Divergent Responses of Hydrophilic CdSe and CdSe@CdS Core–Shell Nanocrystals in Apoptosis and In Vitro Cancer Cell Imaging: A Comparative Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of CdSe Nanocrystals
2.3. Synthesis of CdSe@CdS Core–Shell NCs and Characterization
2.4. In Vitro Biocompatibility and Imaging
2.4.1. Cytotoxicity Assay
2.4.2. Measurement of Intracellular Reactive Oxygen Species (ROS) Level
2.4.3. Electron Paramagnetic Resonance (EPR) Spectroscopy
2.4.4. Microscopic Analysis
2.4.5. Cellular Uptake
2.4.6. Analysis of Mitochondrial Membrane Potential (MMP)
2.4.7. Western Blot Analysis for Protein Extraction
2.4.8. Cytochrome C in the Cytosolic Fraction
2.4.9. FACS Analysis
3. Results
3.1. Characterization of Nanoparticles
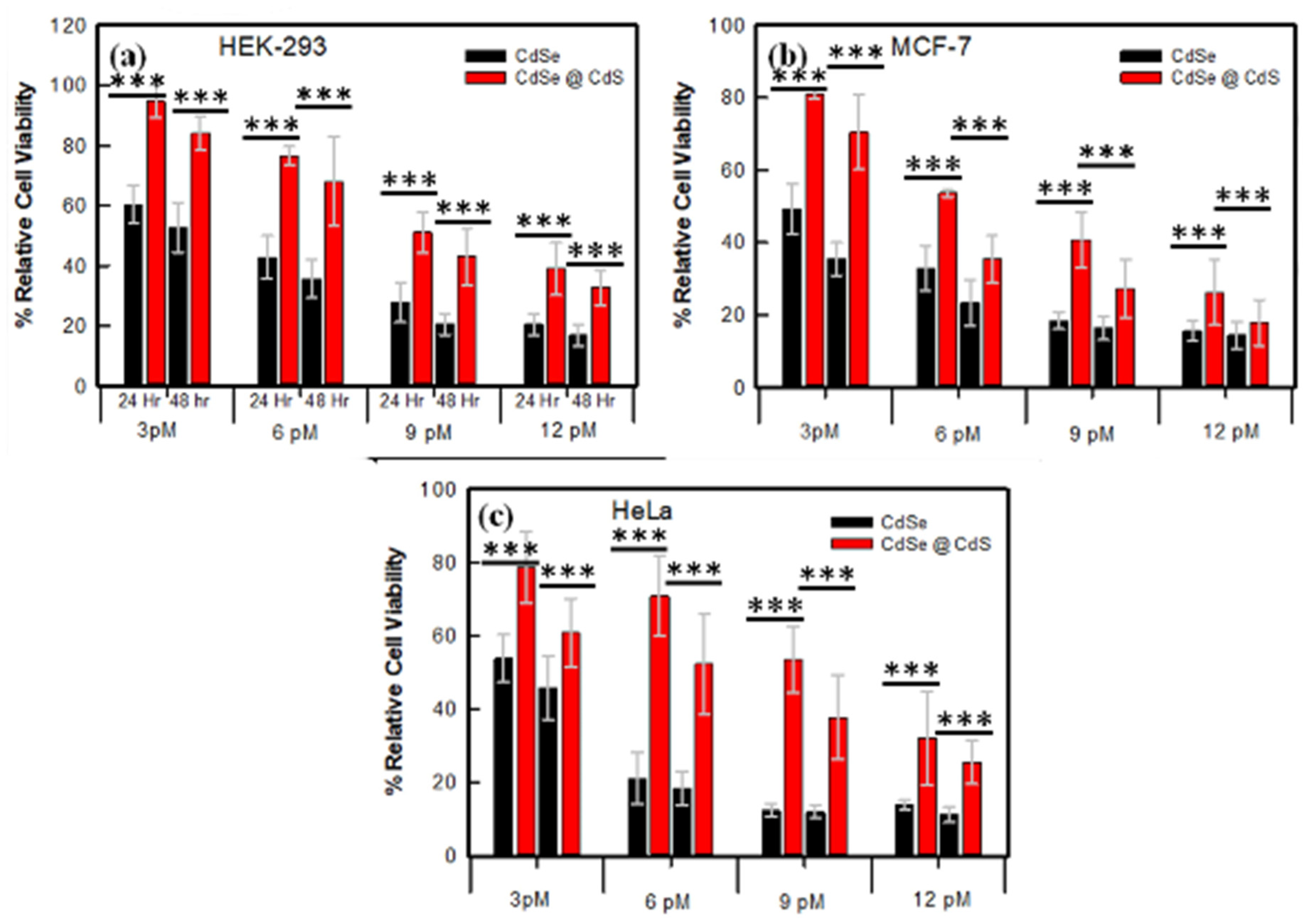
3.2. Cytotoxicity Analysis for Biocompatibility Screening
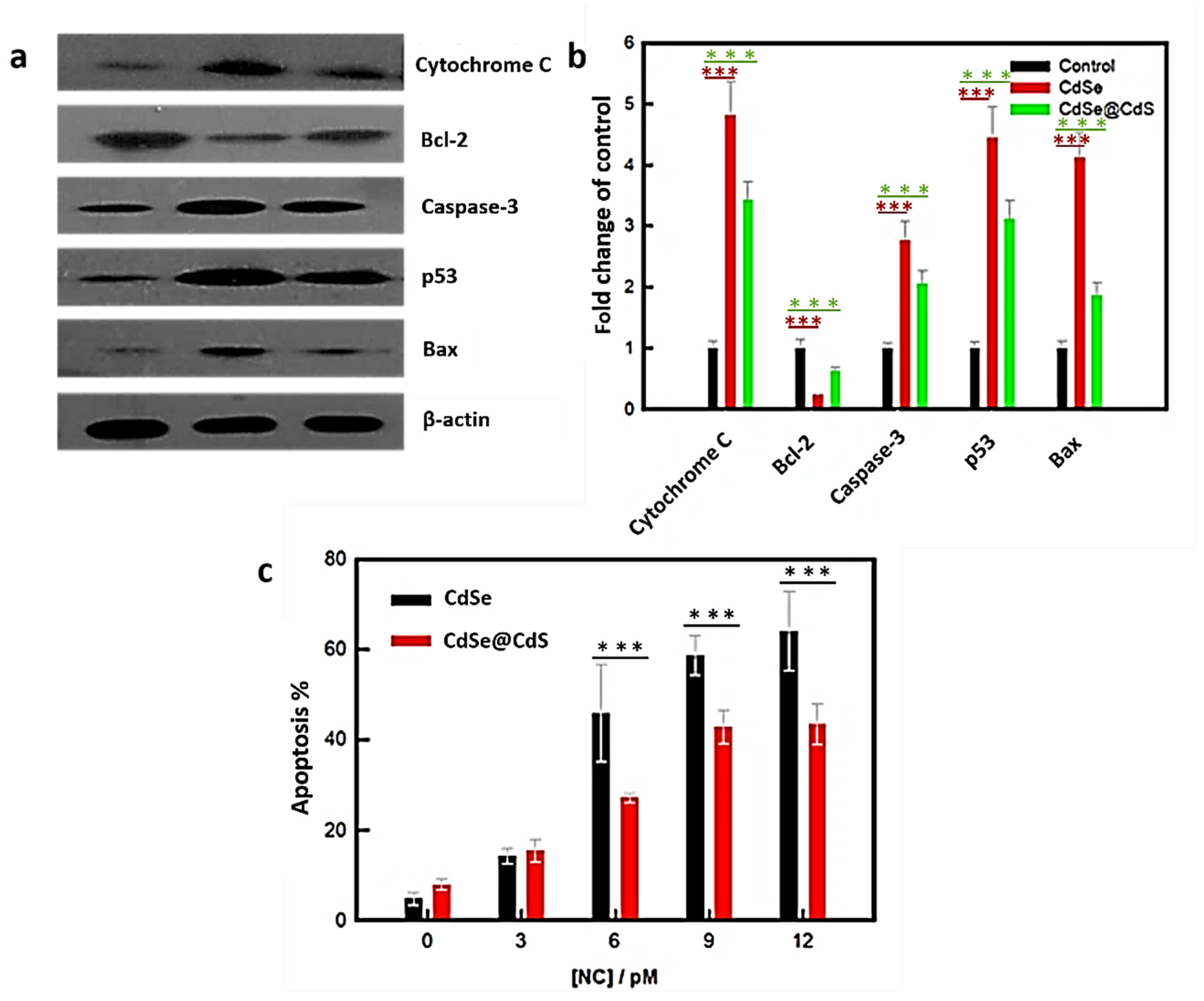
3.3. NC-Induced ROS Production
3.4. Detection of Free Radicals by EPR
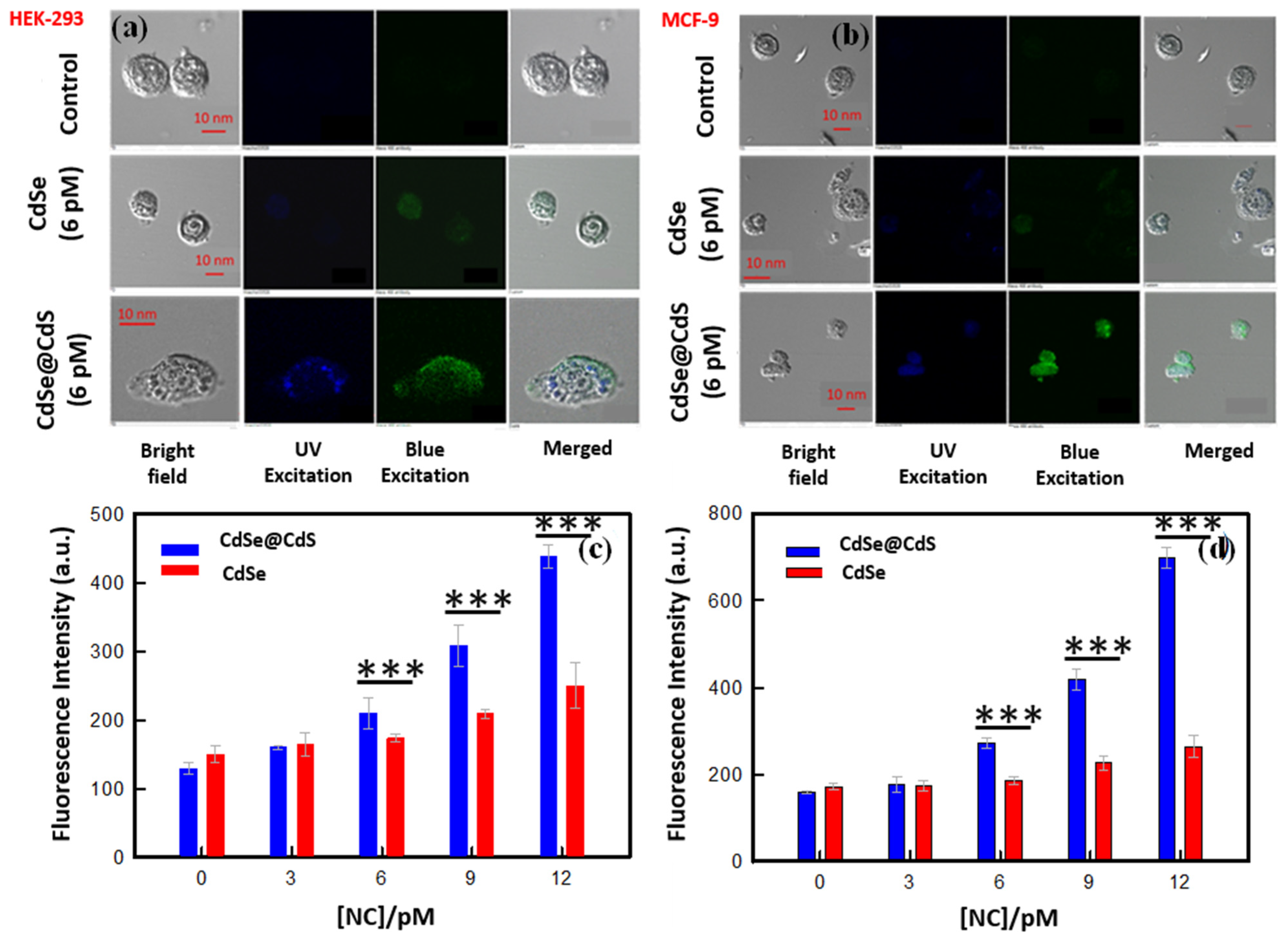
3.5. Effect on Cellular Morphology
3.6. Cellular Uptake
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ekimov, A.I.; Efros, A.L.; Onushchenko, A.A. Quantum size effect in semiconductor microcrystals. Solid State Commun. 1985, 56, 921–924. [Google Scholar] [CrossRef]
- Ajith, M.P.; Pardhiya, S.; Prabhakar, A.K.; Rajamani, P. Ag@CDs nanohybrid: Fabrication, design of a multi-mode chemosensory probe for selective Fe3+ detection and logic gate operation. Chemosphere 2022, 303, 135090. [Google Scholar] [CrossRef]
- Prabhakar, A.K.; Ajith, M.P.; Ananthanarayanan, A.; Routh, P.; Mohan, B.C.; Thamizhchelvan, A.M. Ball-milled graphene quantum dots for enhanced anti-cancer drug delivery. OpenNano 2022, 8, 100072. [Google Scholar] [CrossRef]
- Mattath, M.N.; Ghosh, D.; Dong, C.; Govindaraju, T.; Shi, S. Mercury mediated DNA–Au/Ag nanocluster ensembles to generate a gray code encoder for biocomputing. Mater. Horiz. 2022, 9, 2109–2114. [Google Scholar] [CrossRef] [PubMed]
- Walling, M.A.; Novak, J.A.; Shepard, J.R.E. Quantum Dots for Live Cell and In Vivo Imaging. Int. J. Mol. Sci. 2009, 10, 441–491. [Google Scholar] [CrossRef]
- Bailey, R.E.; Smith, A.M.; Nie, S. Quantum Dots in Biology and Medicine. Phys. E Low-Dimens. Syst. Nanostructures 2004, 25, 1–12. [Google Scholar] [CrossRef]
- Chan, W.C.W.; Nie, S. Quantum Dot Bioconjugates for Ultrasensitive Nonisotopic Detection. Science 1998, 281, 2016–2018. [Google Scholar] [CrossRef]
- Smith, A.M.; Gao, X.; Nie, S. Quantum Dot Nanocrystals for In Vivo Molecular and Cellular Imaging. Photochem. Photobiol. 2004, 80, 377–385. [Google Scholar] [CrossRef]
- Jovin, T.M. Quantum dots finally come of age. Nat. Biotechnol. 2003, 21, 32–33. [Google Scholar] [CrossRef]
- Derfus, A.M.; Chan, W.C.W.; Bhatia, S.N. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004, 4, 11–18. [Google Scholar] [CrossRef]
- Su, Y.; He, Y.; Lu, H.; Sai, L.; Li, Q.; Li, W.; Wang, L.; Shen, P.; Huang, Q.; Fan, C. The cytotoxicity of cadmium based, aqueous phase—Synthesized, quantum dots and its modulation by surface coating. Biomaterials 2009, 30, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; He, Y.; Su, Y.; Li, X.; Huang, Q.; Wang, H.; Zhang, X.; Tai, R.; Fan, C. The cytotoxicity of cadmium-based quantum dots. Biomaterials 2012, 33, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Ajith, M.P.; Rajamani, P. Nanotechnology for Water Purification—Current Trends and Challenges. J. Nanotechnol. Nanomater. 2021, 2, 88–91. [Google Scholar] [CrossRef]
- Hild, W.A.; Breunig, M.; Goepferich, A. Quantum dots—Nano-sized probes for the exploration of cellular and intracellular targeting. Eur. J. Pharm. Biopharm. 2008, 68, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, C.; Liedl, T.; Kudera, S.; Pellegrino, T.; Muñoz Javier, A.; Gaub, H.E.; Stölzle, S.; Fertig, N.; Parak, W.J. Cytotoxicity of Colloidal CdSe and CdSe/ZnS Nanoparticles. Nano Lett. 2005, 5, 331–338. [Google Scholar] [CrossRef]
- Chan, W.-H.; Shiao, N.-H.; Lu, P.-Z. CdSe quantum dots induce apoptosis in human neuroblastoma cells via mitochondrial-dependent pathways and inhibition of survival signals. Toxicol. Lett. 2006, 167, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Seleverstov, O.; Zabirnyk, O.; Zscharnack, M.; Bulavina, L.; Nowicki, M.; Heinrich, J.-M.; Yezhelyev, M.; Emmrich, F.; O’Regan, R.; Bader, A. Quantum Dots for Human Mesenchymal Stem Cells Labeling. A Size-Dependent Autophagy Activation. Nano Lett. 2006, 6, 2826–2832. [Google Scholar] [CrossRef]
- Clift, M.J.D.; Rothen-Rutishauser, B.; Brown, D.M.; Duffin, R.; Donaldson, K.; Proudfoot, L.; Guy, K.; Stone, V. The impact of different nanoparticle surface chemistry and size on uptake and toxicity in a murine macrophage cell line. Toxicol. Appl. Pharmacol. 2008, 232, 418–427. [Google Scholar] [CrossRef]
- Pons, T.; Pic, E.; Lequeux, N.; Cassette, E.; Bezdetnaya, L.; Guillemin, F.; Marchal, F.; Dubertret, B. Cadmium-Free CuInS2/ZnS Quantum Dots for Sentinel Lymph Node Imaging with Reduced Toxicity. ACS Nano 2010, 4, 2531–2538. [Google Scholar] [CrossRef]
- Maity, A.R.; Palmal, S.; Basiruddin, S.K.; Karan, N.S.; Sarkar, S.; Pradhan, N.; Jana, N.R. Doped semiconductor nanocrystal based fluorescent cellular imaging probes. Nanoscale 2013, 5, 5506–5513. [Google Scholar] [CrossRef]
- Raab, R.M.; Stephanopoulos, G. Dynamics of gene silencing by RNA interference. Biotechnol. Bioeng. 2004, 88, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Su, R.; Chen, W.; Han, Q.; Tian, Y.; Han, J.; Wang, X.; Song, S.; Reddy, K.M.; Deng, H.; et al. Oriented attachment interfaces of zeolitic imidazolate framework nanocrystals. Nanoscale 2023, 15, 7703–7709. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Huang, Y.; Parakhonskiy, B.; Skirtach, A.G. Nanoarchitectonics beyond perfect order—Not quite perfect but quite useful. Nanoscale 2022, 14, 15964–16002. [Google Scholar] [CrossRef] [PubMed]
- Ehman, R.L.; Hendee, W.R.; Welch, M.J.; Dunnick, N.R.; Bresolin, L.B.; Arenson, R.L.; Baum, S.; Hricak, H.; Thrall, J.H. Blueprint for Imaging in Biomedical Research. Radiology 2007, 244, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Selvan, S.T.; Tan, T.T.; Ying, J.Y. Robust, Non-Cytotoxic, Silica-Coated CdSe Quantum Dots with Efficient Photoluminescence. Adv. Mater. 2005, 17, 1620–1625. [Google Scholar] [CrossRef]
- Rajamani, P.; Ajith, M.P.; Gautam, R. Impact of Metal and Metal Oxide Nanoparticles on Male Reproductive System: A Comprehensive Review. Indian J. Biochem. Biophys. 2022, 59, 1048–1055. [Google Scholar] [CrossRef]
- Shih, C.-Y.; Wang, P.-T.; Chung, W.-P.; Wang, W.-H.; Chiang, I.-T.; Su, W.-C.; Huang, W.-L.; Teng, H. Concise nanotherapeutic modality for cancer involving graphene oxide dots in conjunction with ascorbic acid. Nanoscale 2023, 15, 10232–10243. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, M.; Basu, S.M.; Qasim, M.; Giri, J. Polypropylene sulphide coating on magnetic nanoparticles as a novel platform for excellent biocompatible, stimuli-responsive smart magnetic nanocarriers for cancer therapeutics. Nanoscale 2023, 15, 7384–7402. [Google Scholar] [CrossRef]
- Manna, L.; Scher, E.C.; Li, L.-S.; Alivisatos, A.P. Epitaxial Growth and Photochemical Annealing of Graded CdS/ZnS Shells on Colloidal CdSe Nanorods. J. Am. Chem. Soc. 2002, 124, 7136–7145. [Google Scholar] [CrossRef]
- Tyagi, A.; Rawat, K.; Verma, A.K.; Bohidar, H.B. Mechanistic evaluation of the size dependent antimicrobial activity of water soluble QDs. Anal. Methods 2016, 8, 1060–1068. [Google Scholar] [CrossRef]
- Liu, J.; Gu, Y.; Wu, Q.; Wang, X.; Zhao, L.; de Mello, A.; Wen, W.; Tong, R.; Gong, X. Synthesis and Study of CdSe QDs by a Microfluidic Method and via a Bulk Reaction. Crystals 2019, 9, 368. [Google Scholar] [CrossRef]
- Su, H.; Dixon, J.; Wang, A.Y.; Low, J.; Xu, J.; Wang, J. Study on Growth Kinetics of CdSe Nanocrystals with a New Model. Nanoscale Res. Lett. 2010, 5, 823–828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, L.; Guo, X.; Li, Y.; Zhong, X. Bifunctional Multidentate Ligand Modified Highly Stable Water-Soluble Quantum Dots. Inorg. Chem. 2010, 49, 3768–3775. [Google Scholar] [CrossRef] [PubMed]
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for Cell Viability Assays. Food Front. 2020, 1, 332–349. [Google Scholar] [CrossRef]
- Kermanizadeh, A.; Jantzen, K.; Brown, D.M.; Møller, P.; Loft, S. A Flow Cytometry-Based Method for the Screening of Nanomaterial-Induced Reactive Oxygen Species Production in Leukocytes Subpopulations in Whole Blood. Basic Clin. Pharmacol. Toxicol. 2018, 122, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, L.; Zhou, Q.; Chen, F.; Stenzel, M.; Gao, F.; Liu, C.; Yuan, H.; Li, H.; Jiang, Y. Self-Assembled Anionic and Cationic Au Nanoparticles with Au Nanoclusters for the Exploration of Different Biological Responsiveness in Cancer Therapy. Nanoscale Adv. 2021, 3, 2812–2821. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Jasieniak, J.; Smith, L.; van Embden, J.; Mulvaney, P.; Califano, M. Re-examination of the Size-Dependent Absorption Properties of CdSe Quantum Dots. J. Phys. Chem. C 2009, 113, 19468–19474. [Google Scholar] [CrossRef]
- Chen, X.; Hutchison, J.L.; Dobson, P.J.; Wakefield, G. A one-step aqueous synthetic route to extremely small CdSe nanoparticles. J. Colloid Interface Sci. 2008, 319, 140–143. [Google Scholar] [CrossRef]
- Peana, M.; Pelucelli, A.; Chasapis, C.T.; Perlepes, S.P.; Bekiari, V.; Medici, S.; Zoroddu, M.A. Biological Effects of Human Exposure to Environmental Cadmium. Biomolecules 2022, 13, 36. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2008, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Boonstra, J.; Post, J.A. Molecular events associated with reactive oxygen species and cell cycle progression in mammalian cells. Gene 2004, 337, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Deng, C.; Tang, S.; Zhang, M. Mitochondria-Dependent Apoptosis Induced by Nanoscale Hydroxyapatite in Human Gastric Cancer SGC-7901 Cells. Biol. Pharm. Bull. 2007, 30, 128–132. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.R.; Singh, B.N.; Khan, W.; Singh, H.B.; Naqvi, A.H. ROS-mediated apoptotic cell death in prostate cancer LNCaP cells induced by biosurfactant stabilized CdS quantum dots. Biomaterials 2012, 33, 5753–5767. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.C.; Willmore, W.G.; Tayabali, A.F. Cadmium telluride quantum dots cause oxidative stress leading to extrinsic and intrinsic apoptosis in hepatocellular carcinoma HepG2 cells. Toxicology 2013, 306, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.; Hayashi, T.; Su, T.-P.; Betenbaugh, M.J. Bcl-2 family in inter-organelle modulation of calcium signaling; roles in bioenergetics and cell survival. J. Bioenerg. Biomembr. 2014, 46, 1–15. [Google Scholar] [CrossRef]
- Li, C.Y.; Lee, J.S.; Ko, Y.G.; Kim, J.I.; Seo, J.S. Heat shock protein 70 inhibits apoptosis downstream of cytochrome c release and upstream of caspase-3 activation. J. Biol. Chem. 2000, 275, 25665–25671. [Google Scholar] [CrossRef]
- Maysinger, D. Nanoparticles and cells: Good companions and doomed partnerships. Org. Biomol. Chem. 2007, 5, 2335–2342. [Google Scholar] [CrossRef]
- Liang, Z.; Khawar, M.B.; Liang, J.; Sun, H. Bio-Conjugated Quantum Dots for Cancer Research: Detection and Imaging. Front. Oncol. 2021, 11, 749970. [Google Scholar] [CrossRef]





| NCs | Absorbance Maxima (nm) | Emission Maxima (nm) | TEM Size (nm) | Average Crystalline Size (nm) | DLS Size (nm) | Zeta Potential (mV) |
|---|---|---|---|---|---|---|
| CdSe | 505 | 548 | 7 ± 1 | 3.10 ± 0.5 | 51 ± 5 | −45 ± 4 |
| CdSe_CdS | 500 | 542 | 12 ± 1 | 2.70 ± 0.5 | 60 ± 7 | −60 ± 5 |
| NC | Plane (hkl) | 2θ (°) | FWHM (°) | Relative Intensity | Inter-Planar Spacing (Å) | Lattice Vector (Å) | Lattice Strain | Crystallite Size (nm) | Average Size (nm) |
|---|---|---|---|---|---|---|---|---|---|
| CdSe | (111) | 25.2 | 3.83 | 0.579 | 3.54 | 6.13 | 0.0553 | 3.01 | 3.10 |
| (230) | 42.1 | 3.00 | 0.253 | 2.15 | 6.08 | 0.0340 | 2.96 | ||
| (311) | 49.4 | 2.75 | 0.154 | 1.84 | 6.10 | 0.0261 | 3.32 | ||
| CdSe @CdS | (111) | 25.3 | 3.86 | 0.996 | 3.52 | 6.10 | 0.0751 | 2.20 | 2.73 |
| (230) | 42.3 | 3.04 | 0.404 | 2.14 | 6.05 | 0.0343 | 2.93 | ||
| (311) | 49.3 | 3.00 | 0.320 | 1.83 | 6.07 | 0.0283 | 3.05 |
| Cd-Based Nano-Architechtonics | Cell Line Used | Targeted Receptor |
|---|---|---|
| Polymer-coated CdSe/ZnS | Human oral squamous carcinoma cells (BcaCDE885) | Integrin αvβ3 |
| PEGylated CdTe | Human glioblastoma cells (U87 MG cells) | Integrin αvβ3 |
| Magnetic and CdTe QDs immobilized on SiO2 | MCF-7 cells | Mucin 1 protein |
| CdTe QDs | MCF-7 cells | MMP-2 |
| Mercapto-succinic acid-coated CdTe QDs | Fibroadenoma and ductal carcinoma | Glycans |
| CdTe QDs and Fe3O4 NPs; | Colon carcinoma cells LS174 | TAG-72 |
| CdTe/MPA QDs | Gastric cancer cell line MGC80-3 | TAG-72 |
| NAC capped and alloyed with CdTeS | Bel-7402 human hepatoma cells | Folate receptor |
| CdSe/ZnS coated with oleylamine poly (aspartate)-graft-PEGdodecylamine | Human liver cancer (HepG2) cells | VEGFR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, K.; Bhatt, N.; Parambil, A.M.; Kumari, K.; Kumar, R.; Rawat, K.; Rajamani, P.; Bohidar, H.B.; Nadeem, A.; Muthupandian, S.; et al. Divergent Responses of Hydrophilic CdSe and CdSe@CdS Core–Shell Nanocrystals in Apoptosis and In Vitro Cancer Cell Imaging: A Comparative Analysis. J. Funct. Biomater. 2023, 14, 448. https://doi.org/10.3390/jfb14090448
Das K, Bhatt N, Parambil AM, Kumari K, Kumar R, Rawat K, Rajamani P, Bohidar HB, Nadeem A, Muthupandian S, et al. Divergent Responses of Hydrophilic CdSe and CdSe@CdS Core–Shell Nanocrystals in Apoptosis and In Vitro Cancer Cell Imaging: A Comparative Analysis. Journal of Functional Biomaterials. 2023; 14(9):448. https://doi.org/10.3390/jfb14090448
Chicago/Turabian StyleDas, Kishan, Neelima Bhatt, Ajith Manayil Parambil, Kajal Kumari, Raj Kumar, Kamla Rawat, Paulraj Rajamani, Himadri B. Bohidar, Ahmed Nadeem, Saravanan Muthupandian, and et al. 2023. "Divergent Responses of Hydrophilic CdSe and CdSe@CdS Core–Shell Nanocrystals in Apoptosis and In Vitro Cancer Cell Imaging: A Comparative Analysis" Journal of Functional Biomaterials 14, no. 9: 448. https://doi.org/10.3390/jfb14090448
APA StyleDas, K., Bhatt, N., Parambil, A. M., Kumari, K., Kumar, R., Rawat, K., Rajamani, P., Bohidar, H. B., Nadeem, A., Muthupandian, S., & Meena, R. (2023). Divergent Responses of Hydrophilic CdSe and CdSe@CdS Core–Shell Nanocrystals in Apoptosis and In Vitro Cancer Cell Imaging: A Comparative Analysis. Journal of Functional Biomaterials, 14(9), 448. https://doi.org/10.3390/jfb14090448









