Delivery of Melittin as a Lytic Agent via Graphene Nanoparticles as Carriers to Breast Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
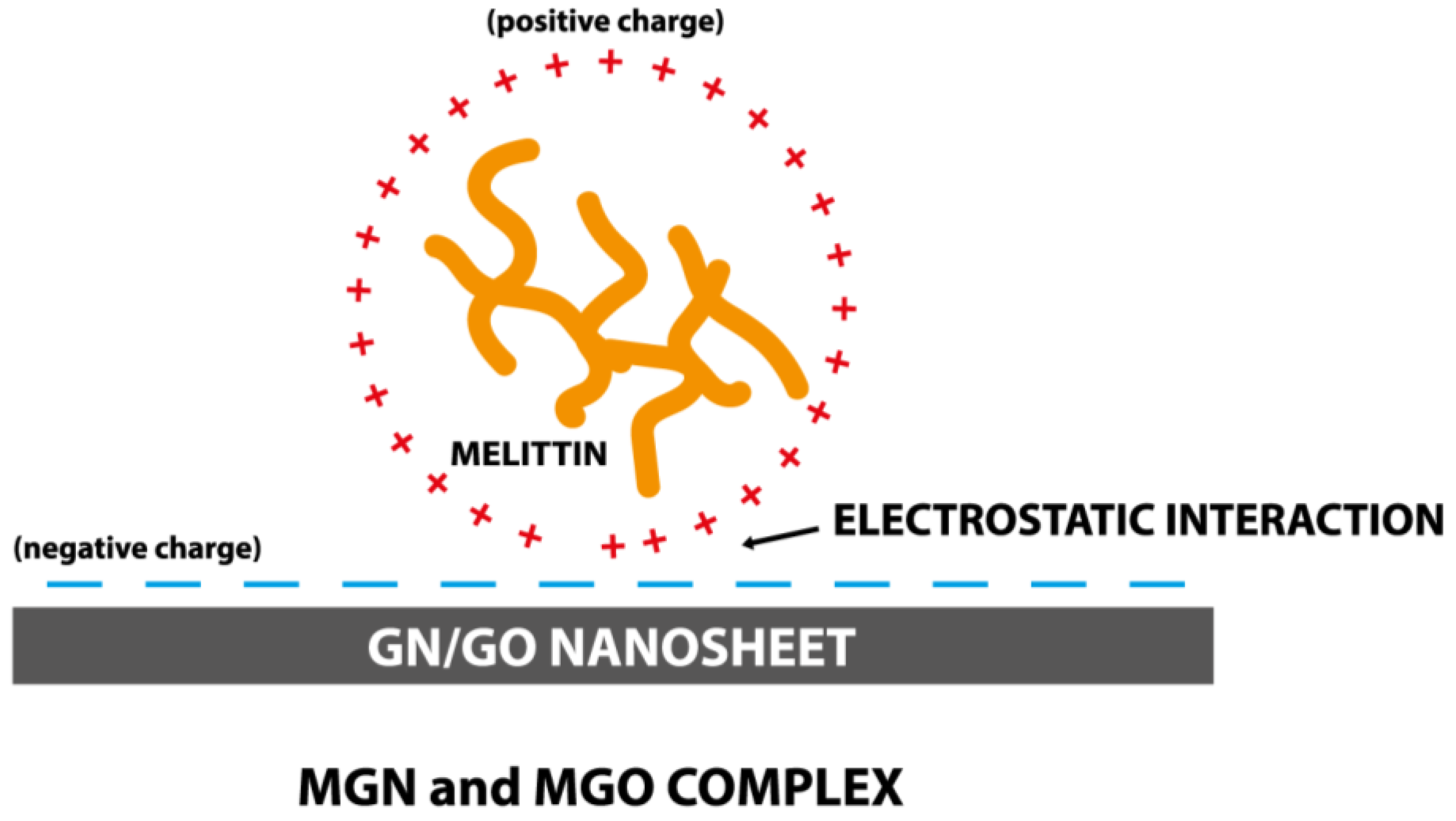
2.1. Complex Preparation and Characterization
2.2. Entrapment Efficiency (EE%) of the MEL in Complexes
2.3. Cell Culture
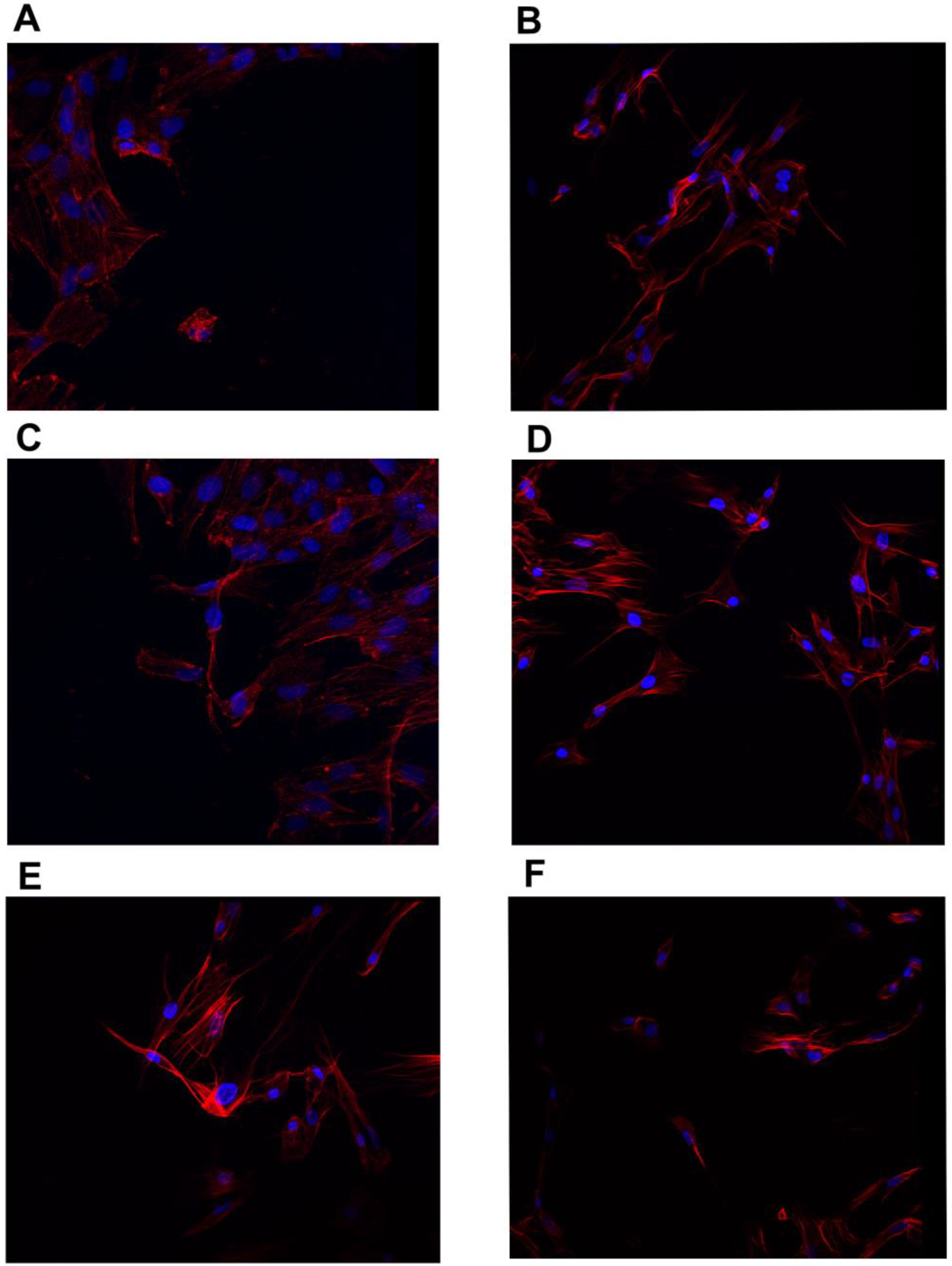
2.4. Confocal Microscopy
2.5. Intracellular pH
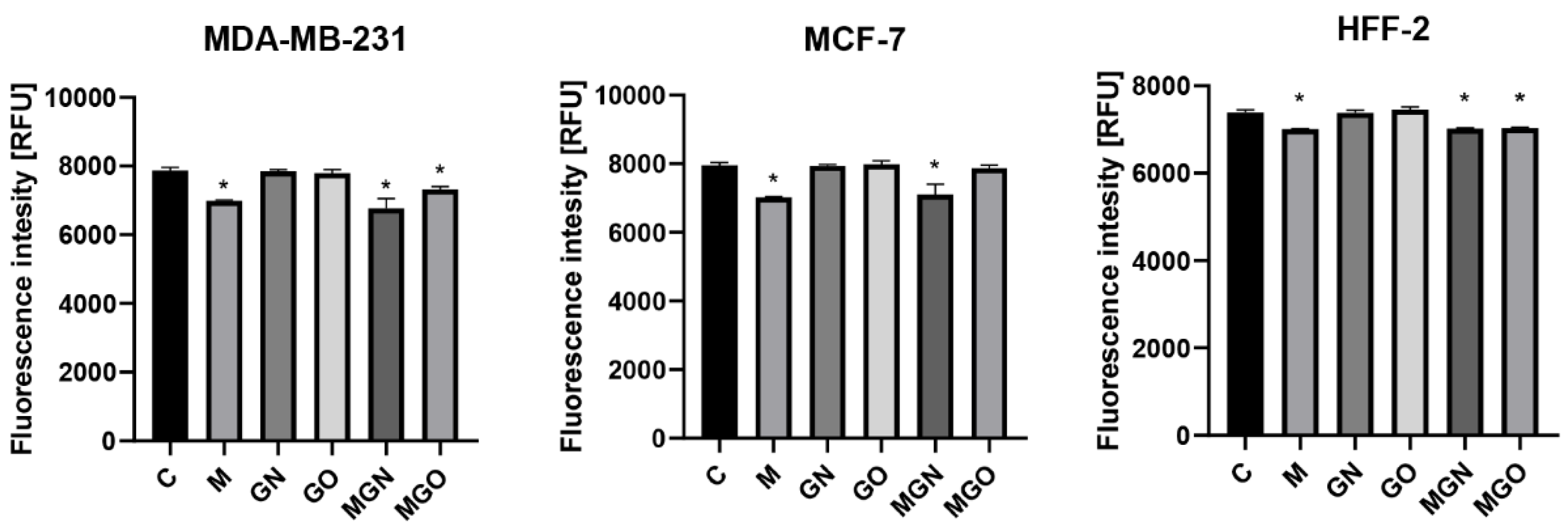
2.6. Potential of Cell Membrane
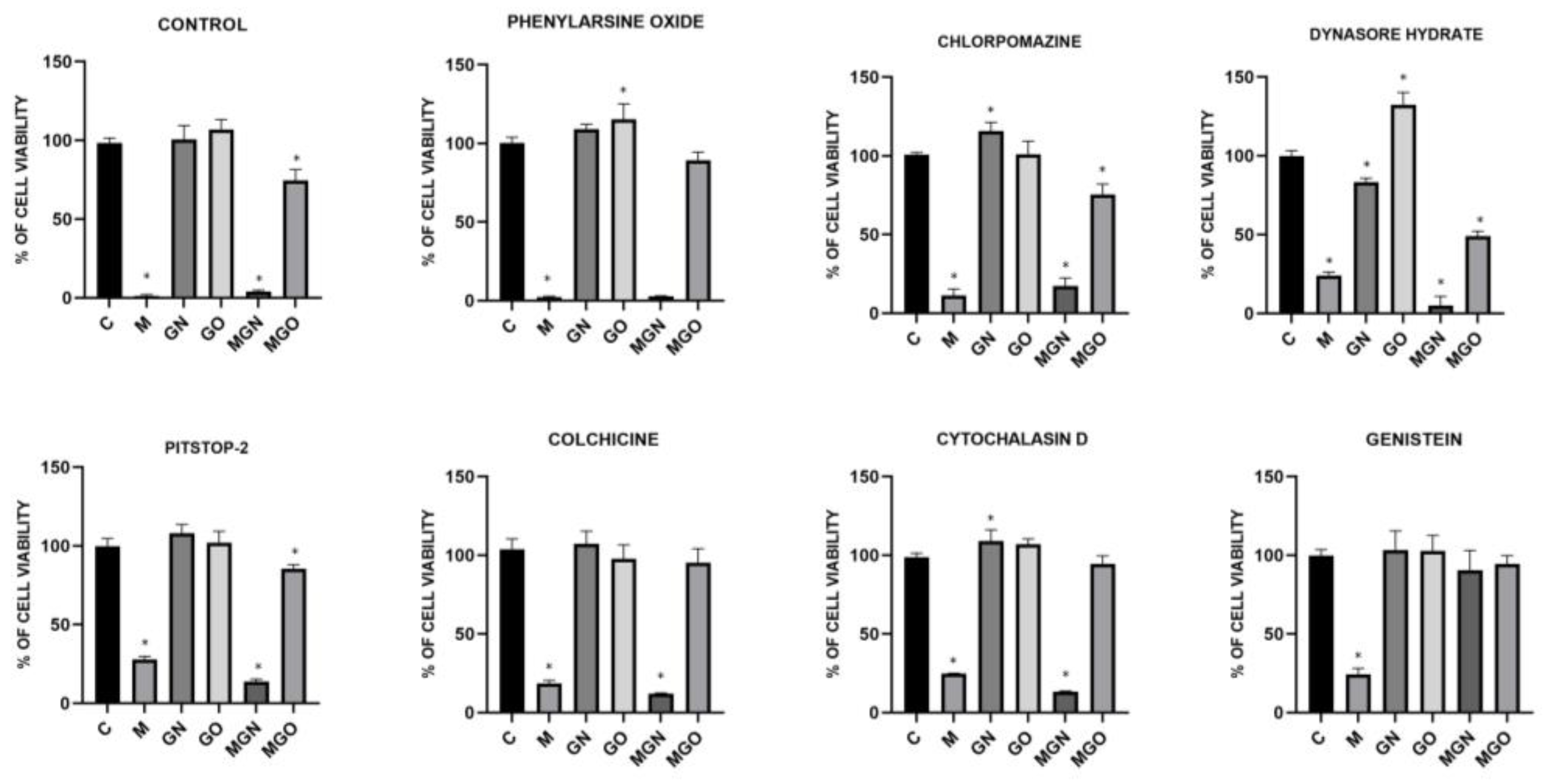
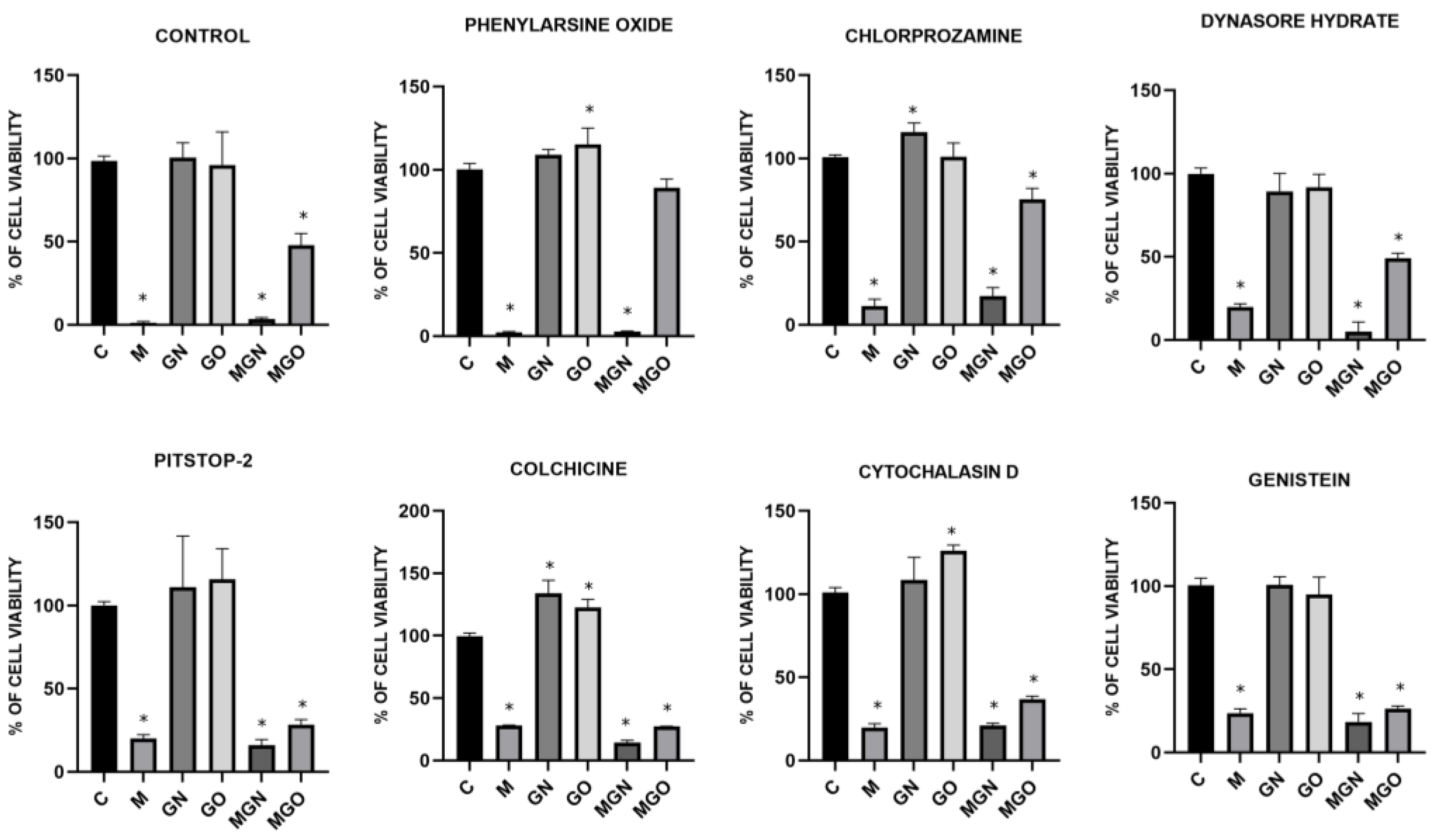
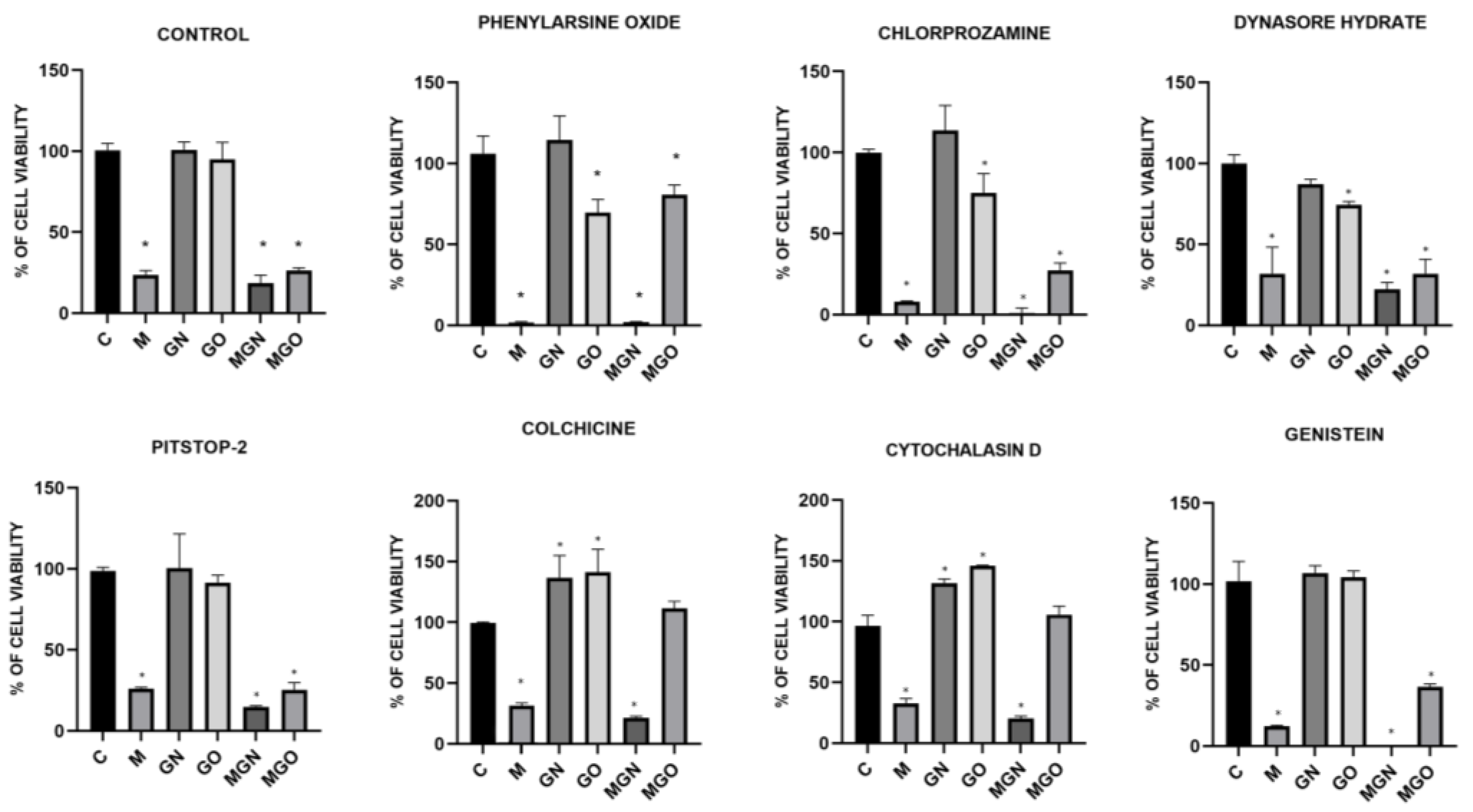
2.7. Transport Inhibitor Test
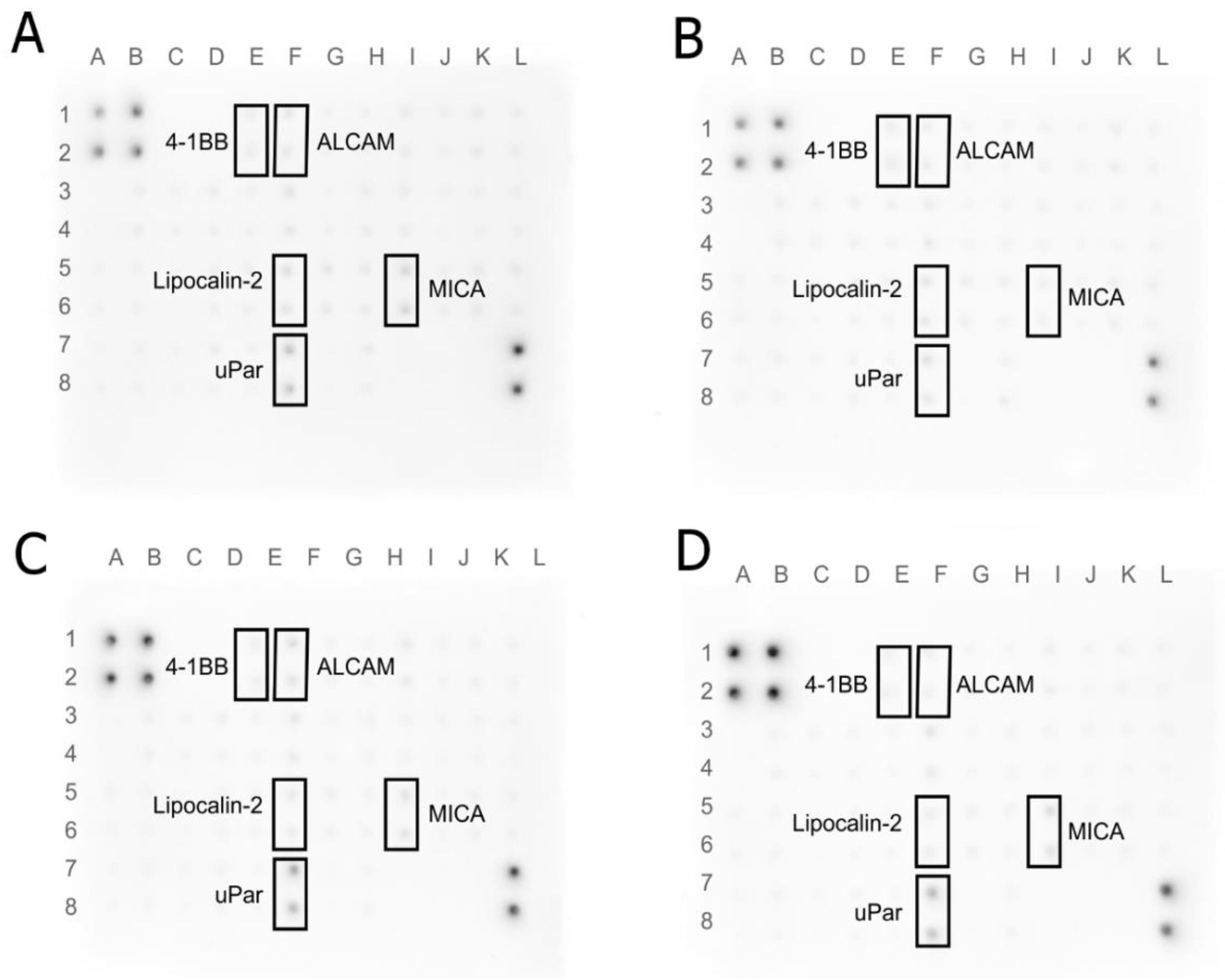
2.8. Human Receptor Antibody Array
2.9. Statistical Analysis
3. Results
3.1. Entrapment Efficiency
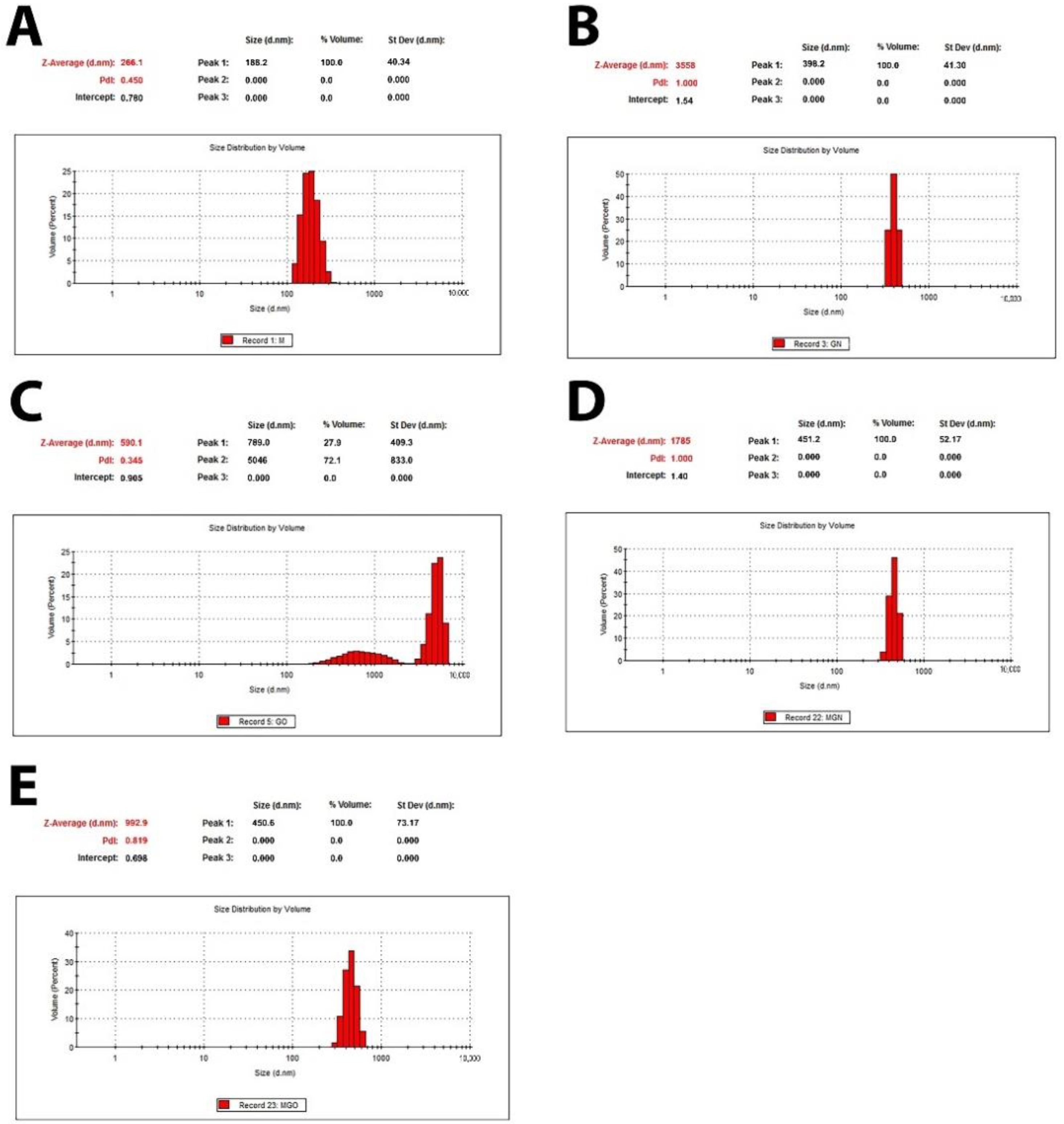
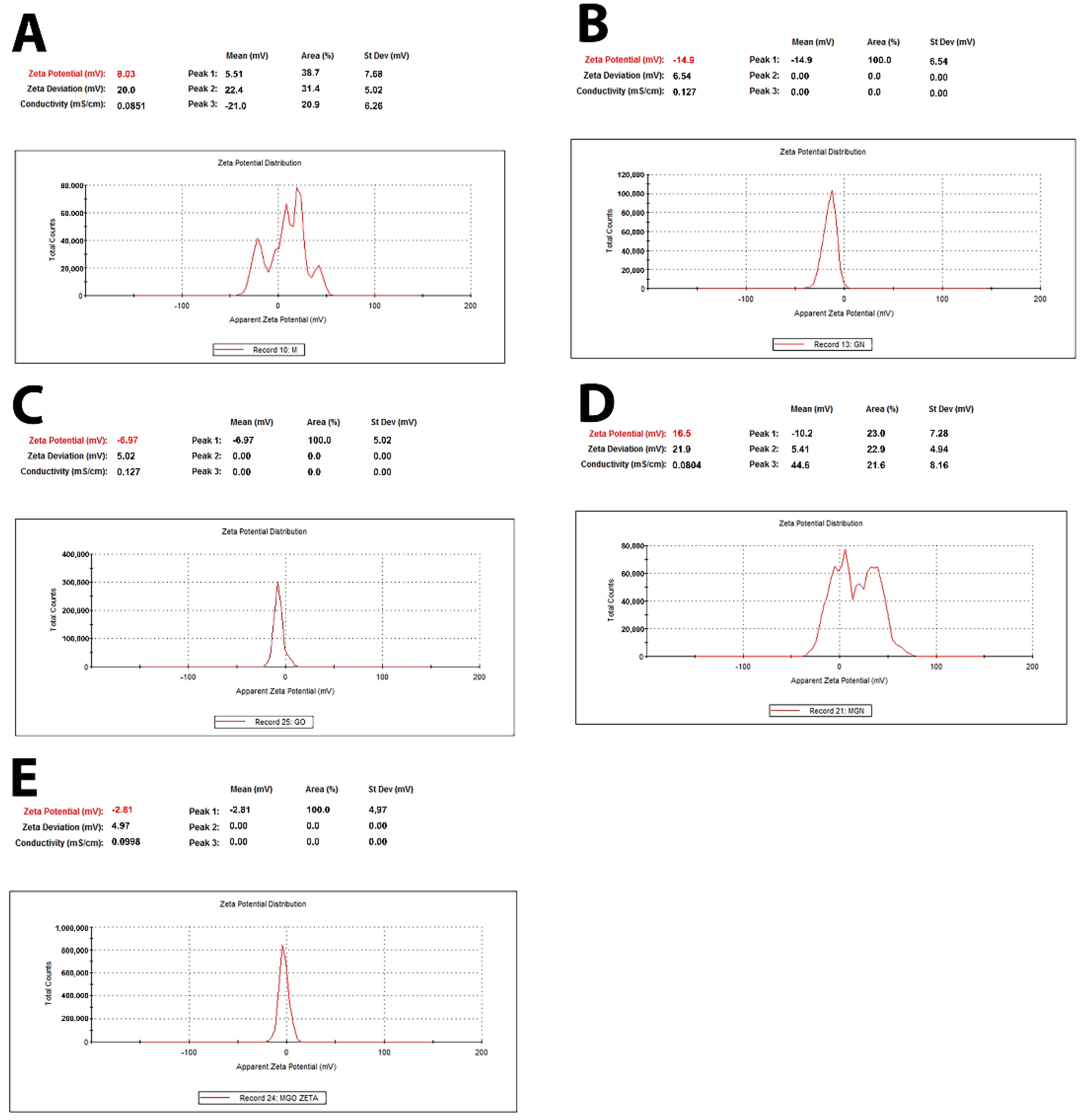
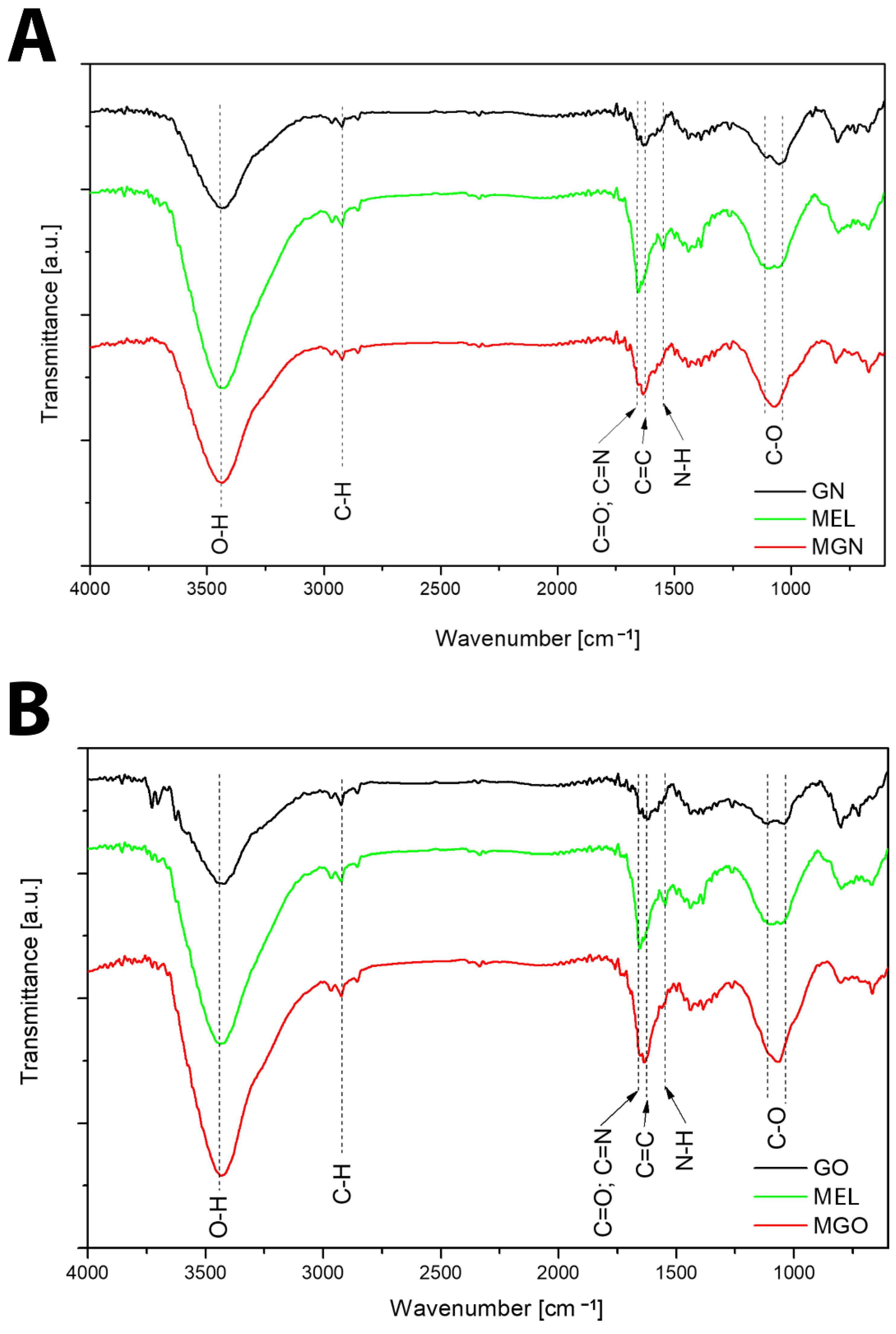
3.2. Characterization of Complexes
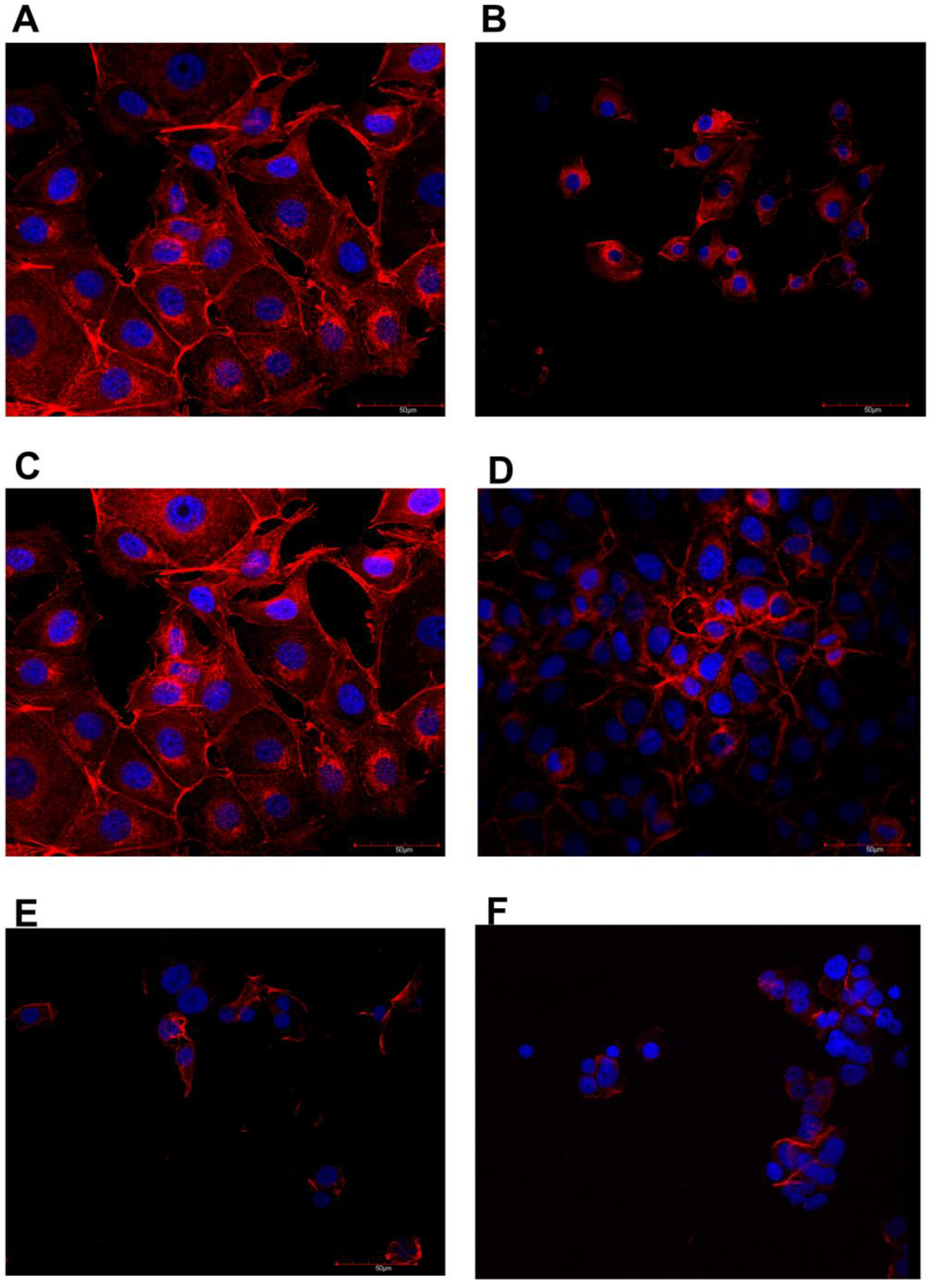
3.3. Confocal Microscopy
3.4. Intracellular pH
3.5. Potential of Cell Membrane
3.6. Transport Inhibitor Test
3.7. Human Receptor Antibody Array
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, X.; Zhang, Q.; Xing, W.; Wang, W. Role of MicroRNA/LncRNA Intertwined With the Wnt/β-Catenin Axis in Regulating the Pathogenesis of Triple-Negative Breast Cancer. Front. Pharm. 2022, 13, 814971. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer: Statistics | Cancer.Net. Available online: https://www.cancer.net/cancer-types/breast-cancer/statistics (accessed on 25 October 2022).
- Rivera-Yañez, N.; Rivera-Yañez, C.R.; Pozo-Molina, G.; Méndez-Catalá, C.F.; Méndez-Cruz, A.R.; Nieto-Yañez, O. Biomedical Properties of Propolis on Diverse Chronic Diseases and Its Potential Applications and Health Benefits. Nutrients 2021, 13, 78. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Fekri, H.S.; Hashemi, F.; Hushmandi, K.; Mohammadinejad, R.; Ashrafizadeh, M.; Zarrabi, A.; Garg, M. Venom Peptides in Cancer Therapy: An Updated Review on Cellular and Molecular Aspects. Pharmacol. Res. 2021, 164, 105327. [Google Scholar] [CrossRef] [PubMed]
- Dutta, P.; Sahu, R.K.; Dey, T.; Lahkar, M.D.; Manna, P.; Kalita, J. Beneficial Role of Insect-Derived Bioactive Components against Inflammation and Its Associated Complications (Colitis and Arthritis) and Cancer. Chem. Biol. Interact 2019, 313, 108824. [Google Scholar] [CrossRef] [PubMed]
- Hixon, K.R.; Klein, R.C.; Eberlin, C.T.; Linder, H.R.; Ona, W.J.; Gonzalez, H.; Sell, S.A. A Critical Review and Perspective of Honey in Tissue Engineering and Clinical Wound Healing. Adv. Wound Care 2019, 8, 403–415. [Google Scholar] [CrossRef]
- Yupanqui Mieles, J.; Vyas, C.; Aslan, E.; Humphreys, G.; Diver, C.; Bartolo, P. Honey: An Advanced Antimicrobial and Wound Healing Biomaterial for Tissue Engineering Applications. Pharmaceutics 2022, 14, 1663. [Google Scholar] [CrossRef]
- Deglovic, J.; Majtanova, N.; Majtan, J. Antibacterial and Antibiofilm Effect of Honey in the Prevention of Dental Caries: A Recent Perspective. Foods 2022, 11, 2670. [Google Scholar] [CrossRef]
- Molan, P.C. Potential of Honey in the Treatment of Wounds and Burns. Am. J. Clin. Derm. 2001, 2, 13–19. [Google Scholar] [CrossRef]
- Raghuraman, H.; Chattopadhyay, A. Melittin: A Membrane-Active Peptide with Diverse Functions. Biosci. Rep. 2007, 27, 189–223. [Google Scholar] [CrossRef]
- Belluati, A.; Mikhalevich, V.; Yorulmaz Avsar, S.; Daubian, D.; Craciun, I.; Chami, M.; Meier, W.P.; Palivan, C.G. How Do the Properties of Amphiphilic Polymer Membranes Influence the Functional Insertion of Peptide Pores? Biomacromolecules 2020, 21, 701–715. [Google Scholar] [CrossRef]
- Lee, M.T.; Sun, T.L.; Hung, W.C.; Huang, H.W. Process of Inducing Pores in Membranes by Melittin. Proc. Natl. Acad. Sci. USA 2013, 110, 14243–14248. [Google Scholar] [CrossRef]
- Wimley, W.C. How Does Melittin Permeabilize Membranes? Biophys. J. 2018, 114, 251. [Google Scholar] [CrossRef]
- Nikodijević, D.D.; Milutinović, M.G.; Cvetković, D.M.; Ćupurdija, M.Đ.; Jovanović, M.M.; Mrkić, I.V.; Jankulović-Gavrović, M.Đ.; Marković, S.D. Impact of Bee Venom and Melittin on Apoptosis and Biotransformation in Colorectal Carcinoma Cell Lines. Toxin Rev. 2019, 40, 1272–1279. [Google Scholar] [CrossRef]
- Galdiero, E.; Siciliano, A.; Gesuele, R.; di Onofrio, V.; Falanga, A.; Maione, A.; Liguori, R.; Libralato, G.; Guida, M. Melittin Inhibition and Eradication Activity for Resistant Polymicrobial Biofilm Isolated from a Dairy Industry after Disinfection. Int. J. Microbiol. 2019, 2019, 125654442. [Google Scholar] [CrossRef]
- Juśkiewicz, J.; Rawicka, A.; Fotschki, B.; Majewski, M.; Zduńczyk, Z. Influence of Supplementation of Lactoferrin, Melittin and Cecropin A to Rat Diet on Changes in Faecal Ammonia Concentrations, Short-Chain Fatty Acid Concentrations and Activities of Bacterial Enzymes. Animals 2021, 11, 1203. [Google Scholar] [CrossRef]
- Hong, J.; Lu, X.; Deng, Z.; Xiao, S.; Yuan, B.; Yang, K. How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane. Molecules 2019, 24, 1775. [Google Scholar] [CrossRef]
- Józefiak, A.; Engberg, R. Insect Proteins as a Potential Source of Antimicrobial Peptides in Livestock Production. A Review. J. Anim. Feed Sci. 2017, 26, 87–99. [Google Scholar] [CrossRef]
- Nguyen, M.H.L.; DiPasquale, M.; Rickeard, B.W.; Yip, C.G.; Greco, K.N.; Kelley, E.G.; Marquardt, D. Time-Resolved SANS Reveals Pore-Forming Peptides Cause Rapid Lipid Reorganization. New J. Chem. 2021, 45, 447–456. [Google Scholar] [CrossRef]
- Gajski, G.; Domijan, A.M.; Žegura, B.; Štern, A.; Gerić, M.; Novak Jovanović, I.; Vrhovac, I.; Madunić, J.; Breljak, D.; Filipič, M.; et al. Melittin Induced Cytogenetic Damage, Oxidative Stress and Changes in Gene Expression in Human Peripheral Blood Lymphocytes. Toxicon 2016, 110, 56–67. [Google Scholar] [CrossRef]
- Carpena, M.; Nuñez-Estevez, B.; Soria-Lopez, A.; Simal-Gandara, J. Bee Venom: An Updating Review of Its Bioactive Molecules and Its Health Applications. Nutrients 2020, 12, 3360. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Y.; Ye, Y.; Wang, X.R.; Lin, L.T.; Xiao, L.Y.; Zhou, P.; Shi, G.X.; Liu, C.Z. Bee Venom Therapy: Potential Mechanisms and Therapeutic Applications. Toxicon 2018, 148, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, O.; Fahim, H.; Mahmoud, A.; Ahmed, E. Bee Venom and Hesperidin Effectively Mitigate Complete Freund’s Adjuvant-Induced Arthritis Via Immunomodulation and Enhancement of Antioxidant Defense System. Arch. Rheumatol. 2017, 33, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Choi, M.S.; Kwak, D.H.; Oh, K.W.; Yoon, D.Y.; Han, S.B.; Song, H.S.; Song, M.J.; Hong, J.T. Anti-Cancer Effect of Bee Venom in Prostate Cancer Cells through Activation of Caspase Pathway via Inactivation of NF-ΚB. Prostate 2011, 71, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Ceremuga, M.; Stela, M.; Janik, E.; Gorniak, L.; Synowiec, E.; Sliwinski, T.; Sitarek, P.; Saluk-Bijak, J.; Bijak, M. Melittin-A Natural Peptide from Bee Venom Which Induces Apoptosis in Human Leukaemia Cells. Biomolecules 2020, 10, 247. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodzadeh, A.; Zarrinnahad, H.; Bagheri, K.P.; Moradia, A.; Shahbazzadeh, D. First Report on the Isolation of Melittin from Iranian Honey Bee Venom and Evaluation of Its Toxicity on Gastric Cancer AGS Cells. J. Chin. Med. Assoc. 2015, 78, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Song, C.-C.; Lu, X.; Cheng, B.-B.; Du, J.; Li, B.; Ling, C.-Q. Effects of Melittin on Growth and Angiogenesis of Human Hepatocellular Carcinoma BEL-7402 Cell Xenografts in Nude Mice. Ai Zheng 2007, 26, 1315–1322. [Google Scholar]
- Sun, D.; Sun, M.; Zhu, W.; Wang, Z.; Li, Y.; Ma, J. The Anti-Cancer Potency and Mechanism of a Novel Tumor-Activated Fused Toxin, DLM. Toxins 2015, 7, 423–438. [Google Scholar] [CrossRef]
- Daniluk, K.; Kutwin, M.; Grodzik, M.; Wierzbicki, M.; Strojny, B.; Szczepaniak, J.; Balaban, J.; Sosnowska, M.; Chwalibog, A.; Sawosz, E.; et al. Use of Selected Carbon Nanoparticles as Melittin Carriers for MCF-7 and MDA-MB-231 Human Breast Cancer Cells. Materials 2020, 13, 90. [Google Scholar] [CrossRef]
- Yao, J.; Zhang, Z.; Li, S.; Li, B.; Wang, X.H. Melittin Inhibits Proliferation, Migration and Invasion of Bladder Cancer Cells by Regulating Key Genes Based on Bioinformatics and Experimental Assays. J. Cell. Mol. Med. 2020, 24, 655. [Google Scholar] [CrossRef]
- Qin, G.; Chen, Y.; Li, H.; Xu, S.; Li, Y.; Sun, J.; Rao, W.; Chen, C.; Du, M.; He, K.; et al. Melittin Inhibits Tumor Angiogenesis Modulated by Endothelial Progenitor Cells Associated with the SDF-1α/CXCR4 Signaling Pathway in a UMR-106 Osteosarcoma Xenograft Mouse Model. Mol. Med. Rep. 2016, 14, 57–68. [Google Scholar] [CrossRef]
- Moreno, M.; Toxins, E.G. Three Valuable Peptides from Bee and Wasp Venoms for Therapeutic and Biotechnological Use: Melittin, Apamin and Mastoparan. Toxins 2015, 7, 1126–1150. [Google Scholar] [CrossRef]
- Hoskin, D.W.; Ramamoorthy, A. Studies on Anticancer Activities of Antimicrobial Peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2008, 1778, 357–375. [Google Scholar] [CrossRef]
- Gajski, G.; Garaj-Vrhovac, V. Melittin: A Lytic Peptide with Anticancer Properties. Environ. Toxicol. Pharm. 2013, 36, 697–705. [Google Scholar] [CrossRef]
- Dey, A.D.; Bigham, A.; Esmaeili, Y.; Ashrafizadeh, M.; Moghaddam, F.D.; Tan, S.C.; Yousefiasl, S.; Sharma, S.; Maleki, A.; Rabiee, N.; et al. Dendrimers as Nanoscale Vectors: Unlocking the Bars of Cancer Therapy. Semin. Cancer Biol. 2022, 86, 396–419. [Google Scholar] [CrossRef]
- Rahimi, S.; Chen, Y.; Zareian, M.; Pandit, S.; Mijakovic, I. Cellular and Subcellular Interactions of Graphene-Based Materials with Cancerous and Non-Cancerous Cells. Adv. Drug Deliv. Rev. 2022, 189, 114467. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, B.; Zheng, J.; Yu, M.; Zhou, T.; Zhao, K.; Jia, Y.; Gao, X.; Chen, C.; Wei, T. The Inhibition of Migration and Invasion of Cancer Cells by Graphene via the Impairment of Mitochondrial Respiration. Biomaterials 2014, 35, 1597–1607. [Google Scholar] [CrossRef]
- Wu, C.; Wang, C.; Zheng, J.; Luo, C.; Li, Y.; Guo, S.; Zhang, J. Vacuolization in Cytoplasm and Cell Membrane Permeability Enhancement Triggered by Micrometer-Sized Graphene Oxide. ACS Nano 2015, 9, 7913–7924. [Google Scholar] [CrossRef]
- Ou, L.; Song, B.; Liang, H.; Liu, J.; Feng, X.; Deng, B.; Sun, T.; Shao, L. Toxicity of Graphene-Family Nanoparticles: A General Review of the Origins and Mechanisms. Part Fibre Toxicol. 2016, 13, 57. [Google Scholar] [CrossRef]
- Contreras-Torres, F.F.; Rodríguez-Galván, A.; Guerrero-Beltrán, C.E.; Martínez-Lorán, E.; Vázquez-Garza, E.; Ornelas-Soto, N.; García-Rivas, G. Differential Cytotoxicity and Internalization of Graphene Family Nanomaterials in Myocardial Cells. Mater. Sci. Eng. C 2017, 73, 633–642. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma Membrane Changes during Programmed Cell Deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef]
- Ricci, J.-E.; Gottlieb, R.A.; Green, D.R. Caspase-Mediated Loss of Mitochondrial Function and Generation of Reactive Oxygen Species during Apoptosis. J. Cell Biol. 2003, 160, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Rukmini, R.; Chattopadhyay, A. Modulation of Tryptophan Environment in Membrane-Bound Melittin by Negatively Charged Phospholipids: Implications in Membrane Organization and Function. Biochemistry 1997, 36, 14291–14305. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.J.; Tilly, H.; Ward, J.H.; Macarthur, D.C.; Lowe, J.; Coyle, B.; Grundy, R.G. CD105 (Endoglin) Exerts Prognostic Effects via Its Role in the Microvascular Niche of Paediatric High Grade Glioma. Acta Neuropathol. 2012, 124, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Aburayan, W.S.; Alajmi, A.M.; Alfahad, A.J.; Alsharif, W.K.; Alshehri, A.A.; Booq, R.Y.; Alsudir, S.A.; Alsulaihem, F.M.; Bukhary, H.A.; Badr, M.Y.; et al. Melittin from Bee Venom Encapsulating Electrospun Fibers as a Potential Antimicrobial Wound Dressing Patches for Skin Infections. Pharmaceutics 2022, 14, 725. [Google Scholar] [CrossRef] [PubMed]
- Lesiak, B.; Trykowski, G.; Tóth, J.; Biniak, S.; Kövér, L.; Rangam, N.; Stobinski, L.; Malolepszy, A. Chemical and Structural Properties of Reduced Graphene Oxide—Dependence on the Reducing Agent. J. Mater. Sci. 2021, 56, 3738–3754. [Google Scholar] [CrossRef]
- Trayes, K.P.; Cokenakes, S.E.H. Breast Cancer Treatment. Am. Fam. Physician 2021, 104, 171–178. [Google Scholar]
- Özbolat, S.N.; Ayna, A. Chrysin Suppresses HT-29 Cell Death Induced by Diclofenac through Apoptosis and Oxidative Damage. Nutr. Cancer 2021, 73, 1419–1428. [Google Scholar] [CrossRef]
- Duffy, C.; Sorolla, A.; Wang, E.; Golden, E.; Woodward, E.; Davern, K.; Ho, D.; Johnstone, E.; Pfleger, K.; Redfern, A.; et al. Honeybee Venom and Melittin Suppress Growth Factor Receptor Activation in HER2-Enriched and Triple-Negative Breast Cancer. NPJ Precis. Oncol. 2020, 4, 24. [Google Scholar] [CrossRef]
- DeGrado, W.F.; Musso, G.F.; Lieber, M.; Kaiser, E.T.; Kézdy, F.J. Kinetics and Mechanism of Hemolysis Induced by Melittin and by a Synthetic Melittin Analogue. Biophys. J. 1982, 37, 329–338. [Google Scholar] [CrossRef]
- Soliman, C.; Eastwood, S.; Truong, V.K.; Ramsland, P.A.; Elbourne, A. The Membrane Effects of Melittin on Gastric and Colorectal Cancer. PLoS ONE 2019, 14, e0224028. [Google Scholar] [CrossRef]
- Lischer, K.; Sitorus, S.R.A.; Guslianto, B.W.; Avila, F.; Khayrani, A.C.; Sahlan, M. Anti-Breast Cancer Activity on MCF-7 Cells of Melittin from Indonesia’s Apis Cerana: An In Vitro Study. Asian Pac. J. Cancer Prev. 2021, 22, 3913. [Google Scholar] [CrossRef]
- Liu, M.; Wang, H.; Liu, L.; Wang, B.; Sun, G. Melittin-MIL-2 Fusion Protein as a Candidate for Cancer Immunotherapy. J. Transl. Med. 2016, 14, 155. [Google Scholar] [CrossRef]
- Mao, J.; Liu, S.; Ai, M.; Wang, Z.; Wang, D.; Li, X.; Hu, K.; Gao, X.; Yang, Y. A Novel Melittin Nano-Liposome Exerted Excellent Anti-Hepatocellular Carcinoma Efficacy with Better Biological Safety. J. Hematol. Oncol. 2017, 10, 71. [Google Scholar] [CrossRef]
- Hematyar, M.; Soleimani, M.; Es-haghi, A.; Rezaei Mokarram, A. Synergistic Co-Delivery of Doxorubicin and Melittin Using Functionalized Magnetic Nanoparticles for Cancer Treatment: Loading and in Vitro Release Study by LC–MS/MS. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1226–1235. [Google Scholar] [CrossRef]
- Wang, H.; Wang, S.; Wang, R.; Wang, X.; Jiang, K.; Xie, C.; Zhan, C.; Wang, H.; Lu, W. Co-Delivery of Paclitaxel and Melittin by Glycopeptide-Modified Lipodisks for Synergistic Anti-Glioma Therapy. Nanoscale 2019, 11, 13069–13077. [Google Scholar] [CrossRef]
- Dang, Y.Q.; Li, H.W.; Wu, Y. Construction of a Supramolecular Förster Resonance Energy Transfer System and Its Application Based on the Interaction between Cy3-Labeled Melittin and Phosphocholine Encapsulated Quantum Dots. ACS Appl. Mater. Interfaces 2012, 4, 1267–1272. [Google Scholar] [CrossRef]
- Dabbagh Moghaddam, F.; Akbarzadeh, I.; Marzbankia, E.; Farid, M.; Khaledi, L.; Reihani, A.H.; Javidfar, M.; Mortazavi, P. Delivery of Melittin-Loaded Niosomes for Breast Cancer Treatment: An in Vitro and in Vivo Evaluation of Anti-Cancer Effect. Cancer Nanotechnol. 2021, 12, 14. [Google Scholar] [CrossRef]
- Ahlgren, S.; Reijmar, K.; Edwards, K. Targeting Lipodisks Enable Selective Delivery of Anticancer Peptides to Tumor Cells. Nanomedicine 2017, 13, 2325–2328. [Google Scholar] [CrossRef]
- Jeong, I.; Kim, B.-S.; Lee, H.; Lee, K.-M.; Shim, I.; Kang, S.-K.; Yin, C.-S.; Hahm, D.-H. Prolonged Analgesic Effect of PLGA-Encapsulated Bee Venom on Formalin-Induced Pain in Rats. Int. J. Pharm. 2009, 380, 62–66. [Google Scholar] [CrossRef]
- Qi, J.; Chen, Y.; Xue, T.; Lin, Y.; Huang, S.; Cao, S.; Wang, X.; Su, Y.; Lin, Z. Graphene Oxide-Based Magnetic Nanocomposites for the Delivery of Melittin to Cervical Cancer HeLa Cells. Nanotechnology 2020, 31, 065102. [Google Scholar] [CrossRef]
- Honary, S.; Zahir, F. Effect of Zeta Potential on the Properties of Nano-Drug Delivery Systems—A Review (Part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. [Google Scholar] [CrossRef]
- Grillo-Hill, B.K.; Choi, C.; Jimenez-Vidal, M.; Barber, D.L. Increased H+ Efflux Is Sufficient to Induce Dysplasia and Necessary for Viability with Oncogene Expression. Elife 2015, 4, e03270. [Google Scholar] [CrossRef] [PubMed]
- Cong, D.; Zhu, W.; Shi, Y.; Pointer, K.B.; Clark, P.A.; Shen, H.; Kuo, J.S.; Hu, S.; Sun, D. Upregulation of NHE1 Protein Expression Enables Glioblastoma Cells to Escape TMZ-Mediated Toxicity via Increased H+ Extrusion, Cell Migration and Survival. Carcinogenesis 2014, 35, 2014–2024. [Google Scholar] [CrossRef] [PubMed]
- Boron, W.F. Regulation of Intracellular PH. Adv. Physiol. Educ. 2004, 28, 160–179. [Google Scholar] [CrossRef]
- Lauritzen, G.; Stock, C.M.; Lemaire, J.; Lund, S.F.; Jensen, M.F.; Damsgaard, B.; Petersen, K.S.; Wiwel, M.; Rønnov-Jessen, L.; Schwab, A.; et al. The Na+/H+ Exchanger NHE1, but Not the Na+, HCO3(-) Cotransporter NBCn1, Regulates Motility of MCF7 Breast Cancer Cells Expressing Constitutively Active ErbB2. Cancer Lett. 2012, 317, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Koltai, T. Triple-Edged Therapy Targeting Intracellular Alkalosis and Extracellular Acidosis in Cancer. Semin. Cancer Biol. 2017, 43, 139–146. [Google Scholar] [CrossRef]
- Binggeli, R.; Weinstein, R.C. Deficits in Elevating Membrane Potential of Rat Fibrosarcoma Cells after Cell Contact. Cancer Res. 1985, 45, 235–241. [Google Scholar]
- Marmo, A.A.; Morris, D.M.; Schwalke, M.A.; Iliev, I.G.; Rogers, S. Electrical Potential Measurements in Human Breast Cancer and Benign Lesions. Tumor. Biol. 1994, 15, 147–152. [Google Scholar] [CrossRef]
- Quicke, P.; Sun, Y.; Arias-Garcia, M.; Acker, C.D.; Djamgoz, M.B.A.; Bakal, C.; Foust, A.J. Membrane Voltage Fluctuations in Human Breast Cancer Cells. bioRxiv 2021. [Google Scholar] [CrossRef]
- Dutta, D.; Williamson, C.D.; Cole, N.B.; Donaldson, J.G. Pitstop 2 Is a Potent Inhibitor of Clathrin-Independent Endocytosis. PLoS ONE 2012, 7, e45799. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, Q.M.; Du, G.H. Colchicine. In Natural Small Molecule Drugs from Plants; Sringer: Berlin/Heidelberg, Germany, 2022; pp. 503–508. [Google Scholar] [CrossRef]
- Leung, Y.Y.; Hui, L.L.Y.; Kraus, V.B. Colchicine—Update on Mechanisms of Action and Therapeutic Uses. Semin Arthritis Rheum 2015, 45, 341. [Google Scholar] [CrossRef]
- Shoji, K.; Ohashi, K.; Sampei, K.; Oikawa, M.; Mizuno, K. Cytochalasin D Acts as an Inhibitor of the Actin–Cofilin Interaction. Biochem. Biophys. Res. Commun. 2012, 424, 52–57. [Google Scholar] [CrossRef]
- Sit, K.H.; Bay, B.H.; Wong, K.P. Effect of Genistein, a Tyrosine-Specific Protein Kinase Inhibitor, on Cell Rounding by PH Upshifting. In Vitro Cell Dev. Biol. Anim. 1993, 29A, 395–402. [Google Scholar] [CrossRef]
- Frost, S.C.; Lane, M.D.; Gibbs, E.M. Effect of Phenylarsine Oxide on Fluid Phase Endocytosis: Further Evidence for Activation of the Glucose Transporter. J. Cell Physiol. 1989, 141, 467–474. [Google Scholar] [CrossRef]
- Massol, P.; Montcourrier, P.; Guillemot, J.C.; Chavrier, P. Fc Receptor-Mediated Phagocytosis Requires CDC42 and Rac1. EMBO J. 1998, 17, 6219–6229. [Google Scholar] [CrossRef]
- Qian, Z.M.; Li, H.; Sun, H.; Ho, K. Targeted Drug Delivery via the Transferrin Receptor-Mediated Endocytosis Pathway. Pharm. Rev. 2002, 54, 561–587. [Google Scholar] [CrossRef]
- Preta, G.; Cronin, J.G.; Sheldon, I.M. Dynasore—Not Just a Dynamin Inhibitor. Cell Commun. Signal 2015, 13, 24. [Google Scholar] [CrossRef]
- Kirchhausen, T.; Owen, D.; Harrison, S.C. Molecular Structure, Function, and Dynamics of Clathrin-Mediated Membrane Traffic. Cold Spring Harb. Perspect. Biol. 2014, 6, a016725. [Google Scholar] [CrossRef]
- Davies, S.R.; Dent, C.; Watkins, G.; King, J.A.; Mokbel, K.; Jiang, W.G. Expression of the Cell to Cell Adhesion Molecule, ALCAM, in Breast Cancer Patients and the Potential Link with Skeletal Metastasis. Oncol. Rep. 2008, 19, 555–561. [Google Scholar] [CrossRef]
- Burandt, E.; Noubar, T.B.; Lebeau, A.; Minner, S.; Burdelski, C.; Jänicke, F.; Müller, V.; Terracciano, L.; Simon, R.; Sauter, G.; et al. Loss of ALCAM Expression Is Linked to Adverse Phenotype and Poor Prognosis in Breast Cancer: A TMA-Based Immunohistochemical Study on 2,197 Breast Cancer Patients. Oncol. Rep. 2014, 32, 2628–2634. [Google Scholar] [CrossRef]
- Piao, D.; Jiang, T.; Liu, G.; Wang, B.; Xu, J.; Zhu, A. Clinical Implications of Activated Leukocyte Cell Adhesion Molecule Expression in Breast Cancer. Mol. Biol. Rep. 2012, 39, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Jezierska, A.; Matysiak, W.; Motyl, T. ALCAM/CD166 Protects Breast Cancer Cells against Apoptosis and Autophagy. Med. Sci. Monit. 2006, 12, BR263–BR273. [Google Scholar] [PubMed]
- Chu, S.T.; Lin, H.J.; Huang, H.L.; Chen, Y.H. The Hydrophobic Pocket of 24p3 Protein from Mouse Uterine Luminal Fluid: Fatty Acid and Retinol Binding Activity and Predicted Structural Similarity to Lipocalins. J. Pept. Res. 1998, 52, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Flower, D.R. The Lipocalin Protein Family: A Role in Cell Regulation. FEBS Lett. 1994, 354, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Asimakopoulou, A.; Weiskirchen, S.; Weiskirchen, R. Lipocalin 2 (LCN2) Expression in Hepatic Malfunction and Therapy. Front. Physiol. 2016, 7, 430. [Google Scholar] [CrossRef]
- Hu, C.; Yang, K.; Li, M.; Huang, W.; Zhang, F.; Wang, H. Lipocalin 2: A Potential Therapeutic Target for Breast Cancer Metastasis. Onco Targets 2018, 11, 8099. [Google Scholar] [CrossRef]
- Leng, X.; Wu, Y.; Arlinghaus, R.B. Relationships of Lipocalin 2 with Breast Tumorigenesis and Metastasis. J. Cell. Physiol. 2011, 226, 309–314. [Google Scholar] [CrossRef]
- Tong, Z.; Wu, X.; Ovcharenko, D.; Zhu, J.; Chen, C.S.; Kehrer, J.P. Neutrophil Gelatinase-Associated Lipocalin as a Survival Factor. Biochem. J. 2005, 391, 441. [Google Scholar] [CrossRef]
- Arango, D.; Laiho, P.; Kokko, A.; Alhopuro, P.; Sammalkorpi, H.; Salovaara, R.; Nicorici, D.; Hautaniemi, S.; Alazzouzi, H.; Mecklin, J.P.; et al. Gene-Expression Profiling Predicts Recurrence in Dukes’ C Colorectal Cancer. Gastroenterology 2005, 129, 874–884. [Google Scholar] [CrossRef]
- Iannetti, A.; Pacifico, F.; Acquaviva, R.; Lavorgna, A.; Crescenzi, E.; Vascotto, C.; Tell, G.; Salzano, A.M.; Scaloni, A.; Vuttariello, E.; et al. The Neutrophil Gelatinase-Associated Lipocalin (NGAL), a NF-KappaB-Regulated Gene, Is a Survival Factor for Thyroid Neoplastic Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 14058–14063. [Google Scholar] [CrossRef]
- Cho, H.B.; Kim, J.H. Lipocalin2 Expressions Correlate Significantly With Tumor Differentiation in Epithelial Ovarian Cancer. J. Histochem. Cytochem. 2009, 57, 513. [Google Scholar] [CrossRef]
- Borkham-Kamphorst, E.; van de Leur, E.; Zimmermann, H.W.; Karlmark, K.R.; Tihaa, L.; Haas, U.; Tacke, F.; Berger, T.; Mak, T.W.; Weiskirchen, R. Protective Effects of Lipocalin-2 (LCN2) in Acute Liver Injury Suggest a Novel Function in Liver Homeostasis. Biochim. Biophys. Acta 2013, 1832, 660–673. [Google Scholar] [CrossRef]
- Borkham-Kamphorst, E.; Drews, F.; Weiskirchen, R. Induction of Lipocalin-2 Expression in Acute and Chronic Experimental Liver Injury Moderated by pro-Inflammatory Cytokines Interleukin-1β through Nuclear Factor-ΚB Activation. Liver Int. 2011, 31, 656–665. [Google Scholar] [CrossRef]
- di Mauro, C.; Pesapane, A.; Formisano, L.; Rosa, R.; D’Amato, V.; Ciciola, P.; Servetto, A.; Marciano, R.; Orsini, R.C.; Monteleone, F.; et al. Urokinase-Type Plasminogen Activator Receptor (UPAR) Expression Enhances Invasion and Metastasis in RAS Mutated Tumors. Sci. Rep. 2017, 7, 9338. [Google Scholar] [CrossRef]
- Mahmood, N.; Mihalcioiu, C.; Rabbani, S.A. Multifaceted Role of the Urokinase-Type Plasminogen Activator (UPA) and Its Receptor (UPAR): Diagnostic, Prognostic, and Therapeutic Applications. Front. Oncol. 2018, 8, 24. [Google Scholar] [CrossRef]
- Nantajit, D.; Chailapakul, P.; Bawornpatarapakorn, S.; Chamchod, S.; Laebua, K. Prognostic Significance of UPA and UPAR Expression in Patients with Cervical Cancer Undergoing Radiotherapy. Oncol. Lett. 2021, 21, 423. [Google Scholar] [CrossRef]
- Persson, M.; Skovgaard, D.; Brandt-Larsen, M.; Christensen, C.; Madsen, J.; Nielsen, C.H.; Thurison, T.; Klausen, T.L.; Holm, S.; Loft, A.; et al. First-in-Human UPAR PET: Imaging of Cancer Aggressiveness. Theranostics 2015, 5, 1303–1316. [Google Scholar] [CrossRef]
- Duffy, M.J.; Duggan, C. The Urokinase Plasminogen Activator System: A Rich Source of Tumour Markers for the Individualised Management of Patients with Cancer. Clin. Biochem. 2004, 37, 541–548. [Google Scholar] [CrossRef]
- Tsai, M.C.; Yen, Y.H.; Chang, K.C.; Hung, C.H.; Chen, C.H.; Lin, M.T.; Hu, T.H. Elevated Levels of Serum Urokinase Plasminogen Activator Predict Poor Prognosis in Hepatocellular Carcinoma after Resection. BMC. Cancer 2019, 19, 1169. [Google Scholar] [CrossRef]
- Almasi, C.E.; Christensen, I.J.; Høyer-Hansen, G.; Danø, K.; Pappot, H.; Dienemann, H.; Muley, T. Urokinase Receptor Forms in Serum from Non-Small Cell Lung Cancer Patients: Relation to Prognosis. Lung Cancer 2011, 74, 510–515. [Google Scholar] [CrossRef]
- Masucci, M.T.; Minopoli, M.; di Carluccio, G.; Motti, M.L.; Carriero, M.V. Therapeutic Strategies Targeting Urokinase and Its Receptor in Cancer. Cancers 2022, 14, 498. [Google Scholar] [CrossRef] [PubMed]
- Vinay, D.S.; Kwon, B.S. 4-1BB Signaling beyond T Cells. Cell. Mol. Immunol. 2011, 8, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Vinay, D.S.; Kwon, B.S. 4-1BB (CD137), an Inducible Costimulatory Receptor, as a Specific Target for Cancer Therapy. BMB Rep. 2014, 47, 122. [Google Scholar] [CrossRef] [PubMed]
- Hangiu, O.; Compte, M.; Dinesen, A.; Navarro, R.; Tapia-Galisteo, A.; Mandrup, O.A.; Erce-Llamazares, A.; Lázaro-Gorines, R.; Nehme-Álvarez, D.; Domínguez-Alonso, C.; et al. Tumor Targeted 4-1BB Agonist Antibody-Albumin Fusions with High Affinity to FcRn Induce Anti-Tumor Immunity without Toxicity. iScience 2022, 25, 104958. [Google Scholar] [CrossRef] [PubMed]
- Stoll, A.; Bruns, H.; Fuchs, M.; Völkl, S.; Nimmerjahn, F.; Kunz, M.; Peipp, M.; Mackensen, A.; Mougiakakos, D. CD137 (4-1BB) Stimulation Leads to Metabolic and Functional Reprogramming of Human Monocytes/Macrophages Enhancing Their Tumoricidal Activity. Leukemia 2021, 35, 3482. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gao, W.; Ma, T.; Wang, R.; Piao, Y.; Dong, X.; Wang, P.; Zhang, X.; Liu, Y.; Su, W.; et al. CD137 Promotes Bone Metastasis of Breast Cancer by Enhancing the Migration and Osteoclast Differentiation of Monocytes/Macrophages. Theranostics 2019, 9, 2950–2966. [Google Scholar] [CrossRef]
- Sarhan, D.; D’Arcy, P.; Lundqvist, A. Regulation of TRAIL-Receptor Expression by the Ubiquitin-Proteasome System. Int. J. Mol. Sci. 2014, 15, 18557–18573. [Google Scholar] [CrossRef]
- Degli-Esposti, M.A.; Dougall, W.C.; Smolak, P.J.; Waugh, J.Y.; Smith, C.A.; Goodwin, R.G. The Novel Receptor TRAIL-R4 Induces NF-KappaB and Protects against TRAIL-Mediated Apoptosis, yet Retains an Incomplete Death Domain. Immunity 1997, 7, 813–820. [Google Scholar] [CrossRef]
- Chaudhary, P.M.; Eby, M.; Jasmin, A.; Bookwalter, A.; Urray, J.M.; Hood, L. Death Receptor 5, a New Member of the TNFR Family, and DR4 Induce FADD-Dependent Apoptosis and Activate the NF-KappaB Pathway. Immunity 1997, 7, 821–830. [Google Scholar] [CrossRef]
- Crosier, P.S.; Freeman, S.A.; Orlic, D.; Bodine, D.M.; Crosier, K.E. The Dtk Receptor Tyrosine Kinase, Which Binds Protein S, Is Expressed during Hematopoiesis. Exp. Hematol. 1996, 24, 318–323. [Google Scholar]
- TYRO3—Tyrosine-Protein Kinase Receptor TYRO3—Homo Sapiens (Human) | UniProtKB | UniProt. Available online: https://www.uniprot.org/uniprotkb/Q06418/entry (accessed on 26 October 2022).
- al Kafri, N.; Hafizi, S. Identification of Signalling Pathways Activated by Tyro3 That Promote Cell Survival, Proliferation and Invasiveness in Human Cancer Cells. Biochem. Biophys. Rep. 2021, 28, 101111. [Google Scholar] [CrossRef]
- Steinle, A.; Li, P.; Morris, D.L.; Groh, V.; Lanier, L.L.; Strong, R.K.; Spies, T. Interactions of Human NKG2D with Its Ligands MICA, MICB, and Homologs of the Mouse RAE-1 Protein Family. Immunogenetics 2001, 53, 279–287. [Google Scholar] [CrossRef]
- Li, P.; Morris, D.L.; Willcox, B.E.; Steinle, A.; Spies, T.; Strong, R.K. Complex Structure of the Activating Immunoreceptor NKG2D and Its MHC Class I-like Ligand MICA. Nat. Immunol. 2001, 2, 443–451. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daniluk, K.; Lange, A.; Pruchniewski, M.; Małolepszy, A.; Sawosz, E.; Jaworski, S. Delivery of Melittin as a Lytic Agent via Graphene Nanoparticles as Carriers to Breast Cancer Cells. J. Funct. Biomater. 2022, 13, 278. https://doi.org/10.3390/jfb13040278
Daniluk K, Lange A, Pruchniewski M, Małolepszy A, Sawosz E, Jaworski S. Delivery of Melittin as a Lytic Agent via Graphene Nanoparticles as Carriers to Breast Cancer Cells. Journal of Functional Biomaterials. 2022; 13(4):278. https://doi.org/10.3390/jfb13040278
Chicago/Turabian StyleDaniluk, Karolina, Agata Lange, Michał Pruchniewski, Artur Małolepszy, Ewa Sawosz, and Sławomir Jaworski. 2022. "Delivery of Melittin as a Lytic Agent via Graphene Nanoparticles as Carriers to Breast Cancer Cells" Journal of Functional Biomaterials 13, no. 4: 278. https://doi.org/10.3390/jfb13040278
APA StyleDaniluk, K., Lange, A., Pruchniewski, M., Małolepszy, A., Sawosz, E., & Jaworski, S. (2022). Delivery of Melittin as a Lytic Agent via Graphene Nanoparticles as Carriers to Breast Cancer Cells. Journal of Functional Biomaterials, 13(4), 278. https://doi.org/10.3390/jfb13040278





