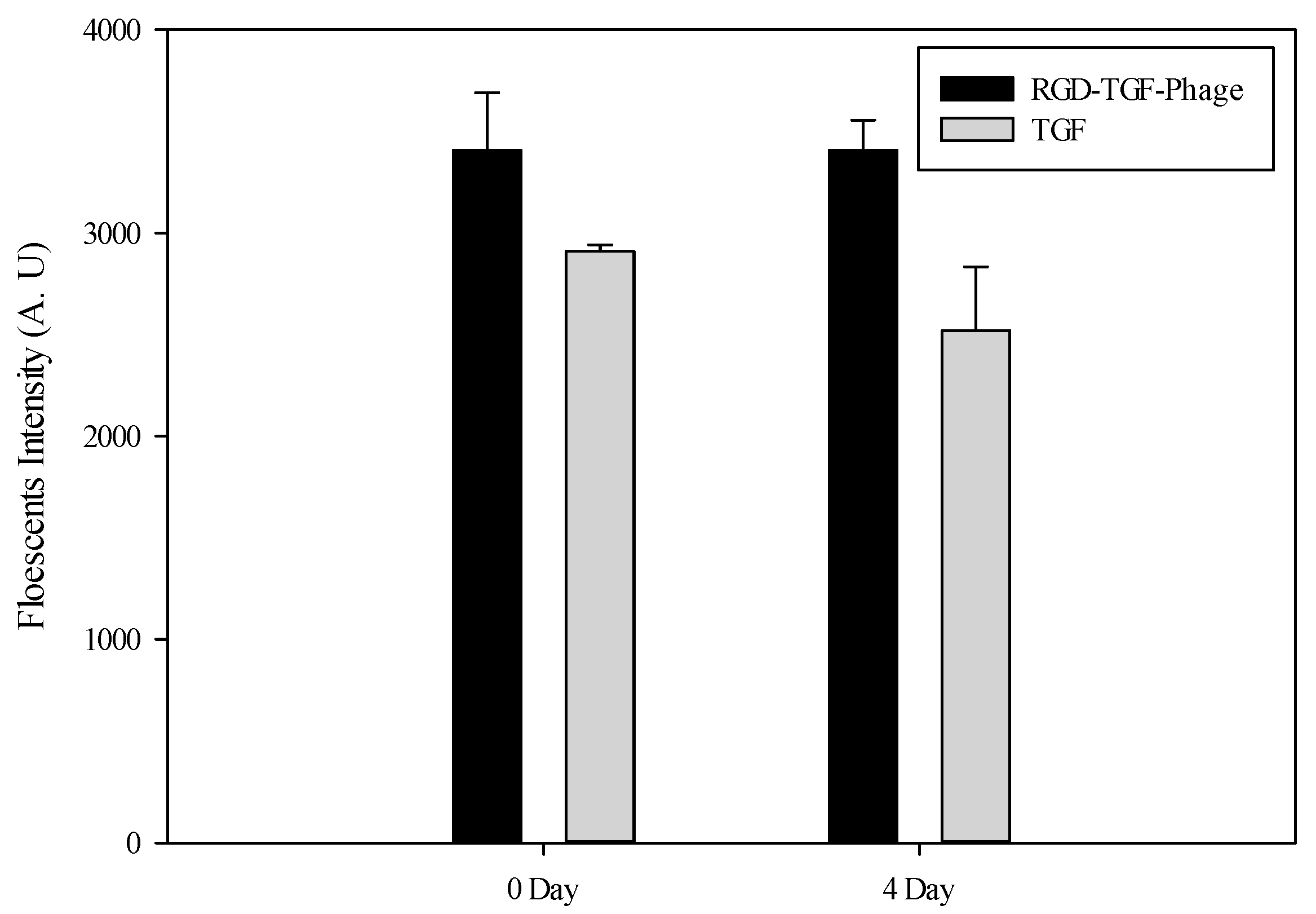
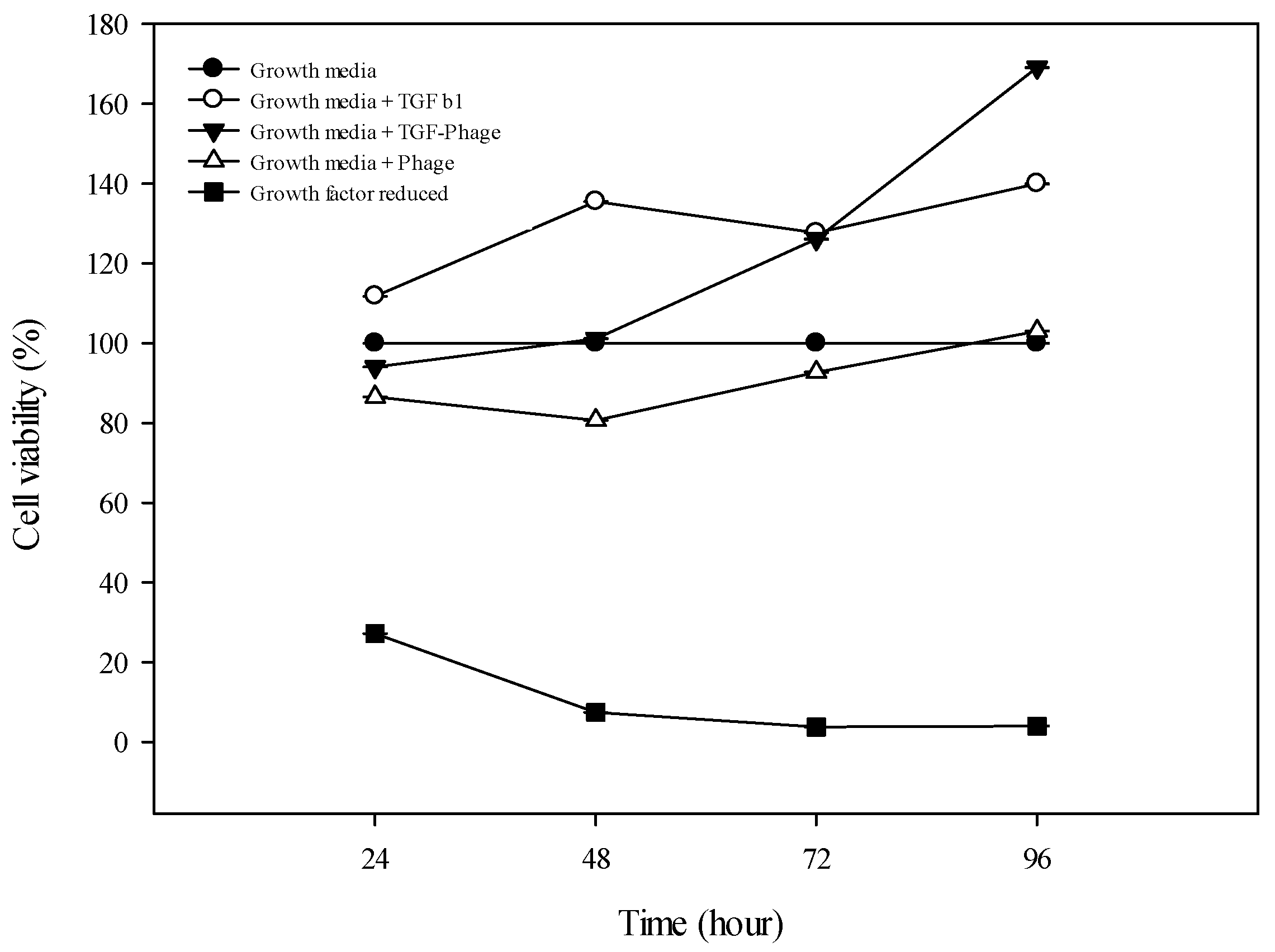
3.3. Viability Test
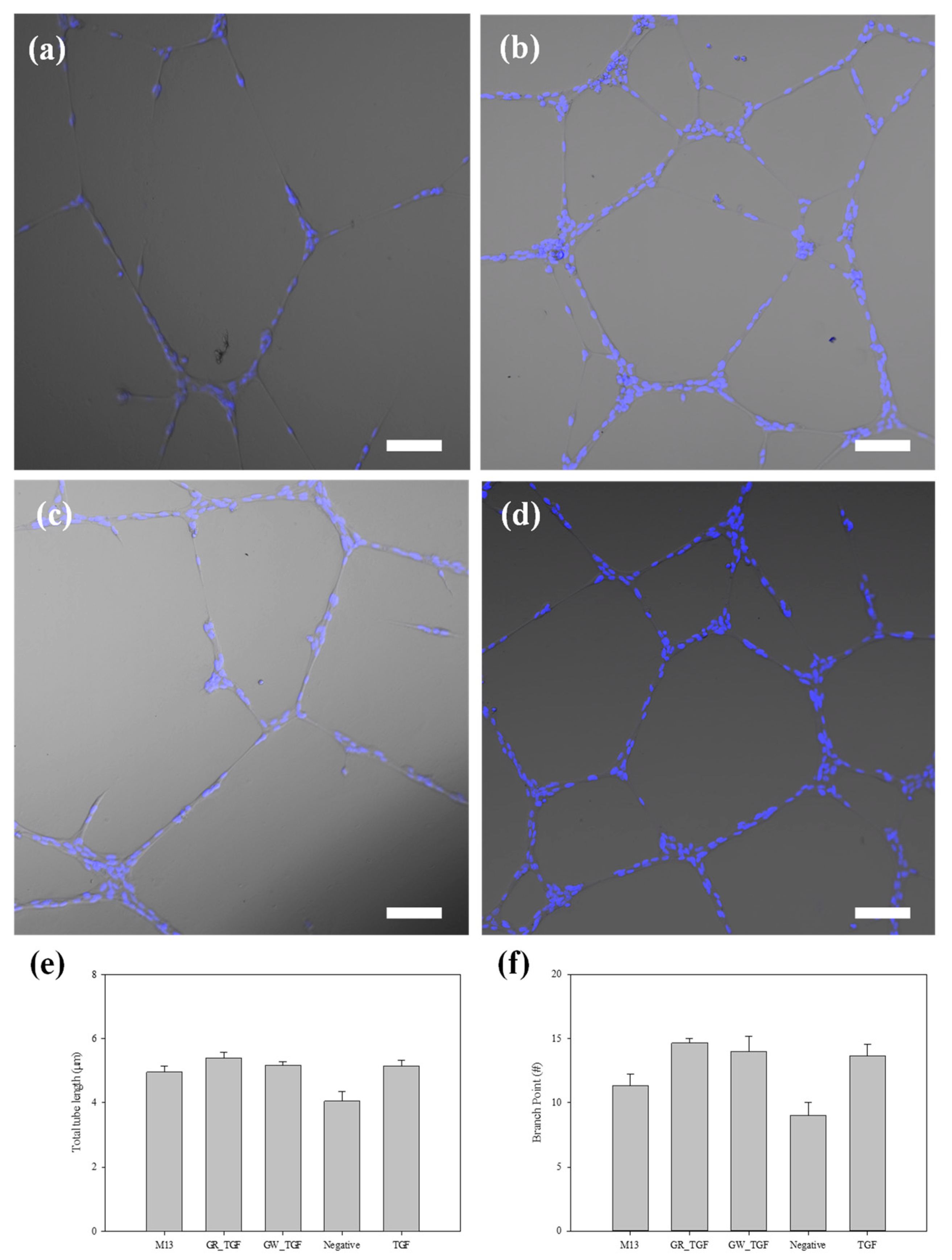
3.4. Tube Formation of ECs
3.5. Migration Test on a Microfluidics Chip

4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer Sequence (5′–3′) | |
|---|---|
| HindIII_RGDGR_KpnI_F# | AGC TTT TGT AGG GGT GAC GGT AGG TGC GGT AC |
| HindIII_RGDGR_KpnI_R# | CGC ACC TAC CGT CAC CCC TAC AAA |
| SfiI_TGFbeta1 F# | GGC CCA GCC GGC CAT GGC CCT GGA CAC |
| TGFbeta1_Not I R# | TGC GGC CGC GCT GCA CTT GCA GGA G |
| VE cadherin F# | TGT GGG CTC TCT GTT TGT TGA G |
| VE cadherin R# | CTT CAT CGT CGA GGC CAC A |
| VE cadherin Probe | CGA GGG CAT CAT CA |
| CD31 F# | AAC AGT GTT GAC ATG AAG AGC CTG |
| CD31 R# | AAG GAT GAC GTG CTG TTT TAC AAC |
| CD31 Probe | CGG ATG TCA GCA CCA C |
| GAPDH F# | CGA GCC ACA TCG CTC AGA C |
| GAPDH R# | CGT ATT GGG CGC CTG GT |
| GAPDH Probe | CGG AGT CAA CGG ATT T |
References
- Gupta, S.; Konradt, C.; Corken, A.; Ware, J.; Nieswandt, B.; Di Paola, J.; Yu, M.; Wang, D.; Nieman, M.T.; Whiteheart, S.W.; et al. Hemostasis vs. homeostasis: Platelets are essential for preserving vascular barrier function in the absence of injury or inflammation. Proc. Natl. Acad. Sci. USA 2020, 117, 24316–24325. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R.; Ruggieri, S.; Annese, T.; Crivellato, E. Surface markers: An identity card of endothelial cells. Microcirculation 2020, 27, e12587. [Google Scholar] [CrossRef]
- Annan, D.A.; Kikuchi, H.; Maishi, N.; Hida, Y.; Hida, K. Tumor Endothelial Cell-A Biological Tool for Translational Cancer Research. Int. J. Mol. Sci. 2020, 21, 3238. [Google Scholar] [CrossRef]
- Kim, S.; Kim, W.; Lim, S.; Jeon, J.S. Vasculature-On-A-Chip for In Vitro Disease Models. Bioengineering 2017, 4, 8. [Google Scholar] [CrossRef]
- Safari, Z.; Soudi, S.; Jafarzadeh, N.; Hosseini, A.Z.; Vojoudi, E.; Sadeghizadeh, M. Promotion of angiogenesis by M13 phage and RGD peptide in vitro and in vivo. Sci. Rep. 2019, 9, 11182. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, G.; Clarke, J.; Picard, F.; Riches, P.; Jia, L.; Han, F.; Li, B.; Shu, W. 3D bioactive composite scaffolds for bone tissue engineering. Bioact. Mater. 2018, 3, 278–314. [Google Scholar] [CrossRef]
- Rybinski, B.; Franco-Barraza, J.; Cukierman, E. The wound healing, chronic fibrosis, and cancer progression triad. Physiol. Genom. 2014, 46, 223–244. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar]
- Chung, J.Y.; Chan, M.K.-K.; Li, J.S.-F.; Chan, A.S.-W.; Tang, P.C.-T.; Leung, K.-T.; To, K.-F.; Lan, H.-Y.; Tang, P.M.-K. TGF-beta Signaling: From Tissue Fibrosis to Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 7575. [Google Scholar] [CrossRef]
- Meulmeester, E.; Ten Dijke, P. The dynamic roles of TGF-beta in cancer. J. Pathol. 2011, 223, 205–218. [Google Scholar] [CrossRef]
- Kurakazu, I.; Akasaki, Y.; Tsushima, H.; Sueishi, T.; Toya, M.; Kuwahara, M.; Uchida, T.; Lotz, M.K.; Nakashima, Y. TGFbeta1 signaling protects chondrocytes against oxidative stress via FOXO1-autophagy axis. Osteoarthr. Cartil. 2021, 29, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Zacher, A.N., 3rd; Stock, C.A.; Golden, J.W.; Smith, G.P. A new filamentous phage cloning vector: Fd-tet. Gene 1980, 9, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Enshell-Seijffers, D.; Smelyanski, L.; Gershoni, J.M. The rational design of a ‘type 88’ genetically stable peptide display vector in the filamentous bacteriophage fd. Nucleic Acids Res. 2001, 29, e50. [Google Scholar] [CrossRef] [PubMed]
- Staquicini, D.I.; Tang, F.H.F.; Markosian, C.; Yao, V.J.; Staquicini, F.I.; Dodero-Rojas, E.; Contessoto, V.G.; Davis, D.; O’Brien, P.; Habib, N.; et al. Design and proof of concept for targeted phage-based COVID-19 vaccination strategies with a streamlined cold-free supply chain. Proc. Natl. Acad. Sci. USA 2021, 118, e2105739118. [Google Scholar] [CrossRef]
- Baek, I.H.; Han, H.-S.; Baik, S.; Helms, V.; Kim, Y. Detection of Acidic Pharmaceutical Compounds Using Virus-Based Molecularly Imprinted Polymers. Polymers 2018, 10, 974. [Google Scholar] [CrossRef]
- Winton, A.J.; Allen, M.A. Rational Design of a Bifunctional Peptide Exhibiting Lithium Titanate Oxide and Carbon Nanotube Affinities for Lithium-Ion Battery Applications. ACS Appl. Mater. Interfaces 2023, 15, 8579–8589. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ghosh, S.; Gul, A.R.; Bhamore, J.R.; Park, J.P.; Park, T.J. Screening of specific binding peptides using phage-display techniques and their biosensing applications. TrAC Trends Anal. Chem. 2021, 137, 116229. [Google Scholar] [CrossRef]
- Dong, X.; Pan, P.; Ye, J.-J.; Zhang, Q.-L.; Zhang, X.-Z. Hybrid M13 bacteriophage-based vaccine platform for personalized cancer immunotherapy. Biomaterials 2022, 289, 121763. [Google Scholar] [CrossRef]
- Mairaville, C.; Broyon, M.; Maurel, M.; Chentouf, M.; Chiavarina, B.; Turtoi, A.; Pirot, N.; Martineau, P. Identification of monoclonal antibodies from naive antibody phage-display libraries for protein detection in formalin-fixed paraffin-embedded tissues. J. Immunol. Methods 2024, 532, 113730. [Google Scholar] [CrossRef]
- Sanmukh, S.G.; Dos Santos, N.J.; Barquilha, C.N.; De Carvalho, M.; Dos Reis, P.P.; Delella, F.K.; Carvalho, H.F.; Latek, D.; Fehér, T.; Felisbino, S.L. Bacterial RNA virus MS2 exposure increases the expression of cancer progression genes in the LNCaP prostate cancer cell line. Oncol. Lett. 2023, 25, 86. [Google Scholar] [CrossRef]
- Szot-Karpinska, K.; Golec, P.; Leśniewski, A.; Pałys, B.; Marken, F.; Niedziółka-Jönsson, J.; Węgrzyn, G.; Łoś, M. Modified Filamentous Bacteriophage as a Scaffold for Carbon Nanofiber. Bioconjug. Chem. 2016, 27, 2900–2910. [Google Scholar] [CrossRef]
- van Houten, N.E.; Zwick, M.B.; Menendez, A.; Scott, J.K. Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine 2006, 24, 4188–4200. [Google Scholar] [CrossRef] [PubMed]
- Samoylova, T.I.; Braden, T.D.; Spencer, J.A.; Bartol, F.F. Immunocontraception: Filamentous Bacteriophage as a Platform for Vaccine Development. Curr. Med. Chem. 2017, 24, 3907–3920. [Google Scholar] [CrossRef]
- Barbu, E.M.; Cady, K.C.; Hubby, B. Phage Therapy in the Era of Synthetic Biology. Cold Spring Harb. Perspect. Biol. 2016, 8, a023879. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Glucksman, M.J.; Makowski, L. Structural polymorphism correlated to surface charge in filamentous bacteriophages. Biophys. J. 1992, 61, 725–735. [Google Scholar] [CrossRef]
- Cuccia, N.L.; Pothineni, S.; Wu, B.; Harper, J.M.; Burton, J.C. Pore-size dependence and slow relaxation of hydrogel friction on smooth surfaces. Proc. Natl. Acad. Sci. USA 2020, 117, 11247–11256. [Google Scholar] [CrossRef] [PubMed]
- Korkmaz, N.; Kim, Y.J.; Nam, C.H. Bacteriophages as Templates for Manufacturing Supramolecular Structures. Macromol. Biosci. 2013, 13, 376–387. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, Y.J.; Nam, C.H. Cyclic RGD peptide incorporation on phage major coat proteins for improved internalization by HeLa cells. Bioconjug. Chem. 2014, 25, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.R.; Alberts, B.M.; Benzinger, R.; Lawhorne, L.; Treiber, G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 1970, 40, 734–744. [Google Scholar] [CrossRef]
- Warner, C.M.; Barker, N.; Lee, S.-W.; Perkins, E.J. M13 bacteriophage production for large-scale applications. Bioprocess. Biosyst. Eng. 2014, 37, 2067–2072. [Google Scholar] [CrossRef]
- Ando, J.; Yamamoto, K. Effects of shear stress and stretch on endothelial function. Antioxid. Redox Signal 2011, 15, 1389–1403. [Google Scholar] [CrossRef]
- Kocherova, I.; Bryja, A.; Mozdziak, P.; Volponi, A.A.; Dyszkiewicz-Konwińska, M.; Piotrowska-Kempisty, H.; Antosik, P.; Bukowska, D.; Bruska, M.; Iżycki, D.; et al. Human Umbilical Vein Endothelial Cells (HUVECs) Co-Culture with Osteogenic Cells: From Molecular Communication to Engineering Prevascularised Bone Grafts. J. Clin. Med. 2019, 8, 1602. [Google Scholar] [CrossRef]
- Quintard, C.; Tubbs, E.; Jonsson, G.; Jiao, J.; Wang, J.; Werschler, N.; Laporte, C.; Pitaval, A.; Bah, T.-S.; Pomeranz, G.; et al. A microfluidic platform integrating functional vascularized organoids-on-chip. Nat. Commun. 2024, 15, 1452. [Google Scholar] [CrossRef]
- Hussey, G.S.; Cramer, M.C.; Badylak, S.F. Extracellular Matrix Bioscaffolds for Building Gastrointestinal Tissue. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 1–13. [Google Scholar] [CrossRef]
- Abbott, A. Cell culture: Biology’s new dimension. Nature 2003, 424, 870–872. [Google Scholar] [CrossRef]
- National Institute of Allergy and Infectious Diseases (NIAID) A Phase 1b/2 Trial of the Safety and Microbiological Activity of Bacteriophage Therapy in Cystic Fibrosis Subjects Colonized with Pseudomonas Aeruginosa. Available online: https://clinicaltrials.gov/ct2/show/NCT05453578 (accessed on 28 February 2023).
- Meixian, L.; Huan, Z.; Limin, Z.; Jinling, Z.; Rizhi, G.; Yanping, Q.; He, N.; Qiqi, Z.; Li, H.; Min, D.; et al. Saponin I from Shuitianqi () inhibits metastasis by negatively regulating the transforming growth factor-beta1/Smad7 network and epithelial-mesenchymal transition in the intrahepatic metastasis Bagg’s Albino/c mouse model. J. Tradit. Chin. Med. 2024, 44, 642–651. [Google Scholar]
- Yoon, J.; Zirpel, N.K.; Park, H.; Han, S.; Hwang, K.H.; Shin, J.; Cho, S.; Nam, C.; Chung, S. Angiogenic Type I Collagen Extracellular Matrix Integrated with Recombinant Bacteriophages Displaying Vascular Endothelial Growth Factors. Adv. Healthc. Mater. 2016, 5, 205–212. [Google Scholar] [CrossRef]
- Doub, J.B. Risk of Bacteriophage Therapeutics to Transfer Genetic Material and Contain Contaminants Beyond Endotoxins with Clinically Relevant Mitigation Strategies. Infect. Drug Resist. 2021, 14, 5629–5637. [Google Scholar] [CrossRef]
- Luong, T.; Salabarria, A.-C.; Edwards, R.A.; Roach, D.R. Standardized bacteriophage purification for personalized phage therapy. Nat. Protoc. 2020, 15, 2867–2890. [Google Scholar] [CrossRef]
- Bohle, A.S.; Kalthoff, H. Molecular mechanisms of tumor metastasis and angiogenesis. Langenbecks Arch. Surg. 1999, 384, 133–140. [Google Scholar] [CrossRef]
- Vajkoczy, P.; Ullrich, A.; Menger, M.D. Intravital fluorescence videomicroscopy to study tumor angiogenesis and microcirculation. Neoplasia 2000, 2, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Choi, D.H.; Liu, H.-W.; Jung, Y.H.; Ahn, J.; Kim, J.-A.; Oh, D.; Jeong, Y.; Kim, M.; Yoon, H.; Kang, B.; et al. Analyzing angiogenesis on a chip using deep learning-based image processing. Lab Chip 2023, 23, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Caballero, D.; Blackburn, S.M.; de Pablo, M.; Samitier, J.; Albertazzi, L. Tumour-vessel-on-a-chip models for drug delivery. Lab Chip 2017, 17, 3760–3771. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Ahn, J.; Kim, S.; Lee, Y.; Lee, J.; Park, D.; Jeon, N.L. Tumor spheroid-on-a-chip: A standardized microfluidic culture platform for investigating tumor angiogenesis. Lab Chip 2019, 19, 2822–2833. [Google Scholar] [CrossRef]
| Materials | r2 | Logistic Model Parameters | ||
|---|---|---|---|---|
| Height | Slope | Slop at Max 50% | ||
| TGF β1 | 0.9987 | 2.0984 | −4.9648 | 1.5817 |
| RGD-TGF-β1 phage | 0.9974 | 0.4074 | −0.5181 | 0.4317 |
| Bacteriophage | 0.9999 | 0.0166 | −0.0395 | 0.0012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, I.-H.; Helms, V.; Kim, Y. Genetically Engineered Filamentous Bacteriophages Displaying TGF-β1 Promote Angiogenesis in 3D Microenvironments. J. Funct. Biomater. 2024, 15, 314. https://doi.org/10.3390/jfb15110314
Baek I-H, Helms V, Kim Y. Genetically Engineered Filamentous Bacteriophages Displaying TGF-β1 Promote Angiogenesis in 3D Microenvironments. Journal of Functional Biomaterials. 2024; 15(11):314. https://doi.org/10.3390/jfb15110314
Chicago/Turabian StyleBaek, In-Hyuk, Volkhard Helms, and Youngjun Kim. 2024. "Genetically Engineered Filamentous Bacteriophages Displaying TGF-β1 Promote Angiogenesis in 3D Microenvironments" Journal of Functional Biomaterials 15, no. 11: 314. https://doi.org/10.3390/jfb15110314
APA StyleBaek, I.-H., Helms, V., & Kim, Y. (2024). Genetically Engineered Filamentous Bacteriophages Displaying TGF-β1 Promote Angiogenesis in 3D Microenvironments. Journal of Functional Biomaterials, 15(11), 314. https://doi.org/10.3390/jfb15110314







