Image Segmentation for Cardiovascular Biomedical Applications at Different Scales
Abstract
:1. Introduction
2. Segmentation Techniques
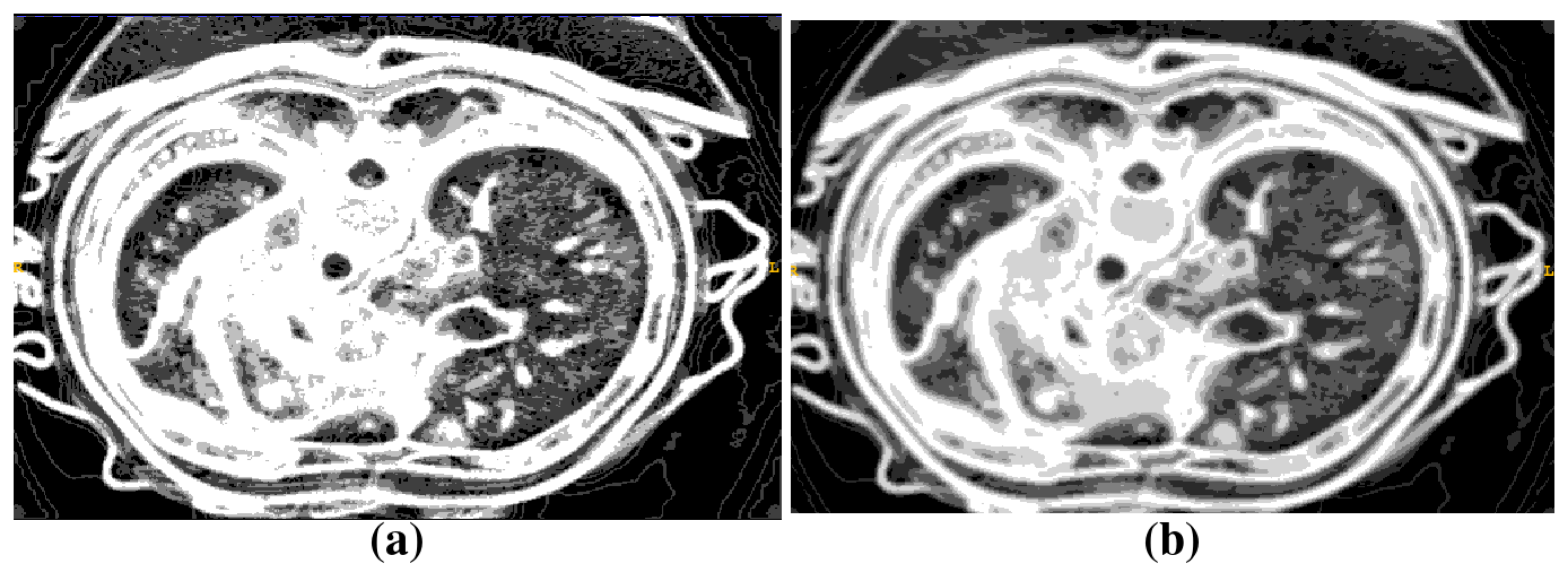
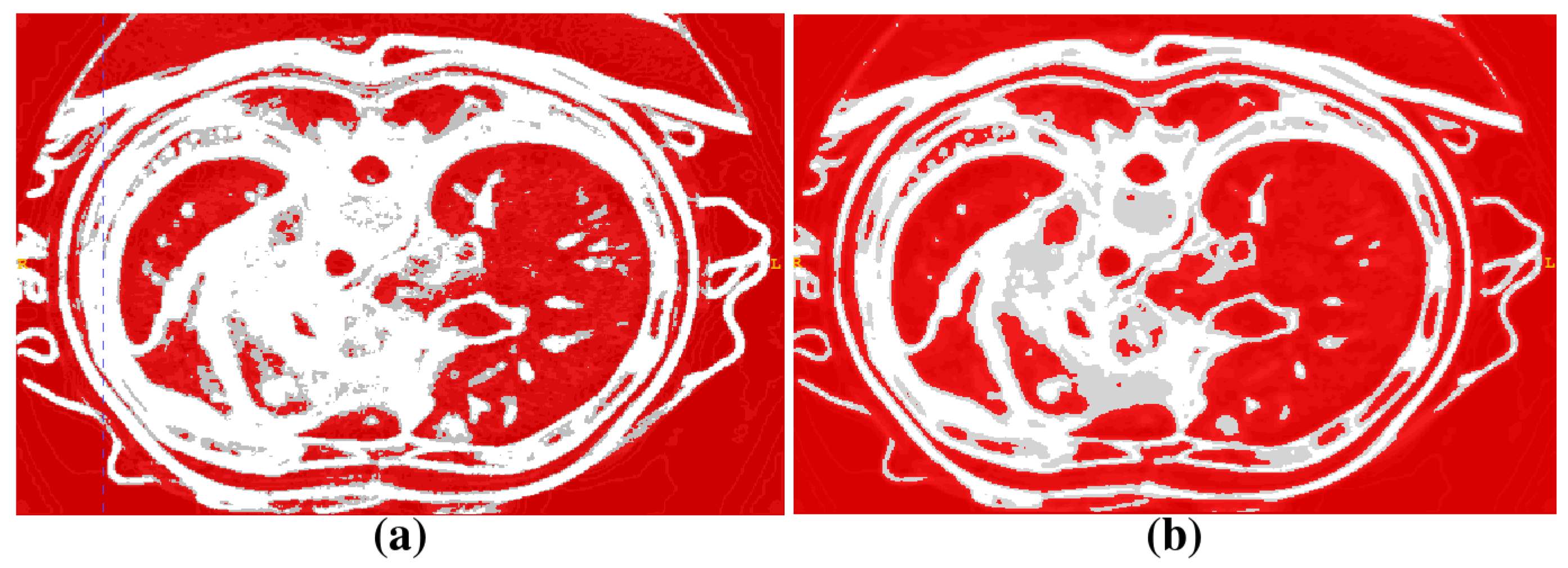
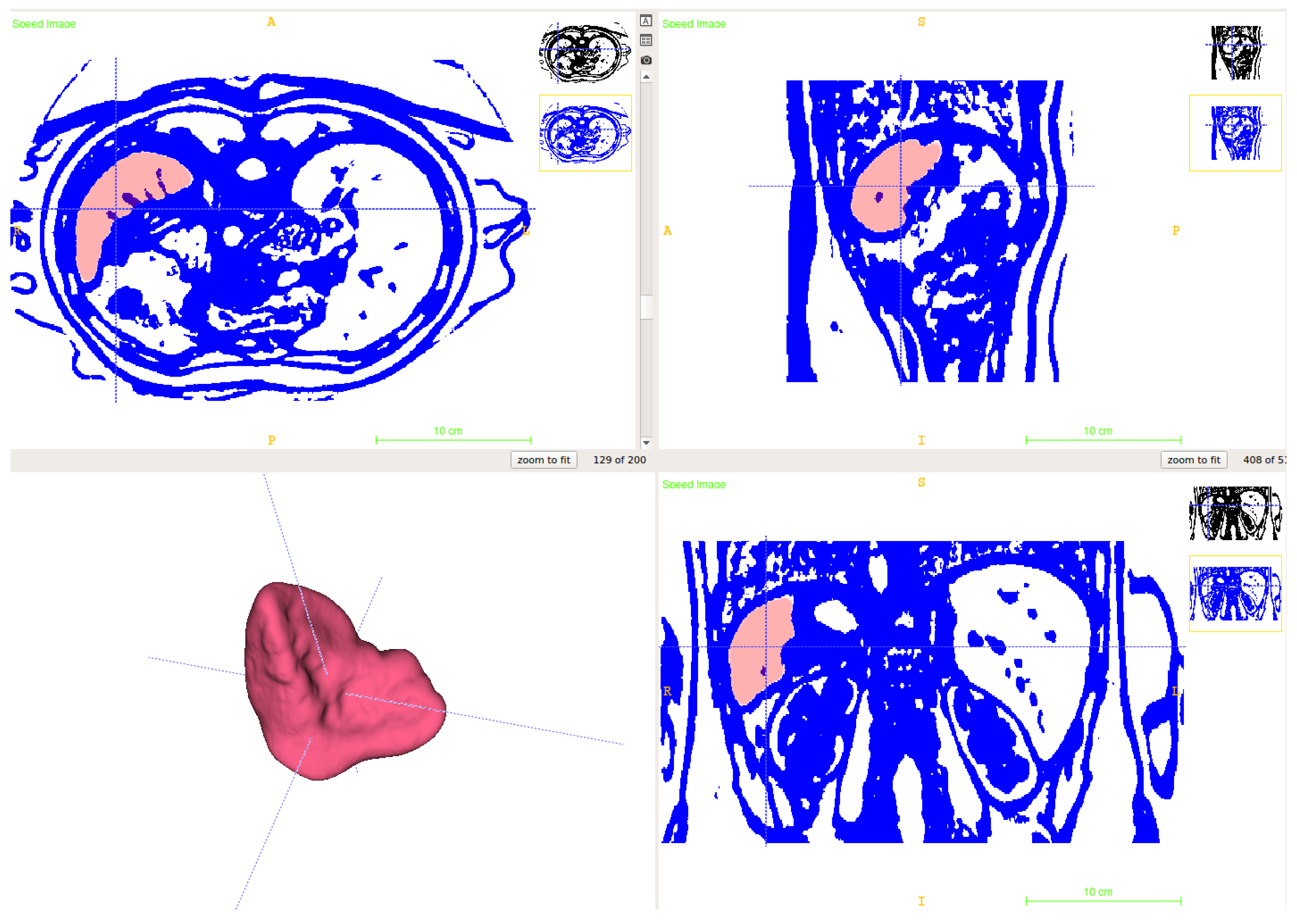
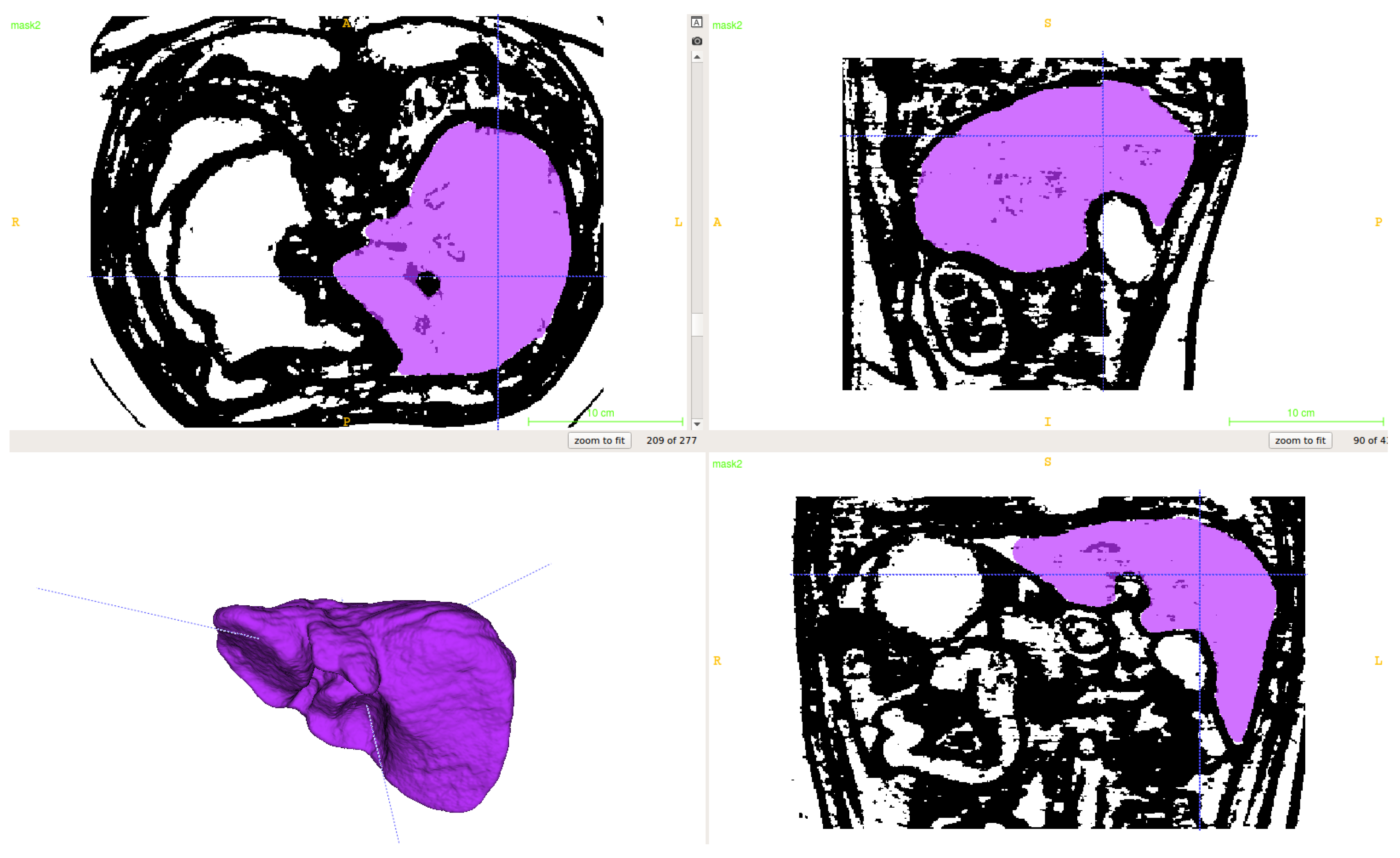
2.1. Segmentation of Abdominal Parenchymal Organs
- Pre-process and smooth the input dataset.
- Compute the spatial-dependence matrix (specified in [18]) for all voxels of the smoothed dataset.
- Compute entropy (1) for each voxel using the spatial-dependence matrix.
- Obtain the binary mask by entropy values thresholding.
- Set the seed points for organs extraction.
- Implement active contours method and extract 3D model.
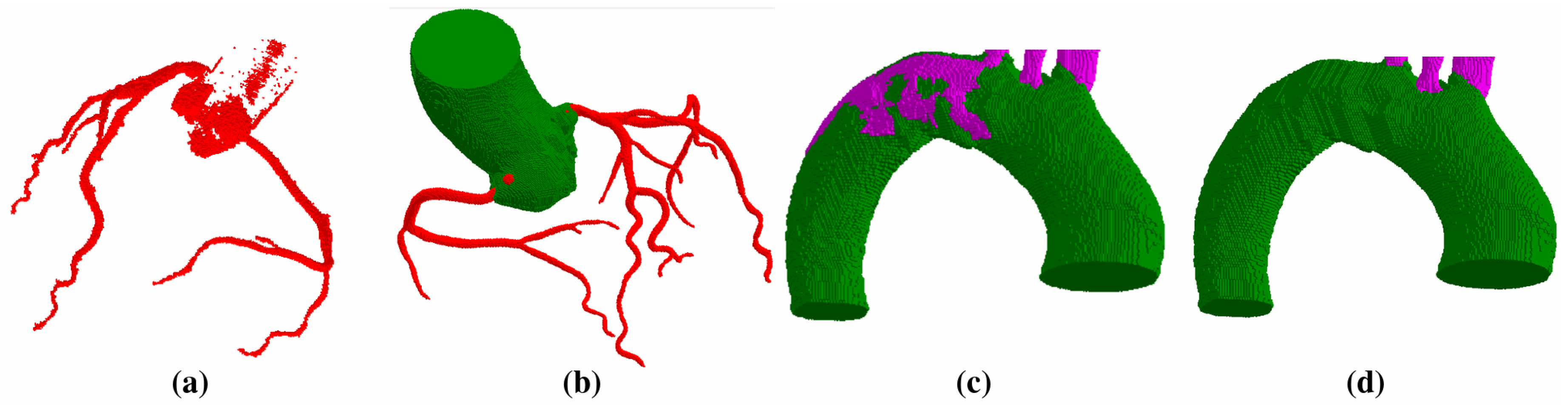
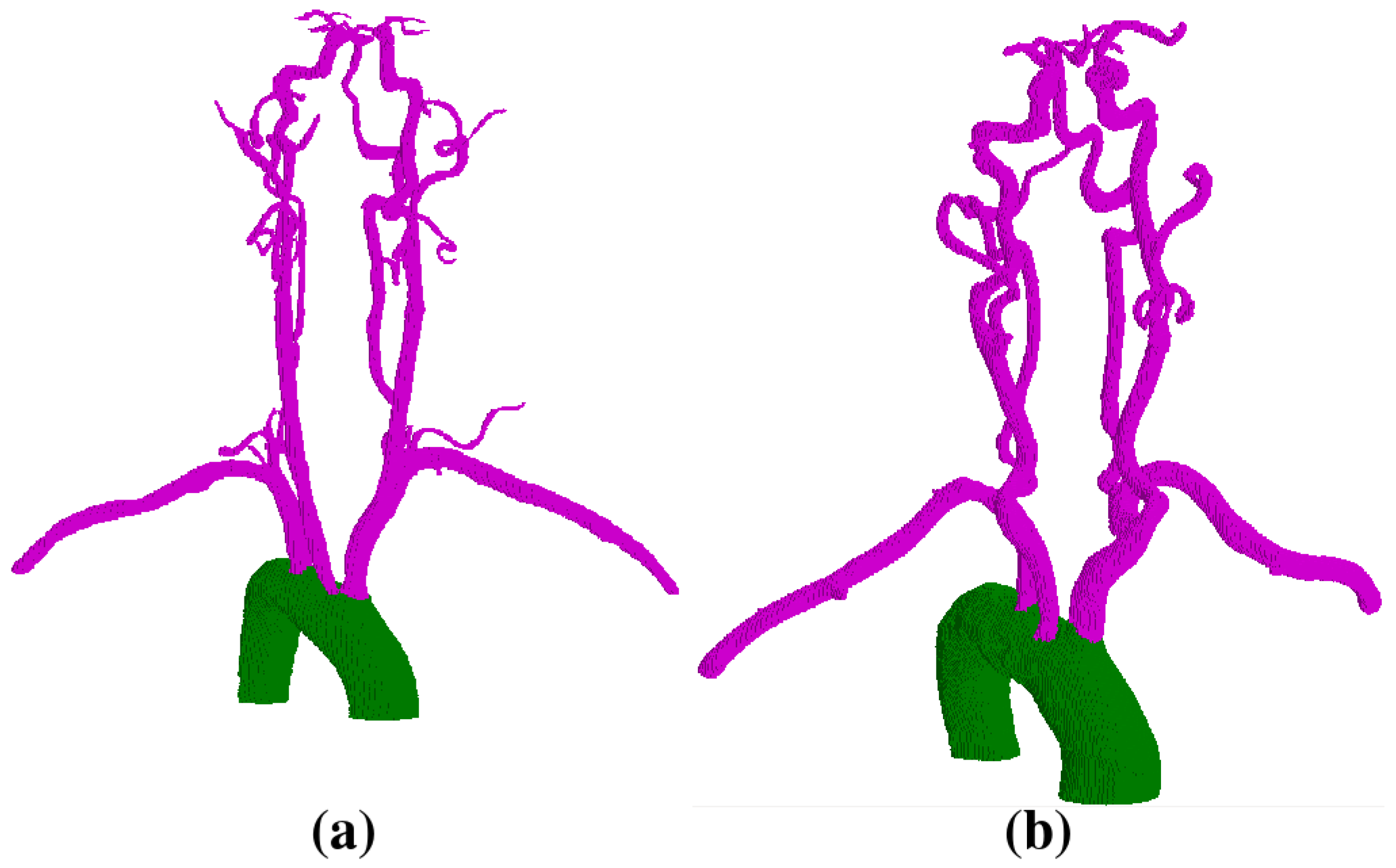
2.2. Vessel Segmentation
- Segment the aorta.
- Remove pulmonary arteries.
- (Cerebral case only) Darken bone intensities.
- Apply Frangi Vesselness filter.
- Locate the ostia points or aortic arch branches and segment the vessels.
- Clean aorta border.
- Compute the radius and the center c of the largest bright disk D on transverse planes using CHT method.
- Construct the connected region mask containing voxel c with the minimal intensity inside of D as the lower threshold.
- Obtain as a result of the IDT method applied to the mask and the seed c.
- Smooth the mask with the parameter r.
- (Cerebral case only) Copy mask to mask . Delete R-border from , then add -border.
- (Cerebral case only) Intersect the mask with the mask .
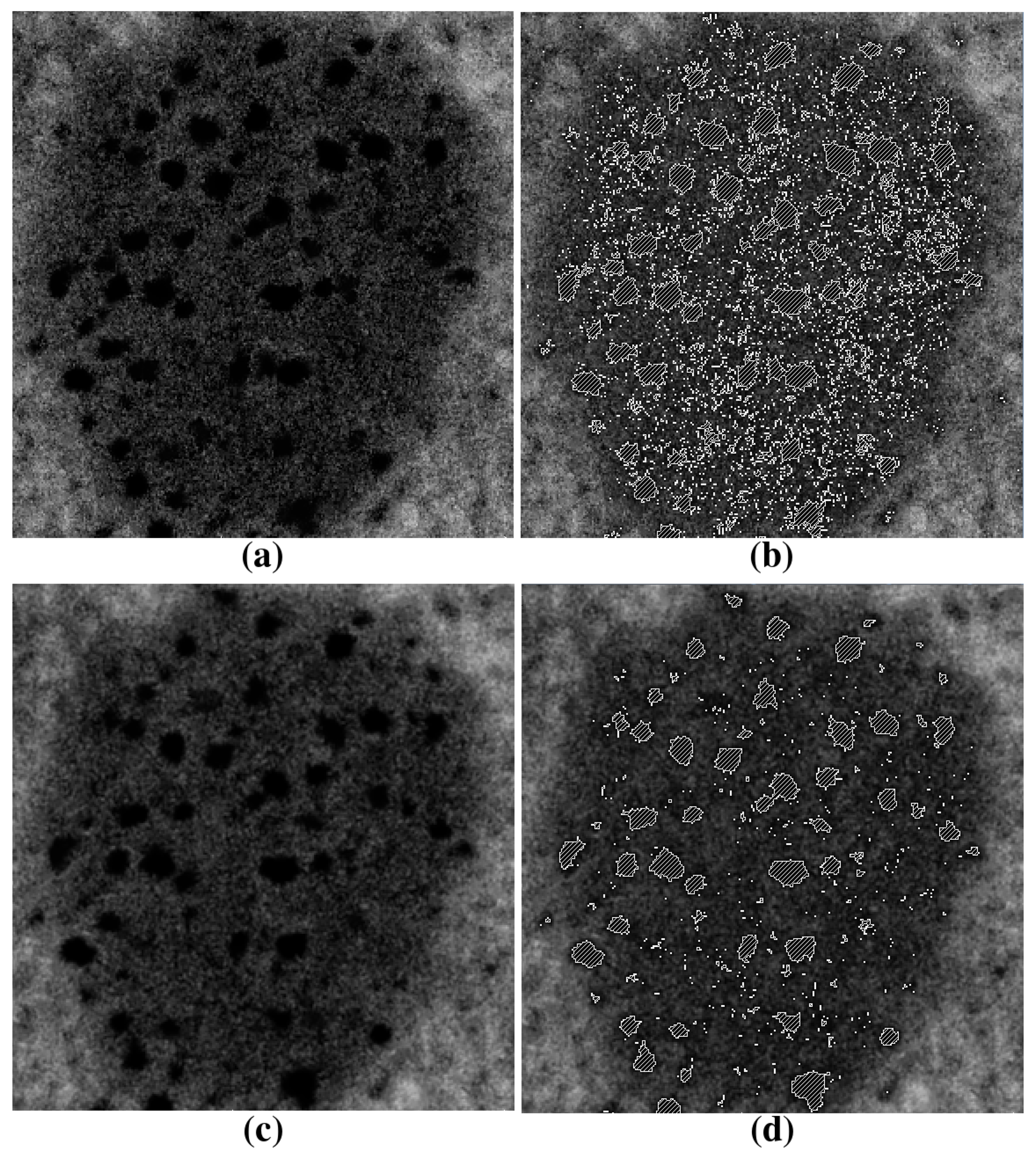
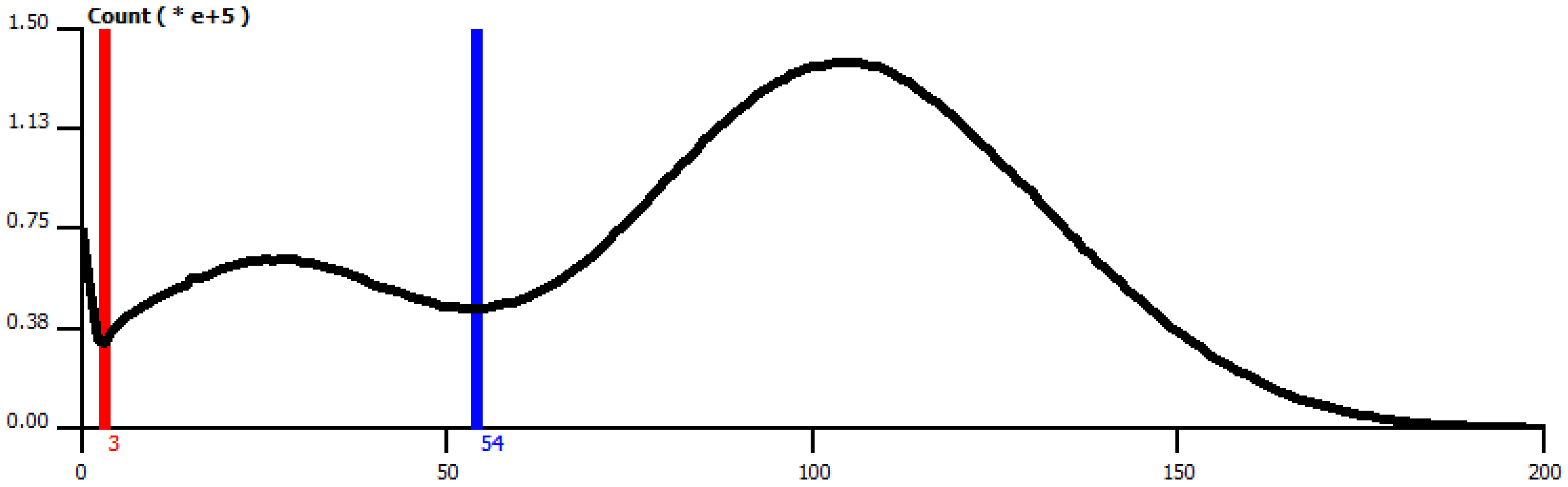
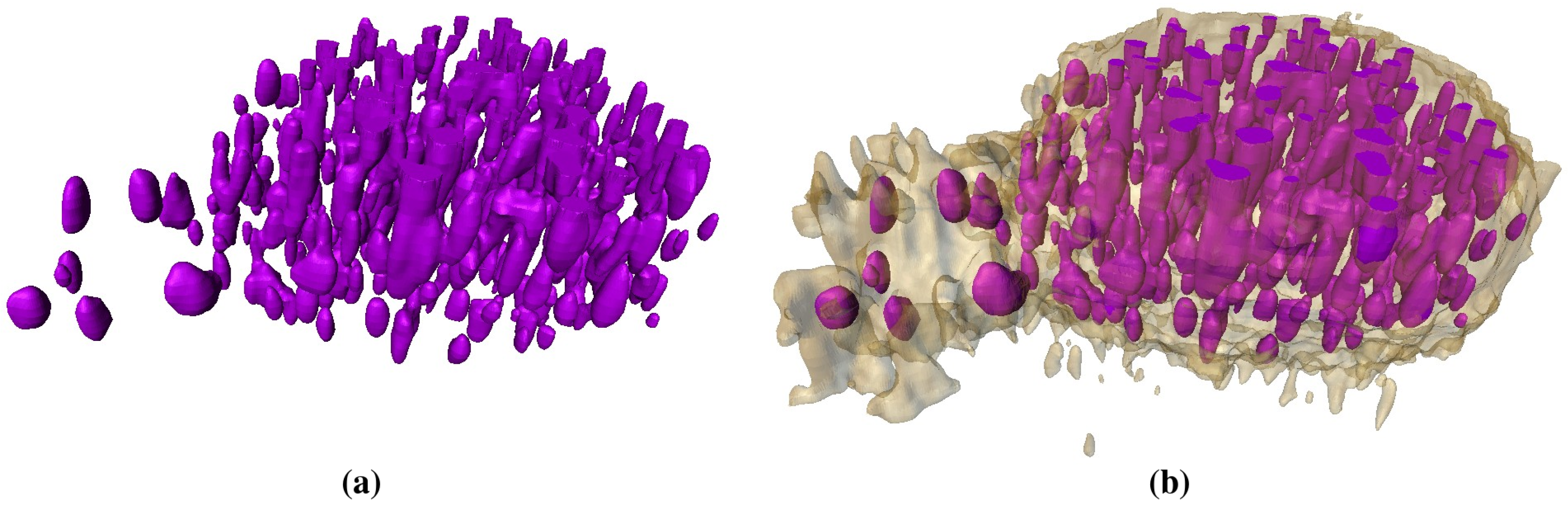
2.3. Segmentation of Lipid Droplets
- Align all 2D slices of the image dataset.
- Detect all lipid droplets using the threshold method (ref. [17]).
- Apply Gaussian smoothing in the vicinity of the lipid droplet.
- Segment inhomogeneous inclusions by thresholding with the lower parameter.
- Apply 3D remove-islands procedure.
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dazzo, F.; Niccum, B. Use of CMEIAS Image Analysis Software to Accurately Compute Attributes of Cell Size, Morphology, Spatial Aggregation and Color Segmentation that Signify in situ Ecophysiological Adaptations in Microbial Biofilm Communities. Computation 2015, 3, 72–98. [Google Scholar] [CrossRef]
- Kislitsyn, A.; Savinkov, R.; Novkovic, M.; Onder, L.; Bocharov, G. Computational Approach to 3D Modeling of the Lymph Node Geometry. Computation 2015, 3, 222–234. [Google Scholar] [CrossRef]
- Sazonov, I.; Yeo, S.Y.; Bevan, R.L.T.; Xie, X.; van Loon, R.; Nithiarasu, P. Modelling pipeline for subject-specific arterial blood flow—A review. Int. J. Numer. Methods Biomed. Eng. 2011, 27, 1868–1910. [Google Scholar] [CrossRef]
- Quarteroni, A.; Tuveri, M.; Veneziani, A. Computational vascular fluid dynamics: Problems, models and methods. Comput. Vis. Sci. 2000, 2, 163–197. [Google Scholar] [CrossRef]
- Gerbeau, J.F.; Vidrascu, M.; Frey, P. Fluid–structure interaction in blood flows on geometries based on medical imaging. Comput. Struct. 2005, 83, 155–165. [Google Scholar] [CrossRef]
- Holtzman-Gazit, M.; Kimmel, R.; Peled, N.; Goldsher, D. Segmentation of thin structures in volumetric medical images. IEEE Trans. Image Proc. 2006, 15, 354–363. [Google Scholar] [CrossRef]
- Radaelli, A.G.; Peiró, J. On the segmentation of vascular geometries from medical images. Int. J. Numer. Methods Biomed. Eng. 2010, 26, 3–34. [Google Scholar] [CrossRef]
- Yeo, S.Y.; Xie, X.; Sazonov, I.; Nithiarasu, P. Segmentation of biomedical images using active contour model with robust image feature and shape prior. Int. J. Numer. Methods Biomed. Eng. 2014, 30, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, T.; Brandt, R.; Menzel, R.; Russakoff, D.B.; Maurer, C.R. Quo Vadis, Atlas-Based Segmentation? In Handbook of Biomedical Image Analysis: Volume III: Registration Models; Springer: Boston, MA, USA, 2005; pp. 435–486. [Google Scholar]
- Isgum, I.; Staring, M.; Rutten, A.; Prokop, M.; Viergever, M.; van Ginneken, B. Multi-Atlas-Based Segmentation with Local Decision Fusion— Application to Cardiac and Aortic Segmentation in CT Scans. IEEE Trans. Med. Imaging 2009, 28, 1000–1010. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Suh, J.W.; Das, S.R.; Pluta, J.B.; Craige, C.; Yushkevich, P.A. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Lines, G.; Buist, M.; Grottum, P.; Pullan, A.; Sundnes, J.; Tveito, A. Mathematical models and numerical methods for the forward problem in cardiac electrophysiology. Comput. Vis Sci. 2002, 5, 215–239. [Google Scholar] [CrossRef]
- Zemzemi, N.; Bernabeu, M.O.; Saiz, J.; Cooper, J.; Pathmanathan, P.; Mirams, G.R.; Pitt-Francis, J.; Rodriguez, B. Computational assessment of drug-induced effects on the electrocardiogram: From ion channel to body surface potentials. Br. J. Pharmacol. 2013, 168, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.J.; Lubliner, J. On the one-dimensional theory of blood flow in the larger vessels. Math. Biosci. 1973, 18, 161–170. [Google Scholar] [CrossRef]
- Zarins, C.K.; Taylor, C.A.; Min, J.K. Computed Fractional Flow Reserve (FFTCT) Derived from Coronary CT Angiography. J. Cardiovasc. Transl. Res. 2013, 6, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Morris, P.D.; Ryan, D.; Morton, A.C.; Lycett, R.; Lawford, P.V.; Hose, D.R.; Gunn, J.P. Virtual Fractional Flow Reserve from Coronary Angiography: Modeling the Significance of Coronary Lesions. JACC Cardiovasc. Interv. 2013, 6, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Sulkin, M.S.; Yang, F.; Holzem, K.M.; Leer, B.V.; Bugge, C.; Laughner, J.I.; Green, K.; Efimov, I.R. Nanoscale three-dimensional imaging of the human myocyte. J. Struct. Biol. 2014, 188, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Frangi, A.; Niessen, W.; Vincken, K.; Viergever, M. Multiscale Vessel Enhancement Filtering. In Medical Image Computing and Computer-Assisted Interventation – MICCAI’98; Springer: Berlin/Heidelberg, Germany, 1998; pp. 130–137. [Google Scholar]
- Sethian, J. Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science; Number 3 in Cambridge Monographs on Applied and Computational Mathematics; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
- Grady, L. Fast, Quality, Segmentation of Large Volumes—Isoperimetric Distance Trees. In Computer Vision—ECCV 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 449–462. [Google Scholar]
- Pudney, C. Distance-Ordered Homotopic Thinning: A Skeletonization Algorithm for 3D Digital Images. Comput. Vis. Image Underst. 1998, 72, 404–413. [Google Scholar] [CrossRef]
- Danilov, A.; Ivanov, Y.; Pryamonosov, R.; Vassilevski, Y. Methods of Graph Network Reconstruction in Personalized Medicine. Int. J. Numer. Methods Biomed. Eng. 2016, 32, e02754. [Google Scholar] [CrossRef] [PubMed]
- Danilov, A.A.; Nikolaev, D.V.; Rudnev, S.G.; Salamatova, V.Y.; Vassilevski, Y.V. Modelling of bioimpedance measurements: Unstructured mesh application to real human anatomy. Russ. J. Numer. Anal. Math. Model. 2012, 27, 431–440. [Google Scholar] [CrossRef]
- Vassilevski, Y.V.; Danilov, A.A.; Simakov, S.S.; Gamilov, T.M.; Ivanov, Y.A.; Pryamonosov, R.A.; Simakov, S.S. Patient-specific anatomical models in human physiology. Russ. J. Numer. Anal. Math. Model. 2015, 30, 185–201. [Google Scholar] [CrossRef]
- Danilov, A.A.; Pryamonosov, R.A.; Yurova, A.S. Image segmentation techniques for biomedical modeling: Electrophysiology and hemodynamics. In Proceedings of the 7th European Congress on Computational Methods in Applied Sciences and Engineering (ECCOMAS 2016), Crete Island, Greece, 5–10 June 2016.
- Yang, G.; Kitslaar, P.; Frenay, M.; Broersen, A.; Boogers, M.J.; Bax, J.J.; Reiber, J.H.C.; Dijkstra, J. Automatic Centerline Extraction of Coronary Arteries in Coronary Computed Tomographic Angiography. Int. J. Cardiovasc. Imaging 2012, 28, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Tek, H. Automatic Coronary Tree Modeling. The Midas Journal—2008 MICCAI Workshop Grand Challenge Coronary Artery Tracking. 2008. Available online: http://hdl.handle.net/10380/1426 (accessed on 7 September 2016).
- Manniesing, R.; Viergever, M.A.; van der Lugt, A.; Niessen, W.J. Cerebral Arteries: Fully Automated Segmentation from CT Angiography—A Feasibility Study 1. Radiology 2008, 247, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Uchiyama, Y.; Zhou, X.; Hara, T.; Asano, T.; Fujita, H. A Fast and Fully Automatic Method for Cerebrovascular Segmentation on Time-of-Flight (TOF) MRA Image. J. Digit. Imaging 2011, 24, 609–625. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.; Bier, P.; Sands, G.; Hunter, P. Cerebral artery segmentation with level set methods. In Proceedings of Image and Vision Computing New Zealand 2007, Hamilton, New Zealand, 5–7 December 2007; pp. 300–304.
- Cuisenaire, O. Fully automated segmentation of carotid and vertebral arteries from CTA. The Midas Journal—Carotid Lumen Segmentation and Stenosis Grading (Grand Challenge). 2009. Available online: http://hdl.handle.net/10380/3100 (accessed on 7 September 2016).
- Lucchi, A.; Smith, K.; Achanta, R.; Knott, G.; Fua, P. Supervoxel-Based Segmentation of Mitochondria in EM Image Stacks with Learned Shape Features. IEEE Trans. Med. Imaging 2012, 31, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Keller, D.U.J.; Weber, F.M.; Seemann, G.; Dössel, O. Ranking the Influence of Tissue Conductivities on Forward-Calculated ECGs. IEEE Trans. Biomed. Eng. 2010, 57, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Colville, J.; Kalaigian, J.; Curran, S.; Jiang, L.; Kijewski, P.; Schwartz, L.H. Automated Quantification of Body Fat Distribution on Volumetric Computed Tomography. J. Comput. Assist. Tomogr. 2006, 30, 777–783. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G.; Sensakovic, W.F. Automated lung segmentation for thoracic CT. Acad. Radiol. 2004, 11, 1011–1021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, C.H.; Chui, C.K.; Ong, S.H. Fast segmentation of bone in CT images using 3D adaptive thresholding. Comput. Biol. Med. 2010, 40, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Cobzas, D.; Birdsell, L.; Lieffers, J.; Baracos, V. Automated segmentation of muscle and adipose tissue on CT images for human body composition analysis. In Proceedings of the Medical Imaging 2009: Visualization, Image-Guided Procedures, and Modeling, Lake Buena Vista, FL, USA, 7 February 2009.
- Danilov, A.; Kramarenko, V.; Nikolaev, D.; Yurova, A. Personalized model adaptation for bioimpedance measurements optimization. Russ. J. Numer. Anal. Math. Model. 2013, 28, 459–470. [Google Scholar] [CrossRef]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Campadelli, P.; Casiraghi, E.; Pratissoli, S.; Lombardi, G. Automatic Abdominal Organ Segmentation from CT images. Electron. Lett. Comput. Vis. Image Anal. 2009, 8, 1–14. [Google Scholar]
- Chan, T.F.; Vese, L.A. Active Contours without Edges. IEEE Trans. Image Proc. 2001, 10, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Marquez-Neila, P.; Baumela, L.; Alvarez, L. A Morphological Approach to Curvature-Based Evolution of Curves and Surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 36, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Yushkevich, P.A.; Piven, J.; Cody Hazlett, H.; Gimpel Smith, R.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Van Andel, H.A.F.G.; Venema, H.W.; Streekstra, G.J.; van Straten, M.; Majoie, C.B.L.M.; den Heeten, G.J.; Grimbergen, C.A. Removal of Bone in CT Angiography by Multiscale Matched Mask Bone Elimination. Med. Phys. 2007, 34, 449–462. [Google Scholar]
- Duda, R.O.; Hart, P.E. Use of the Hough Transformation to Detect Lines and Curves in Pictures. Commun. ACM 1972, 15, 11–15. [Google Scholar] [CrossRef]
- Johnson, H.J.; McCormick, M.M.; Ibanez, L. The ITK Software Guide Book 2: Design and Functionality; Kitware, Inc.: Clifton Park, NY, USA, 2015; Volume 2. [Google Scholar]
- OsiriX. DICOM Image Sample Sets. Available online: http://www.osirix-viewer.com/datasets (accessed on 9 September 2016).
- Gamilov, T.; Pryamonosov, R.; Simakov, S. Modeling of patient-specific cases of atherosclerosis in carotid arteries. In Proceedings of the 7th European Congress on Computational Methods in Applied Sciences and Engineering (ECCOMAS 2016), Crete Island, Greece, 5–10 June 2016.
- Gamilov, T.; Kopylov, P.; Pryamonosov, R.; Simakov, S. Virtual Fractional Flow Reserve Assesment in Patient-Specific Coronary Networks by the 1D Model of Haemodynamics. Russ. J. Numer. Anal. Math. Model. 2015, 30, 269–276. [Google Scholar] [CrossRef]
- Lucchi, A.; Smith, K.; Achanta, R.; Lepetit, V.; Fua, P. A Fully Automated Approach to Segmentation of Irregularly Shaped Cellular Structures in EM Images. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2010; Springer: Berlin/Heidelberg, Germany, 2010; pp. 463–471. [Google Scholar]
| Dataset Size, Voxels | CPU Time, s | GPU Time, s |
|---|---|---|
| 735 | 139 | |
| 1189 | 198 |
| Dataset | Dataset 1 | Dataset 2 |
|---|---|---|
| Resolution | ||
| Spacing | mm | mm |
| Pulmonary removal | s | s |
| Aorta segmentation | s | s |
| Frangi Vesselness | s | s |
| Aortic arch branches | s | s |
| Aorta border cleaning | s | s |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilov, A.; Pryamonosov, R.; Yurova, A. Image Segmentation for Cardiovascular Biomedical Applications at Different Scales. Computation 2016, 4, 35. https://doi.org/10.3390/computation4030035
Danilov A, Pryamonosov R, Yurova A. Image Segmentation for Cardiovascular Biomedical Applications at Different Scales. Computation. 2016; 4(3):35. https://doi.org/10.3390/computation4030035
Chicago/Turabian StyleDanilov, Alexander, Roman Pryamonosov, and Alexandra Yurova. 2016. "Image Segmentation for Cardiovascular Biomedical Applications at Different Scales" Computation 4, no. 3: 35. https://doi.org/10.3390/computation4030035
APA StyleDanilov, A., Pryamonosov, R., & Yurova, A. (2016). Image Segmentation for Cardiovascular Biomedical Applications at Different Scales. Computation, 4(3), 35. https://doi.org/10.3390/computation4030035







