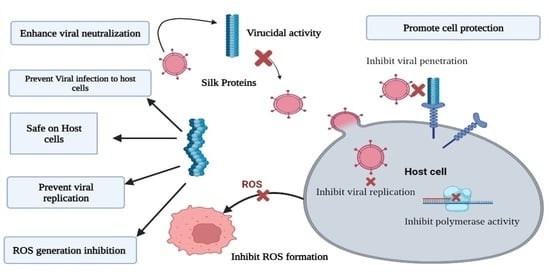
Inhibitory Effects of Bacterial Silk-like Biopolymer on Herpes Simplex Virus Type 1, Adenovirus Type 7 and Hepatitis C Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Whole Bacterial Polymeric Silk-like Protein Isolation and Silk-like Protein Preparation
2.2. Serum Sample Collection
2.3. Human Peripheral Blood Mononuclear Cell Separation
2.4. Cytotoxicity Assays
2.5. The Antiviral Activity of Silk Proteins against ADV7 and HSV-1
2.5.1. MTT and RT-qPCR Assays for Investigation of Antiviral Activity of Silk Protein
2.5.2. Evaluation of the Inhibitory Impact of Silk Protein on DNA Polymerase Activity of ADV7 and HSV-1
2.5.3. The Antiviral Activity of Silk Protein against HCV
2.5.4. Quantification of HCV Genomic and Antigenomic RNA Strands Using RT-PCR
2.5.5. Quantification of the Induced ROS Using Oxidized DCFDA and Flow Cytometry
2.6. Protein Modeling and Validation
2.7. Molecular Docking Analysis
2.8. Assessment of the Binding Affinity in the Docked Complexes
2.9. Active Site Prediction
2.10. Statistical Analysis
3. Results
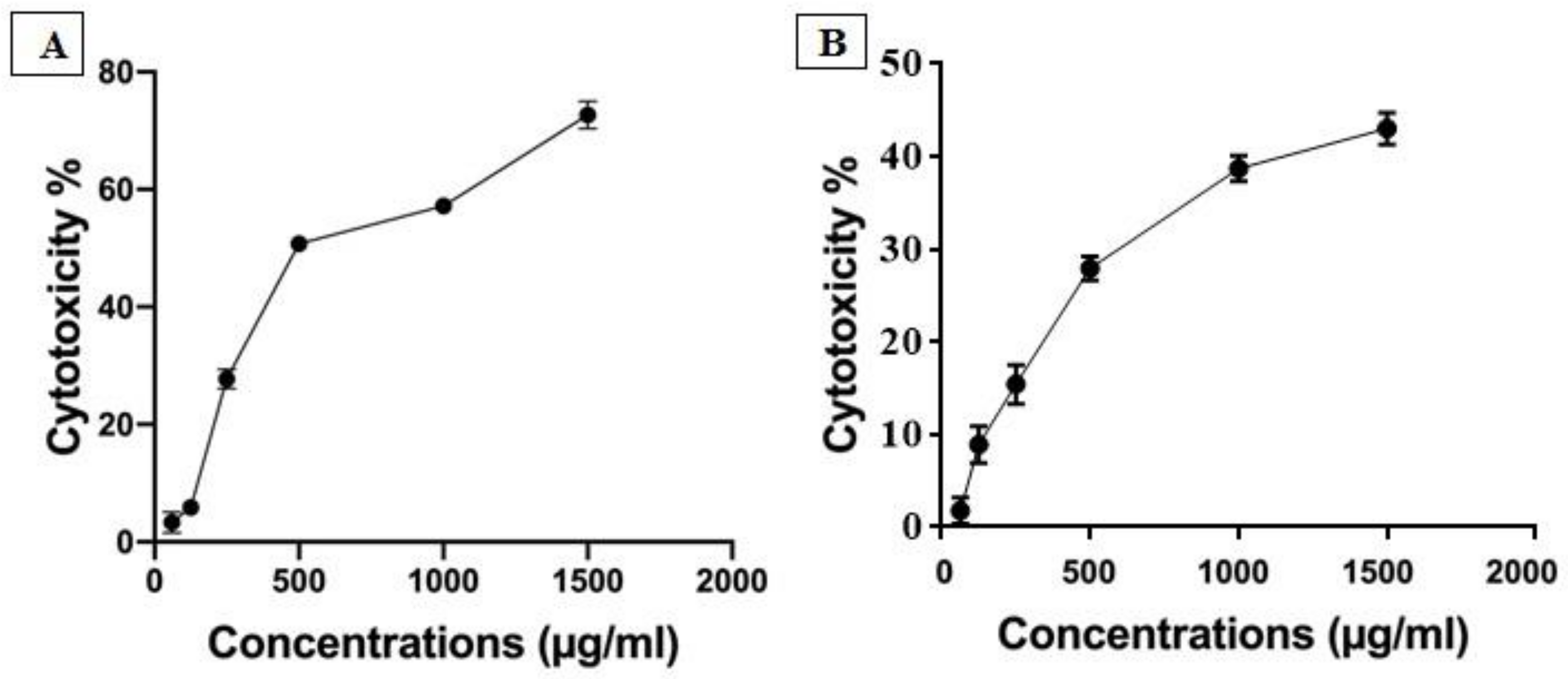
3.1. Cytotoxicity Assay
3.2. Antiviral Assays
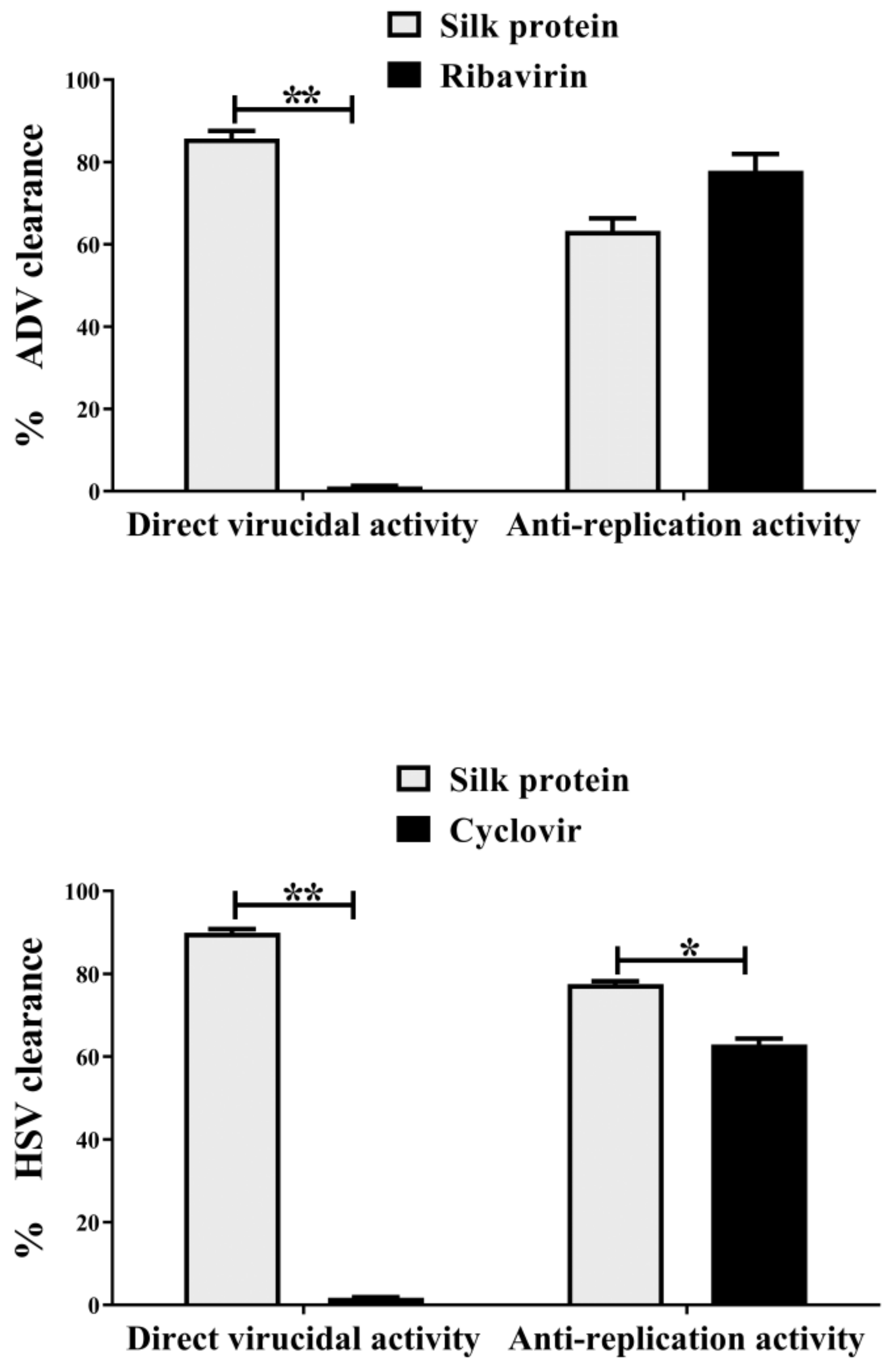
3.2.1. Antiviral Assays on ADV7 and HSV1
3.2.2. Anti-HCV Activity of Silk Protein
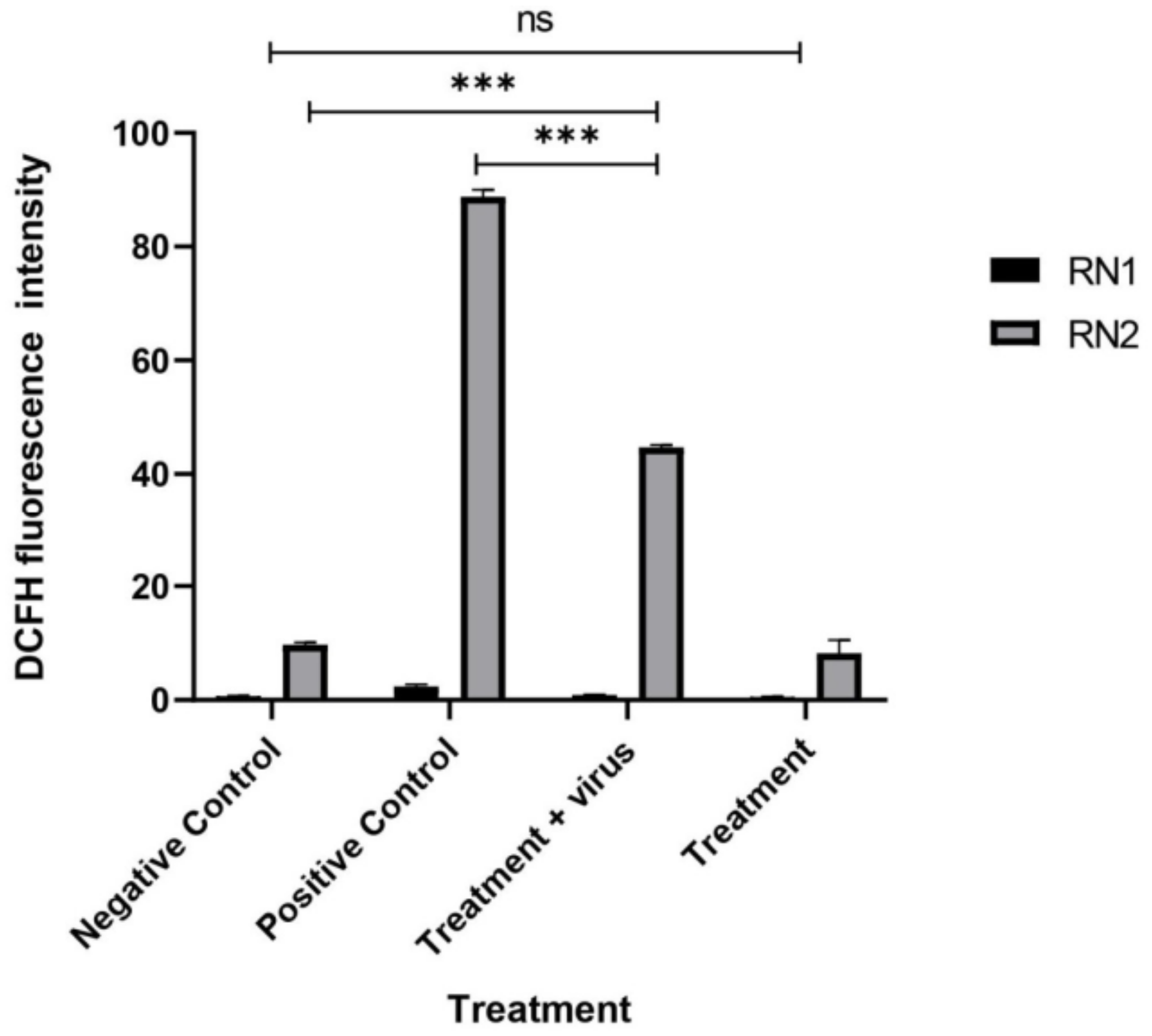
3.2.3. Quantification of the Induced ROS Using Flow Cytometry in HCV-Infected Model
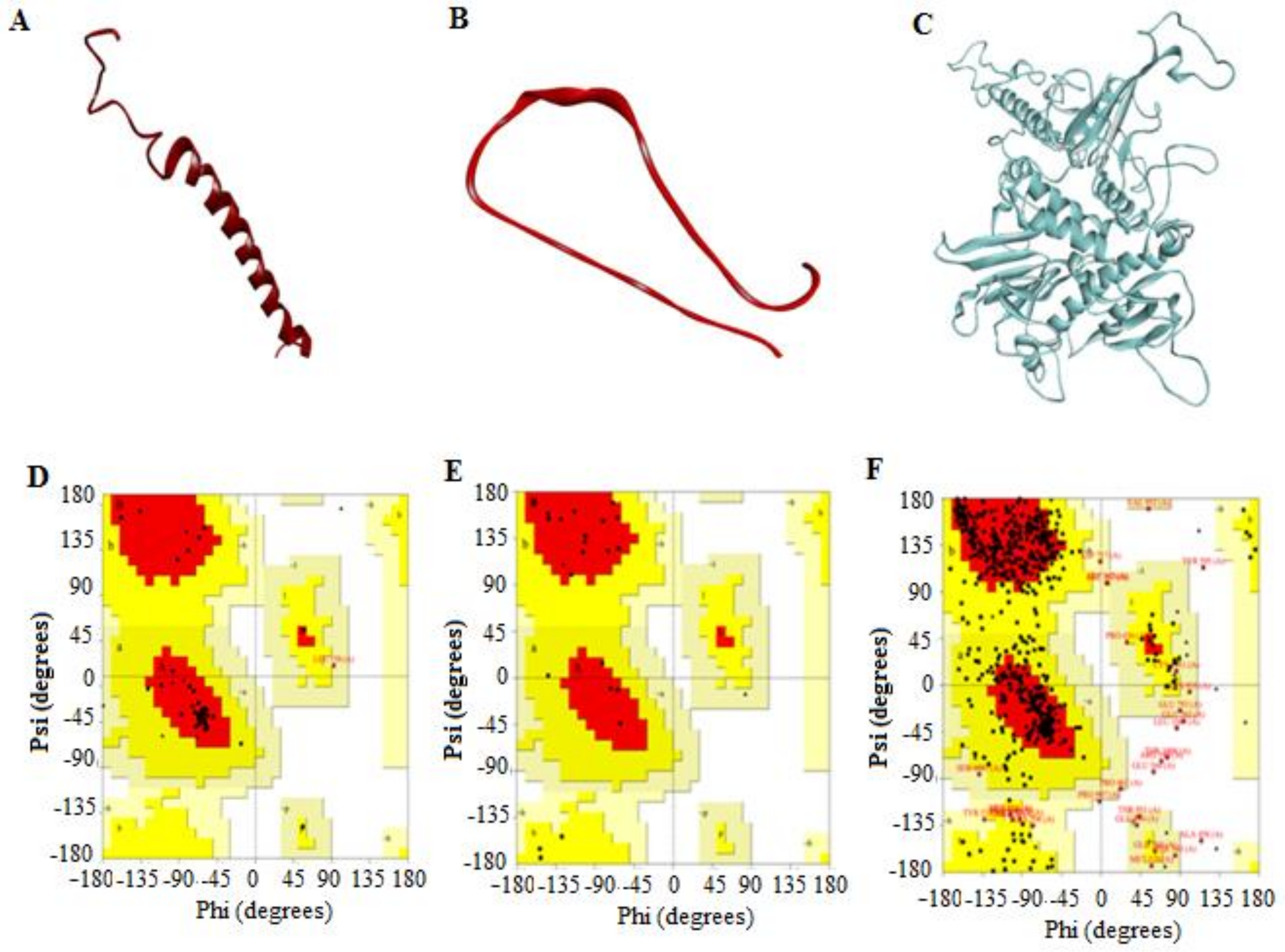
3.2.4. 3D Predicted Structure Models
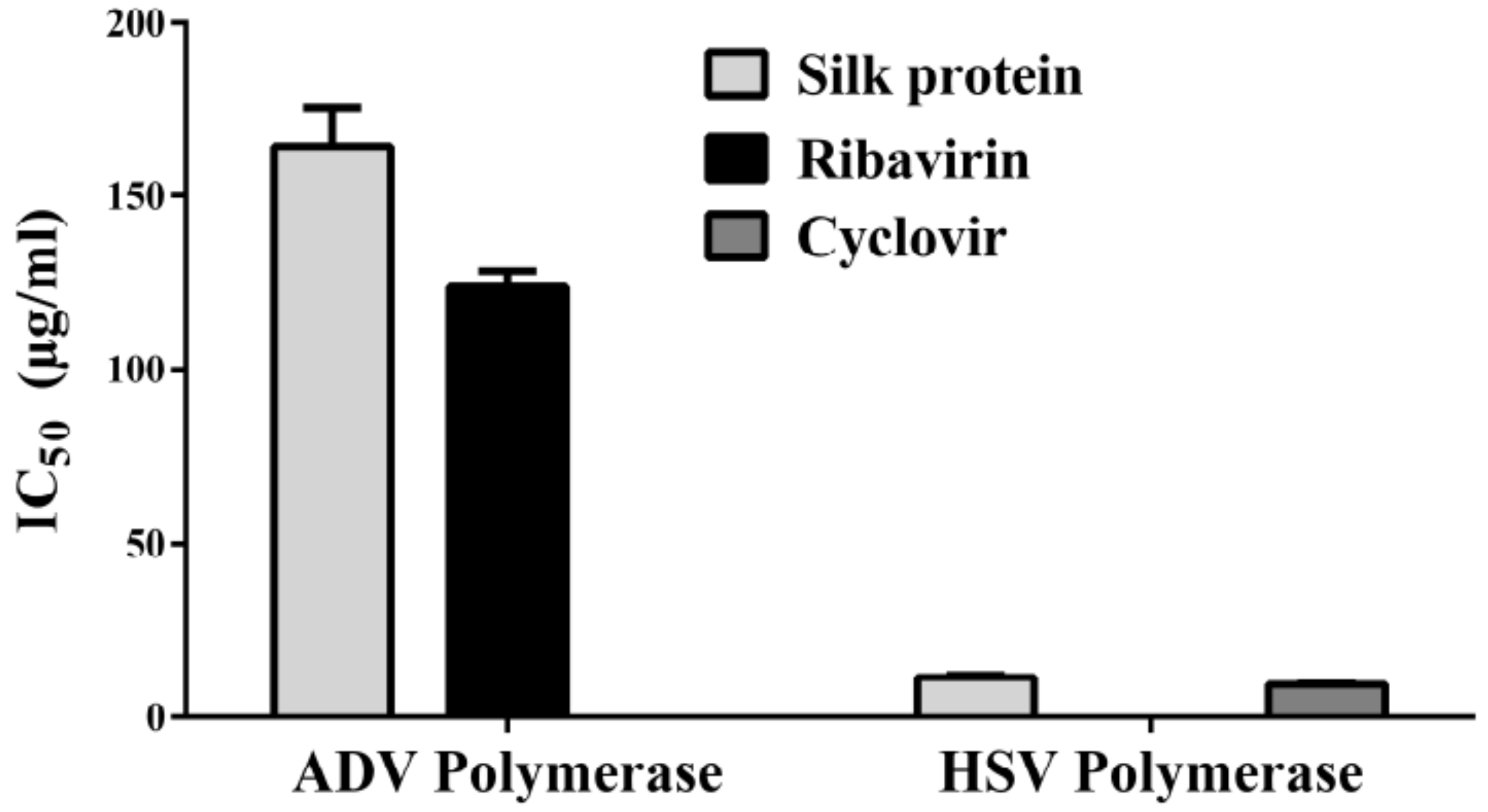
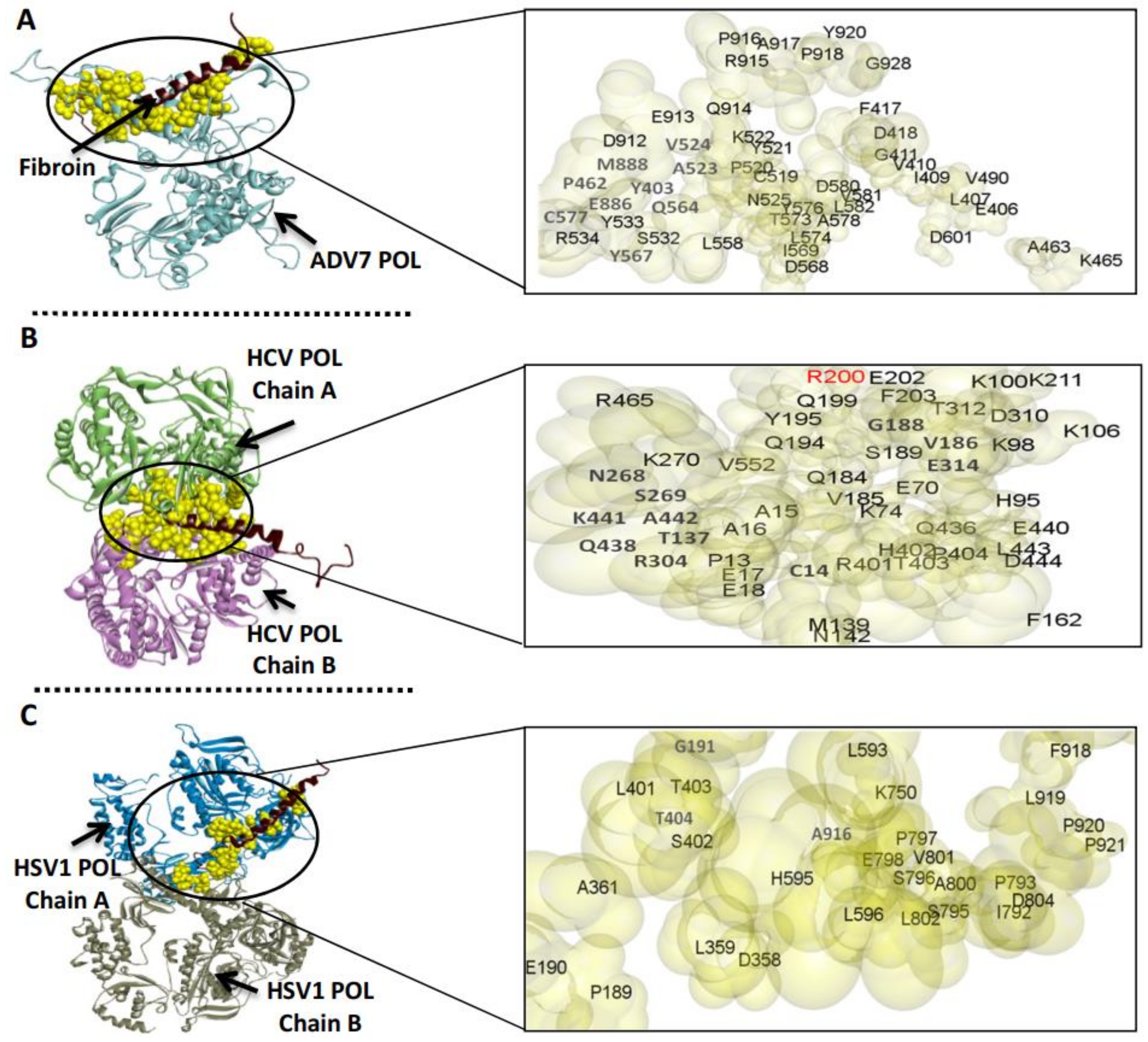
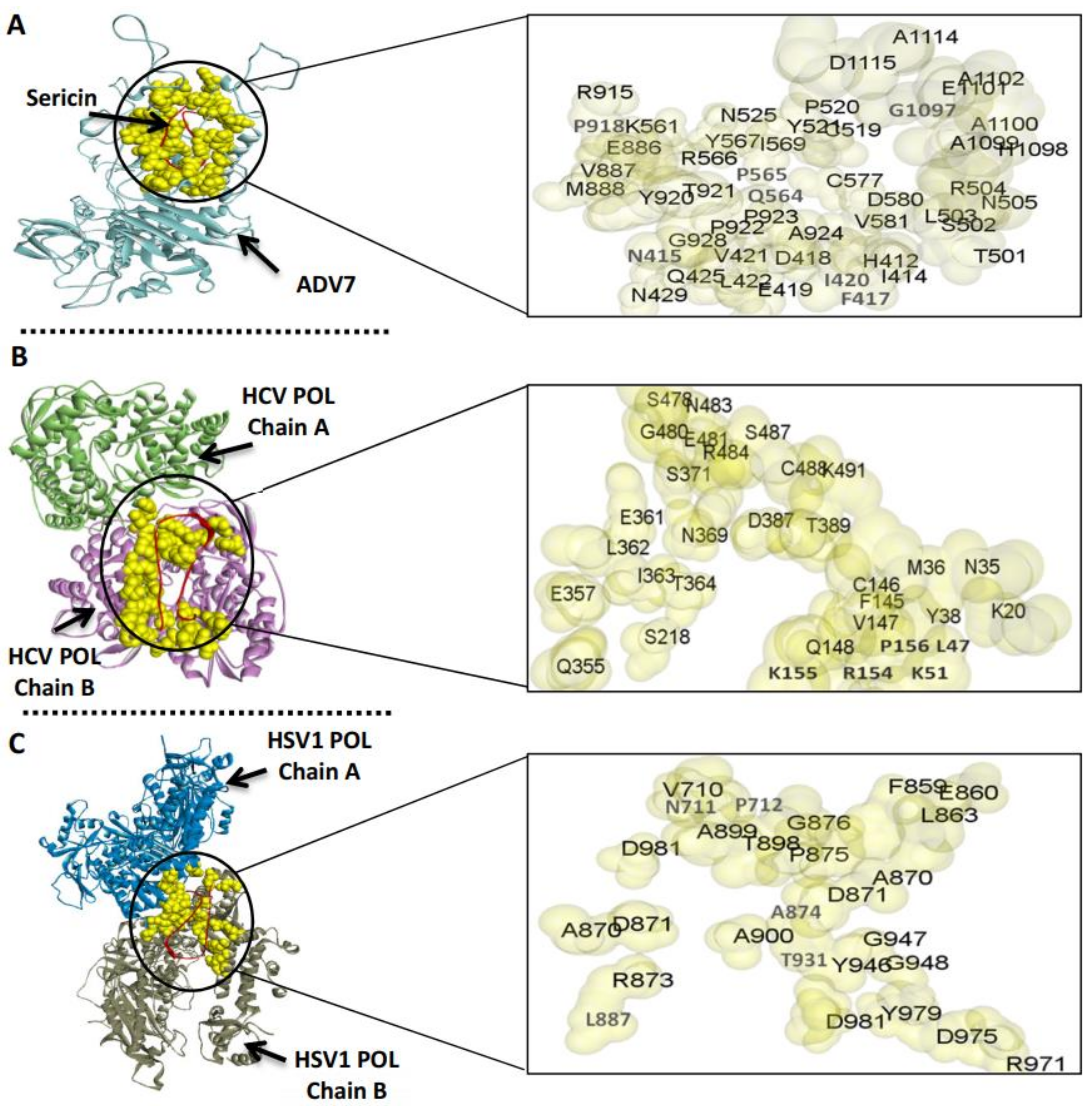
3.3. The Predicted Inhibitory Mechanism of Silk Fibroin and Sericin on ADV7, HCV, and HSV-1 Polymerases
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.; Jiang, X.; Bao, J.; Wang, Y.; Liu, H.; Tang, L. Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions. Front. Immunol. 2018, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Saad, M.H.; Badierah, R.; Redwan, E.M.; El-Fakharany, E.M. A Comprehensive Insight into the Role of Exosomes in Viral Infection: Dual Faces Bearing Different Functions. Pharmaceutics 2021, 13, 1405. [Google Scholar] [CrossRef] [PubMed]
- Holland, C.; Numata, K.; Rnjak-Kovacina, J.; Seib, F.P. The Biomedical Use of Silk: Past, Present, Future. Adv. Health Mater. 2019, 8, e1800465. [Google Scholar] [CrossRef] [Green Version]
- Pereira, A.M.; Machado, R.; da Costa, A.; Ribeiro, A.; Collins, T.; Gomes, A.; Leonor, I.B.; Kaplan, D.L.; Reis, R.L.; Casal, M. Silk-based biomaterials functionalized with fibronectin type II promotes cell adhesion. Acta Biomater. 2017, 47, 50–59. [Google Scholar] [CrossRef]
- Silva, V.R.; Ribani, M.; Gimenes, M.L.; Scheer, A.P. High Molecular Weight Sericin Obtained by High Temperature and Ultrafiltration Process. Procedia Eng. 2012, 42, 833–841. [Google Scholar] [CrossRef] [Green Version]
- Zhaorigetu, S.; Sasaki, M.; Kato, N. Consumption of Sericin Suppresses Colon Oxidative Stress and Aberrant Crypt Foci in 1,2-Dimethylhydrazine-Treated Rats by Colon Undigested Sericin. J. Nutr. Sci. Vitaminol. 2007, 53, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuchadomrong, S.; Senakoon, W.; Sirimungkararat, S.; Senawong, T.; Kitikoon, P. Antibacterial and antioxidant activities of sericin powder from Eri Silkworm Cocoons Correlating to Degumming processes. Int. J. Wilk Silkmoth Silk 2008, 13, 69–78. [Google Scholar]
- Kumar, J.P.; Mandal, B.B. Antioxidant potential of mulberry and non-mulberry silk sericin and its implications in biomedicine. Free. Radic. Biol. Med. 2017, 108, 803–818. [Google Scholar] [CrossRef] [PubMed]
- Chanu, S.B.; Devi, S.K.; Singh, L.R. Silk Protein Sericin: Structure, Secretion, Composition and Antimicrobial Potential. Front. Anti-Infect. Agents 2019, 1, 183–194. [Google Scholar] [CrossRef]
- El-Fakharany, E.M.; Abu-Elreesh, G.M.; Kamoun, E.A.; Zaki, S.; Abd-El-Haleem, D.A. In vitro assessment of the bioactivities of sericin protein extracted from a bacterial silk-like biopolymer. RSC Adv. 2020, 10, 5098–5107. [Google Scholar] [CrossRef] [Green Version]
- Chlapanidas, T.; Faragò, S.; Lucconi, G.; Perteghella, S.; Galuzzi, M.; Mantelli, M.; Avanzini, M.A.; Tosca, M.C.; Marazzi, M.; Vigo, D.; et al. Sericins exhibit ROS-scavenging, anti-tyrosinase, anti-elastase, and in vitro immunomodulatory activities. Int. J. Biol. Macromol. 2013, 58, 47–56. [Google Scholar] [CrossRef]
- Ahamad, M.S.I.; Vootla, S. Extraction and evaluation of antimicrobial potential of antheraeamylitta silk sericin. Inter J Recent Sci Res 2018, 9, 32019–32022. [Google Scholar]
- Kunz, R.I.; Brancalhão, R.M.C.; Ribeiro, L.D.F.C.; Natali, M.R.M. Silkworm Sericin: Properties and Biomedical Applications. BioMed Res. Int. 2016, 2016, 8175701. [Google Scholar] [CrossRef] [Green Version]
- Chithrashree, G.C.; Kumar, M.S.; Sharada, A.C. Sericin, a Versatile Protein from Silkworm—Biomedical Applications. Shanlax Int. J. Arts, Sci. Humanit. 2021, 8, 6–11. [Google Scholar] [CrossRef]
- Griffin, D.W. The Quest for Extraterrestrial Life: What About the Viruses? Astrobiology 2013, 13, 774–783. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation (WHO). Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections 2019: Accountability for the Global Health Sector Strategies, 2016–2021; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Forsythe, S.S.; McGreevey, W.; Whiteside, A.; Shah, M.; Cohen, J.; Hecht, R.; Bollinger, L.A.; Kinghorn, A. Twenty Years of Antiretroviral Therapy for People Living With HIV: Global Costs, Health Achievements, Economic Benefits. Health Aff. 2019, 38, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Ehwarieme, R.; Agarwal, A.N.; Alkhateb, R.; Bowling, J.E.; Anstead, G.M. A Surprising Cause of Liver Abscesses in a Post-Chemotherapy Patient: Herpes Simplex Virus. Cureus 2021, 13. [Google Scholar] [CrossRef]
- El-Tantawy, W.H.; Temraz, A. Natural products for the management of the hepatitis C virus: A biochemical review. Arch. Physiol. Biochem. 2018, 126, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Tovo, P.-A.; Calitri, C.; Scolfaro, C.; Gabiano, C.; Garazzino, S. Vertically acquired hepatitis C virus infection: Correlates of transmission and disease progression. World J. Gastroenterol. 2016, 22, 1382–1392. [Google Scholar] [CrossRef]
- El Fakharany, E.; El-Baky, N.A.; Linjawi, M.H.; AlJaddawi, A.A.; Saleem, T.H.; Nassar, A.Y.; Osman, A.; Redwan, E.M. Influence of camel milk on the hepatitis C virus burden of infected patients. Exp. Ther. Med. 2017, 13, 1313–1320. [Google Scholar] [CrossRef]
- Redwan, E.M.; Uversky, V.N.; El-Fakharany, E.M.; Al-Mehdar, H. Potential lactoferrin activity against pathogenic viruses. Comptes Rendus. Biol. 2014, 337, 581–595. [Google Scholar] [CrossRef] [PubMed]
- El-Fakharany, E.M. Nanoformulation of lactoferrin potentiates its activity and enhances novel biotechnological applications. Int. J. Biol. Macromol. 2020, 165, 970–984. [Google Scholar] [CrossRef]
- Saad, M.H.; El-Fakharany, E.M.; Salem, M.S.; Sidkey, N.M. The use of cyanobacterial metabolites as natural medical and biotechnological tools: Review article. J. Biomol. Struct. Dyn. 2020, 1–23. [Google Scholar] [CrossRef] [PubMed]
- El-Fakharany, E.M. Nanoformulation approach for improved stability and efficiency of lactoperoxidase. Preparative Biochemistry and Biotechnology 2020, 51, 629–641. [Google Scholar] [CrossRef]
- El-Fakharany, E.M.; Saad, M.H.; Salem, M.S.; Sidkey, N.M. Biochemical characterization and application of a novel lectin from the cyanobacterium Lyngabya confervoides MK012409 as an antiviral and anticancer agent. Int. J. Biol. Macromol. 2020, 161, 417–430. [Google Scholar] [CrossRef]
- Saad, M.H.; El-Fakharany, E.M.; Salem, M.S.; Sidkey, N.M. In vitro assessment of dual (antiviral and antitumor) activity of a novel lectin produced by the newly cyanobacterium isolate, Oscillatoria acuminate MHM-632 MK014210.1. J. Biomol. Struct. Dyn. 2020, 1–21. [Google Scholar] [CrossRef]
- El-Maradny, Y.A.; El-Fakharany, E.M.; Abu-Serie, M.M.; Hashish, M.H.; Selim, H.S. Lectins purified from medicinal and edible mushrooms: Insights into their antiviral activity against pathogenic viruses. Int. J. Biol. Macromol. 2021, 179, 239–258. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, E.A.; Abu-Elreesh, G.M.; El-Fakharany, E.M.; Abd-El-Haleem, D. A Novel Bacterial Polymeric Silk-Like Protein from a Petroleum Origin Bacillus sp. strain NE: Isolation and Characterization. J. Polym. Environ. 2019, 27, 1629–1641. [Google Scholar] [CrossRef]
- Zaki, S.; Farag, S.; Abu Elreesh, G.; Elkady, M.; Nosier, M.; El Abd, D. Characterization of bioflocculants produced by bacteria isolated from crude petroleum oil. Int. J. Environ. Sci. Technol. 2011, 8, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Lohr, H.F.; Goergen, B.; Biischenfelde, K.-H.M.Z.; Gerken, G. HCV replication in mononuclear cells stimulates anti-HCV-secreting B cells and reflects nonresponsiveness to interferon-α. J. Med. Virol. 1995, 46, 314–320. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Redwan, E.M.; Almehdar, H.A.; El-Fakharany, E.M.; Baig, A.-W.K.; Uversky, V.N. Potential antiviral activities of camel, bovine, and human lactoperoxidases against hepatitis C virus genotype 4. RSC Adv. 2015, 5, 60441–60452. [Google Scholar] [CrossRef]
- Abdallah, A.E.; Alesawy, M.S.; Eissa, S.I.; El-Fakharany, E.M.; Kalaba, M.H.; Sharaf, M.H.; Shama, N.M.A.; Mahmoud, S.H.; Mostafa, A.; Al-Karmalawy, A.A.; et al. Design and synthesis of new 4-(2-nitrophenoxy)benzamide derivatives as potential antiviral agents: Molecular modeling and in vitro antiviral screening. New J. Chem. 2021, 45, 16557–16571. [Google Scholar] [CrossRef]
- Ramakrishnan, M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016, 5, 85–86. [Google Scholar] [CrossRef]
- Kessler, H.H.; Mühlbauer, G.; Rinner, B.; Stelzl, E.; Berger, A.; Dörr, H.-W.; Santner, B.; Marth, E.; Rabenau, H. Detection of Herpes Simplex Virus DNA by Real-Time PCR. J. Clin. Microbiol. 2000, 38, 2638–2642. [Google Scholar] [CrossRef] [Green Version]
- Heim, A.; Ebnet, C.; Harste, G.; Pring-Åkerblom, P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J. Med. Virol. 2003, 70, 228–239. [Google Scholar] [CrossRef]
- Mentel, R.; Kurek, S.; Wegner, U.; Janta-Lipinski, M.V.; Gürtler, L.; Matthes, E. Inhibition of adenovirus DNA polymerase by modified nucleoside triphosphate analogs correlate with their antiviral effects on cellular level. Med. Microbiol. Immunol. 2000, 189, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Schnute, M.E.; Anderson, D.J.; Brideau, R.J.; Ciske, F.L.; Collier, S.A.; Cudahy, M.M.; Eggen, M.; Genin, M.J.; Hopkins, T.A.; Judge, T.M.; et al. 2-Aryl-2-hydroxyethylamine substituted 4-oxo-4,7-dihydrothieno[2,3-b]pyridines as broad-spectrum inhibitors of human herpesvirus polymerases. Bioorganic Med. Chem. Lett. 2007, 17, 3349–3353. [Google Scholar] [CrossRef]
- Knopf, K.-W. Properties of Herpes Simplex Virus DNA Polymerase and Characterization of Its Associated Exonuclease Activity. JBIC J. Biol. Inorg. Chem. 1979, 98, 231–244. [Google Scholar] [CrossRef]
- Redwan, E.M.; El-Fakharany, E.M.; Uversky, V.N.; Linjawi, M.H. Screening the anti infectivity potentials of native N- and C-lobes derived from the camel lactoferrin against hepatitis C virus. BMC Complement. Altern. Med. 2014, 14, 219. [Google Scholar] [CrossRef] [Green Version]
- Pradel, F.; Paranhos-Baccalà, G.; Sodoyer, M.; Chevallier, P.; Mandrand, B.; Lotteau, V.; André, P. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 2001, 95, 111–119. [Google Scholar] [CrossRef]
- Anticoli, S.; Amatore, D.; Matarrese, P.; De Angelis, M.; Palamara, A.T.; Nencioni, L.; Ruggieri, A. Counteraction of HCV-Induced Oxidative Stress Concurs to Establish Chronic Infection in Liver Cell Cultures. Oxid. Med. Cell. Longev. 2019, 2019, 6452390. [Google Scholar] [CrossRef]
- Jastrzebska, K.; Kucharczyk, K.; Florczak, A.; Dondajewska, E.; Mackiewicz, A.; Dams-Kozlowska, H. Silk as an innovative biomaterial for cancer therapy. Rep. Pr. Oncol. Radiother. 2015, 20, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, N.; Hoagland, D.A.; Farris, R.J. Effect of moisture absorption on the thermal properties of Bombyx mori silk fibroin films. J. Appl. Polym. Sci. 1997, 63, 401–410. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Jabłońska, J.; Pravda, L.; Vařeková, R.S.; Thornton, J. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.-C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. In 2-D Proteome Analysis Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 1999; Volume 112, pp. 531–552. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.P.A.; Pappa, G.L.; Pires, D.E.V.; Izidoro, S.C. GASS-WEB: A web server for identifying enzyme active sites based on genetic algorithms. Nucleic Acids Res. 2017, 45, W315–W319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fahnestock, S.R.; Yao, Z.; Bedzyk, L.A. Microbial production of spider silk proteins. Rev. Mol. Biotechnol. 2000, 74, 105–119. [Google Scholar] [CrossRef]
- Yang, Y.J.; Choi, Y.S.; Jung, D.; Park, B.R.; Hwang, W.B.; Kim, H.W.; Cha, H.J. Production of a novel silk-like protein from sea anemone and fabrication of wet-spun and electrospun marine-derived silk fibers. NPG Asia Mater. 2013, 5, e50. [Google Scholar] [CrossRef]
- Antony, V.A.R.; Chinnamal, S.K. Enzymatic degumming of silk using Bacillus sp. Int. J. Sci. Tech. Mang. 2015, 4, 458–465. [Google Scholar]
- Sharma, R. Enzyme Inhibition: Mechanisms and Scope. In Enzyme Inhibition and Bioapplications; IntechOpen: London, UK, 2012. [Google Scholar]
- Fan, J.-B.; Wu, L.-P.; Chen, L.-S.; Mao, X.-Y.; Ren, F.-Z. Antioxidant Activities of Silk Sericin from Silkwormbombyx MorI. J. Food Biochem. 2009, 33, 74–88. [Google Scholar] [CrossRef]
- Da Silva, T.L.; Da Silva, A.C.; Ribani, M.; Vieira, M.G.A.; Gimenes, M.L.; Da Silva, M.G.C. Evaluation of molecular weight distribution of sericin in solutions concentrated via precipitation by ethanol and precipitation by freezing/thawing. Chem. Eng. Trans. 2014, 38, 103–108. [Google Scholar]
- Turbiani, F.R.B.; Tomadon, J.; Seixas, F.L.; Gimenes, M.L. Properties and structure of sericin films: Effect of the crosslinking degree. In Proceedings of the 10th International Conference on Chemical and Process Engineering, Moscow, Russian, 28–30 April 2011. [Google Scholar]


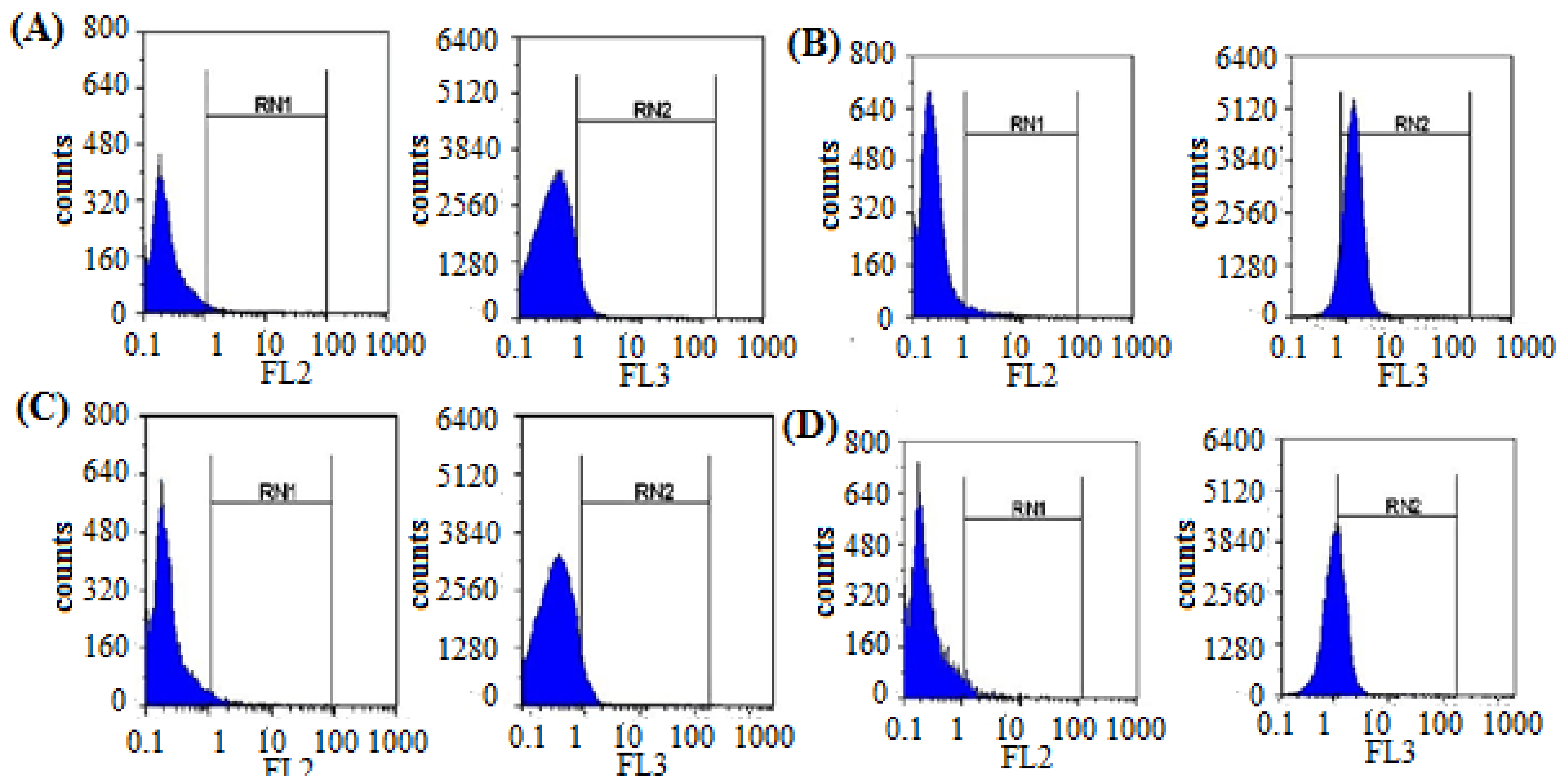
| Samples | Fluor | Cq | Virus Conc. (Copies/mL) | Inhibition% |
|---|---|---|---|---|
| Treated cells | SYBR | 23.89 | 0.9 × 105 | 66.2 |
| Positive control | SYBR | 28.33 | 1.36 × 106 | 0.0 |
| Negative control | SYBR | 4.69 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Fakharany, E.M.; Abu-Serie, M.M.; Habashy, N.H.; El-Deeb, N.M.; Abu-Elreesh, G.M.; Zaki, S.; Abd-EL-Haleem, D. Inhibitory Effects of Bacterial Silk-like Biopolymer on Herpes Simplex Virus Type 1, Adenovirus Type 7 and Hepatitis C Virus Infection. J. Funct. Biomater. 2022, 13, 17. https://doi.org/10.3390/jfb13010017
El-Fakharany EM, Abu-Serie MM, Habashy NH, El-Deeb NM, Abu-Elreesh GM, Zaki S, Abd-EL-Haleem D. Inhibitory Effects of Bacterial Silk-like Biopolymer on Herpes Simplex Virus Type 1, Adenovirus Type 7 and Hepatitis C Virus Infection. Journal of Functional Biomaterials. 2022; 13(1):17. https://doi.org/10.3390/jfb13010017
Chicago/Turabian StyleEl-Fakharany, Esmail M., Marwa M. Abu-Serie, Noha H. Habashy, Nehal M. El-Deeb, Gadallah M. Abu-Elreesh, Sahar Zaki, and Desouky Abd-EL-Haleem. 2022. "Inhibitory Effects of Bacterial Silk-like Biopolymer on Herpes Simplex Virus Type 1, Adenovirus Type 7 and Hepatitis C Virus Infection" Journal of Functional Biomaterials 13, no. 1: 17. https://doi.org/10.3390/jfb13010017
APA StyleEl-Fakharany, E. M., Abu-Serie, M. M., Habashy, N. H., El-Deeb, N. M., Abu-Elreesh, G. M., Zaki, S., & Abd-EL-Haleem, D. (2022). Inhibitory Effects of Bacterial Silk-like Biopolymer on Herpes Simplex Virus Type 1, Adenovirus Type 7 and Hepatitis C Virus Infection. Journal of Functional Biomaterials, 13(1), 17. https://doi.org/10.3390/jfb13010017