Challenges in the Computational Modeling of the Protein Structure—Activity Relationship
Abstract
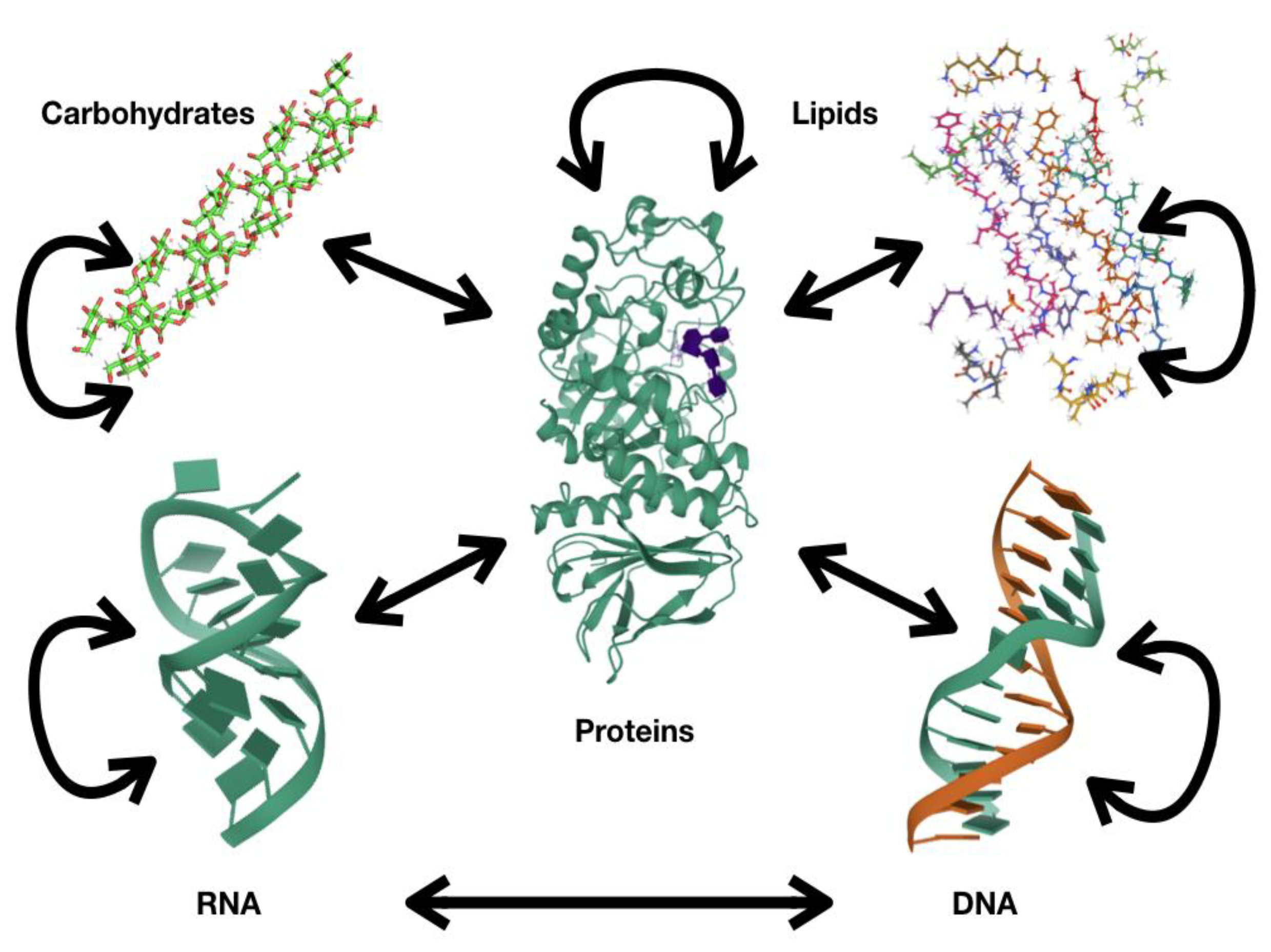
1. Proteins: Fundamental Polymers for Biological Systems
2. Protein Structure
3. Modeling Protein Structure–Activity Relationship
4. Databases and Prediction Methods
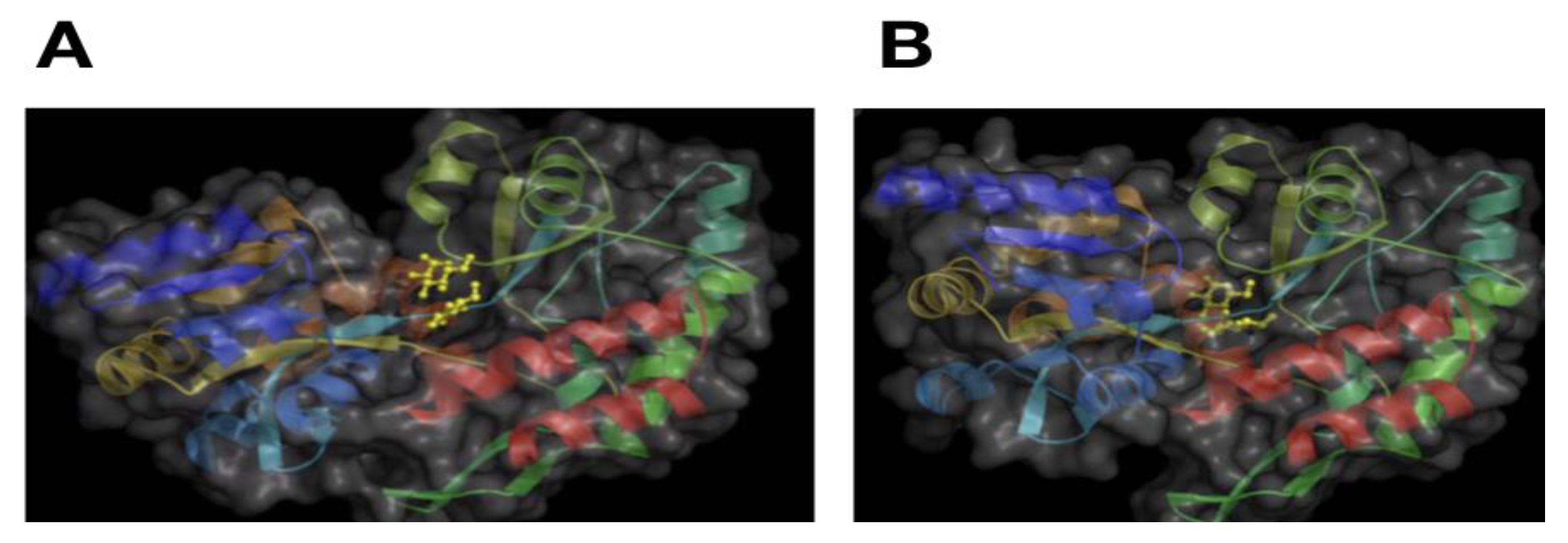
5. Predicting Critical Residues for Protein Activity
6. Prediction of Protein Activity
7. Prediction of Protein Structure from Protein Sequence
8. Protein Design
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crick, F.H. On protein synthesis—PubMed. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar]
- Jeffery, C.J. Multifunctional proteins: Examples of gene sharing. Ann. Med. 2003, 35, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Yoshimura, A.; Sumizawa, T.; Haraguchi, M.; Akiyama, S.I.; Fukui, K.; Ishizawa, M.; Yamada, Y. Angiogenic factor. Nature 1992, 356, 668. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.-P. Allostery and the Monod-Wyman-Changeux Model after 50 Years. Annu. Rev. Biophys. 2012, 41, 103–133. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Nussinov, R. Allostery: An Overview of Its History, Concepts, Methods, and Applications. PLoS Comput. Biol. 2016, 12, e1004966. [Google Scholar] [CrossRef]
- Ahmed, M.H.; Ghatge, M.S.; Safo, M.K. Hemoglobin: Structure, Function and Allostery. Subcell. Biochem. 2020, 94, 345–382. [Google Scholar] [CrossRef]
- Mittal, S.; Saluja, D. Protein Post-translational Modifications: Role in Protein Structure, Function and Stability. In Proteostasis and Chaperone Surveillance; Springer: New Delhi, India, 2015; pp. 25–37. Available online: https://link.springer.com/chapter/10.1007/978-81-322-2467-9_2 (accessed on 29 January 2021).
- Brinkjost, T.; Ehrt, C.; Koch, O.; Mutzel, P. SCOT: Rethinking the classification of secondary structure elements. Bioinformatics 2019, 36, 2417–2428. [Google Scholar] [CrossRef]
- Flores, S.; Echols, N.; Milburn, D.; Hespenheide, B.; Keating, K.; Lu, J.; Wells, S.; Yu, E.Z.; Thorpe, M.; Gerstein, M. The Database of Macromolecular Motions: New features added at the decade mark. Nucleic Acids Res. 2006, 34, D296–D301. [Google Scholar] [CrossRef]
- Vila, J.A. Metamorphic Proteins in Light of Anfinsen’s Dogma. J. Phys. Chem. Lett. 2020, 11, 4998–4999. [Google Scholar] [CrossRef]
- Porter, L.L.; Looger, L.L. Extant fold-switching proteins are widespread. Proc. Natl. Acad. Sci. USA 2018, 115, 5968–5973. [Google Scholar] [CrossRef]
- Jain, V.; Yogavel, M.; Oshima, Y.; Kikuchi, H.; Touquet, B.; Hakimi, M.A.; Sharma, A. Structure of prolyl-tRNA synthetase-halofuginone complex provides basis for development of drugs against malaria and toxoplasmosis. Structure 2015, 23, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 932–951. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Ambriz-Rivas, M.; Pastor, N.; Del Rio, G. Relating Protein Structure and Function Through a Bijection and Its Implications on Protein Structure Prediction. In Protein Interactions; Cai, J., Wang, R., Eds.; InTech: London, UK, 2012; pp. 350–368. Available online: www.intechopen.com (accessed on 29 January 2021).
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProtKB/Swiss-Prot 2020_06. 2020. Available online: https://www.uniprot.org/statistics/Swiss-Prot (accessed on 29 January 2021).
- Consortium, T.G.O. Gene Ontology Resource. 2021. Available online: http://geneontology.org/stats.html (accessed on 29 January 2021).
- Noble, K. Artificial Intelligence Solution to a 50-Year-Old Science Challenge Could ‘Revolutionise’ Medical Research. 2020. Available online: https://predictioncenter.org/casp14/doc/CASP14_press_release.html (accessed on 29 January 2021).
- Budowski-Tal, I.; Nov, Y.; Kolodny, R. FragBag, an accurate representation of protein structure, retrieves structural neighbors from the entire PDB quickly and accurately. Proc. Natl. Acad. Sci. USA 2010, 107, 3481–3486. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, S.C.; He, L.; Li, M. Fingerprinting protein structures effectively and efficiently. Bioinformatics 2014, 30, 949–955. [Google Scholar] [CrossRef][Green Version]
- Corral-Corral, R.; Chávez, E.; Del Rio, G. Machine Learnable Fold Space Representation based on Residue Cluster Classes. Comput. Biol. Chem. 2015, 59, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Fontove, F.; Del Rio, G. Residue Cluster Classes: A Unified Protein Representation for Efficient Structural and Functional Classification. Entropy 2020, 22, 472. [Google Scholar] [CrossRef]
- Zhou, N.; Jiang, Y.; Bergquist, T.R.; Lee, A.J.; Kacsoh, B.Z.; Crocker, A.W.; Lewis, K.A.; Georghiou, G.; Nguyen, H.N.; Hamid, N.; et al. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 2019, 20, 1–23. [Google Scholar] [CrossRef]
- Ziegler, S.J.; Mallinson, S.J.; John, P.C.S.; Bomble, Y.J. Advances in integrative structural biology: Towards understanding protein complexes in their cellular context. Comput. Struct. Biotechnol. J. 2021, 19, 214–225. [Google Scholar] [CrossRef]
- Vakser, I.A. Challenges in protein docking. Curr. Opin. Struct. Biol. 2020, 64, 160–165. [Google Scholar] [CrossRef]
- Verkhivker, G.M.; Agajanian, S.; Hu, G.; Tao, P. Allosteric Regulation at the Crossroads of New Technologies: Multiscale Modeling, Networks, and Machine Learning. Front. Mol. Biosci. 2020, 7, 136. [Google Scholar] [CrossRef]
- Khatun, M.S.; Shoombuatong, W.; Hasan, M.; Kurata, H. Evolution of Sequence-based Bioinformatics Tools for Protein-protein Interaction Prediction. Curr. Genom. 2020, 21, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Haspel, N.; Jagodzinski, F. Methods for Detecting Critical Residues in Proteins. In In Vitro Mutagenesis; Humana Press: New York, NY, USA, 2016; Volume 1498, pp. 227–242. Available online: https://pubmed.ncbi.nlm.nih.gov/27709579/ (accessed on 3 February 2021).
- Corral-Corral, R.; Beltrán, J.A.; Brizuela, C.A.; Del Rio, G. Systematic Identification of Machine-Learning Models Aimed to Classify Critical Residues for Protein Function from Protein Structure. Molecules 2017, 22, 1673. [Google Scholar] [CrossRef]
- Molina, H.M.M.; Millán-Pacheco, C.; Pastor, N.; Del Rio, G. Computer-based screening of functional conformers of proteins. PLoS Comput. Biol. 2008, 4, e1000009. [Google Scholar]
- Gray, V.E.; Hause, R.J.; Fowler, D.M. Analysis of Large-Scale Mutagenesis Data to Assess the Impact of Single Amino Acid Substitutions. Genetics 2017, 207, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.L.; Beuning, P.J.; Ondrechen, M.J. Biochemical functional predictions for protein structures of unknown or uncertain function. Comput. Struct. Biotechnol. J. 2015, 13, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, P.; Kumar, S.; Bachhawat, A.K.; Pandit, S.B. CSmetaPred: A consensus method for prediction of catalytic residues. BMC Bioinform. 2017, 18, 583. [Google Scholar] [CrossRef]
- Das, K.; Bauman, J.D.; Clark, A.D.; Frenkel, Y.V.; Lewi, P.J.; Shatkin, A.J.; Hughes, S.H.; Arnold, E. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations. Proc. Natl. Acad. Sci. USA 2008, 105, 1466–1471. [Google Scholar] [CrossRef]
- Loeb, D.D.; Swanstrom, R.; Everitt, L.; Manchester, M.; Stamper, S.E.; Hutchison, C.A. Complete mutagenesis of the HIV-1 protease. Nat. Cell Biol. 1989, 340, 397–400. [Google Scholar] [CrossRef]
- Dubreuil, B.; Sass, E.; Nadav, Y.; Heidenreich, M.; Georgeson, J.M.; Weill, U.; Duan, Y.; Meurer, M.; Schuldiner, M.; Knop, M.; et al. YeastRGB: Comparing the abundance and localization of yeast proteins across cells and libraries. Nucleic Acids Res. 2019, 47, D1245–D1249. [Google Scholar] [CrossRef]
- You, R.; Yao, S.; Xiong, Y.; Huang, X.; Sun, F.; Mamitsuka, H.; Zhu, S. NetGO: Improving large-scale protein function prediction with massive network information. Nucleic Acids Res. 2019, 47, W379–W387. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.K.; Bhuiyan, M.; Kihara, D. DextMP: Deep dive into text for predicting moonlighting proteins. Bioinformatics 2017, 33, i83–i91. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; McGraw, J.; Kihara, D. MPFit: Computational Tool for Predicting Moonlighting Proteins. In Protein Function Prediction; Humana Press: New York, NY, USA, 2017; Volume 1611, pp. 45–57. Available online: https://pubmed.ncbi.nlm.nih.gov/28451971/ (accessed on 3 February 2021).
- Chen, C.; Liu, H.; Zabad, S.; Rivera, N.; Rowin, E.; Hassan, M.; De Jesus, S.M.G.; Santos, P.S.L.; Kravchenko, K.; Mikhova, M.; et al. MoonProt 3.0: An update of the moonlighting proteins database. Nucleic Acids Res. 2021, 49, D368–D372. [Google Scholar] [CrossRef]
- Laskowski, R.A. Integrated Servers for Structure-Informed Function Prediction. In From Protein Structure to Function with Bioinformatics, 2nd ed.; Springer: Heidelberg, The Netherlands, 2017; pp. 427–448. Available online: https://link.springer.com/chapter/10.1007/978-94-024-1069-3_13 (accessed on 3 February 2021).
- Callaway, E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nat. Cell Biol. 2020, 588, 203–204. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Adhikari, B.; Cheng, J. Improved protein structure reconstruction using secondary structures, contacts at higher distance thresholds, and non-contacts. BMC Bioinform. 2017, 18, 380. [Google Scholar] [CrossRef]
- Breu, H.; Kirkpatrick, D.G. Unit disk graph recognition is NP-hard. Comput. Geom. 1998, 9, 3–24. [Google Scholar] [CrossRef]
- Torrisi, M.; Pollastri, G.; Le, Q. Deep learning methods in protein structure prediction. Comput. Struct. Biotechnol. J. 2020, 18, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, B. A fully open-source framework for deep learning protein real-valued distances. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Reggiani, F.; Gobbi, G.; Ciarrocchi, A.; Sancisi, V. YAP and TAZ Are Not Identical Twins. Trends Biochem. Sci. 2021, 46, 154–168. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Ortega, E.; Aguirre-López, B.; Reyes-Vivas, H.; González-Andrade, M.; Campero-Basaldúa, J.C.; Pardo, J.P.; González, A. Saccharomyces cerevisiae Differential Functionalization of Presumed ScALT1 and ScALT2 Alanine Transaminases Has Been Driven by Diversification of Pyridoxal Phosphate Interactions. Front. Microbiol. 2018, 9, 944. [Google Scholar] [CrossRef] [PubMed]
- Stamboulian, M.; Guerrero, R.F.; Hahn, M.W.; Radivojac, P. The ortholog conjecture revisited: The value of orthologs and paralogs in function prediction. Bioinformatics 2020, 36, i219–i226. [Google Scholar] [CrossRef] [PubMed]
- Bourgeat, L.; Serghei, A.; Lesieur, C. Experimental Protein Molecular Dynamics: Broadband Dielectric Spectroscopy coupled with nanoconfinement. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Kuhlman, B.; Bradley, P. Advances in protein structure prediction and design. Nat. Rev. Mol. Cell Biol. 2019, 20, 681–697. [Google Scholar] [CrossRef]
- Tinberg, C.E.; Khare, S.D.; Dou, J.; Doyle, L.; Nelson, J.W.; Schena, A.; Jankowski, W.; Kalodimos, C.G.; Johnsson, K.; Stoddard, B.L.; et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nat. Cell Biol. 2013, 501, 212–216. [Google Scholar] [CrossRef]
- Röthlisberger, D.; Khersonsky, O.; Wollacott, A.M.; Jiang, L.; DeChancie, J.; Betker, J.; Gallaher, J.L.; Althoff, E.A.; Zanghellini, A.; Dym, O.; et al. Kemp elimination catalysts by computational enzyme design. Nat. Cell Biol. 2008, 453, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Song, Y.; DiMaio, F.; Wang, R.Y.-R.; Kim, D.; Miles, C.; Brunette, T.; Thompson, J.; Baker, D. High-Resolution Comparative Modeling with RosettaCM. Structure 2013, 21, 1735–1742. [Google Scholar] [CrossRef]
- Makigaki, S.; Ishida, T. Sequence Alignment Using Machine Learning for Accurate Template-Based Protein Structure Prediction. BIO-PROTOCOL 10; 2020. Available online: https://pubmed.ncbi.nlm.nih.gov/33659566/ (accessed on 5 March 2021).
- Qin, X.; Liu, M.; Zhang, L.; Liu, G. Structural protein fold recognition based on secondary structure and evolutionary information using machine learning algorithms. Comput. Biol. Chem. 2021, 91, 107456. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Johannissen, L.O.; Hay, S. Predicting new protein conformations from molecular dynamics simulation conformational landscapes and machine learning. Proteins Struct. Funct. Bioinform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Duarte, J.M.; Sathyapriya, R.; Stehr, H.; Filippis, I.; Lappe, M. Optimal contact definition for reconstruction of Contact Maps. BMC Bioinform. 2010, 11, 283. [Google Scholar] [CrossRef] [PubMed]
- Thibert, B.; Bredesen, D.E.; Del Rio, G. Improved prediction of critical residues for protein function based on network and phylogenetic analyses. BMC Bioinform. 2005, 6, 213. [Google Scholar] [CrossRef] [PubMed]
- Perkel, J.M. Ten computer codes that transformed science. Nat. Cell Biol. 2021, 589, 344–348. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Del Río, G. Challenges in the Computational Modeling of the Protein Structure—Activity Relationship. Computation 2021, 9, 39. https://doi.org/10.3390/computation9040039
Del Río G. Challenges in the Computational Modeling of the Protein Structure—Activity Relationship. Computation. 2021; 9(4):39. https://doi.org/10.3390/computation9040039
Chicago/Turabian StyleDel Río, Gabriel. 2021. "Challenges in the Computational Modeling of the Protein Structure—Activity Relationship" Computation 9, no. 4: 39. https://doi.org/10.3390/computation9040039
APA StyleDel Río, G. (2021). Challenges in the Computational Modeling of the Protein Structure—Activity Relationship. Computation, 9(4), 39. https://doi.org/10.3390/computation9040039





