Evolution by Pervasive Gene Fusion in Antibiotic Resistance and Antibiotic Synthesizing Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Case 1
2.2. Case 2
2.3. BLAST Analysis and Network Construction
2.4. Identification of Composite Genes
2.5. Investigation of Domain Architecture
2.6. BLAST Analysis and Sequence Alignments
2.7. N-Rooted Fusion Graph Construction
3. Results and Discussion
3.1. Case 1: Polyketide Biosynthetic Genes
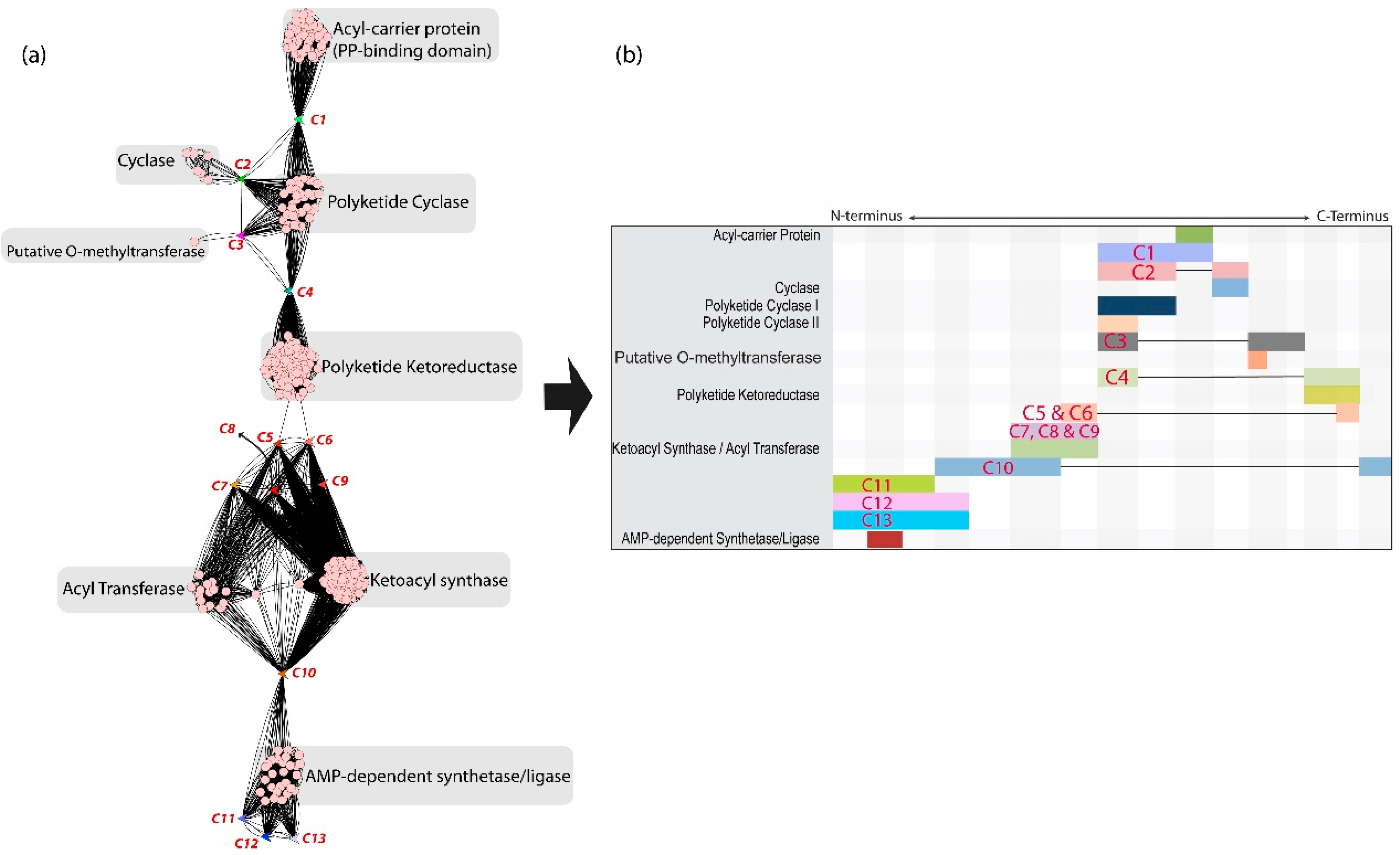
3.2. Composite Gene 1 Analysis
3.3. GCC Analysis
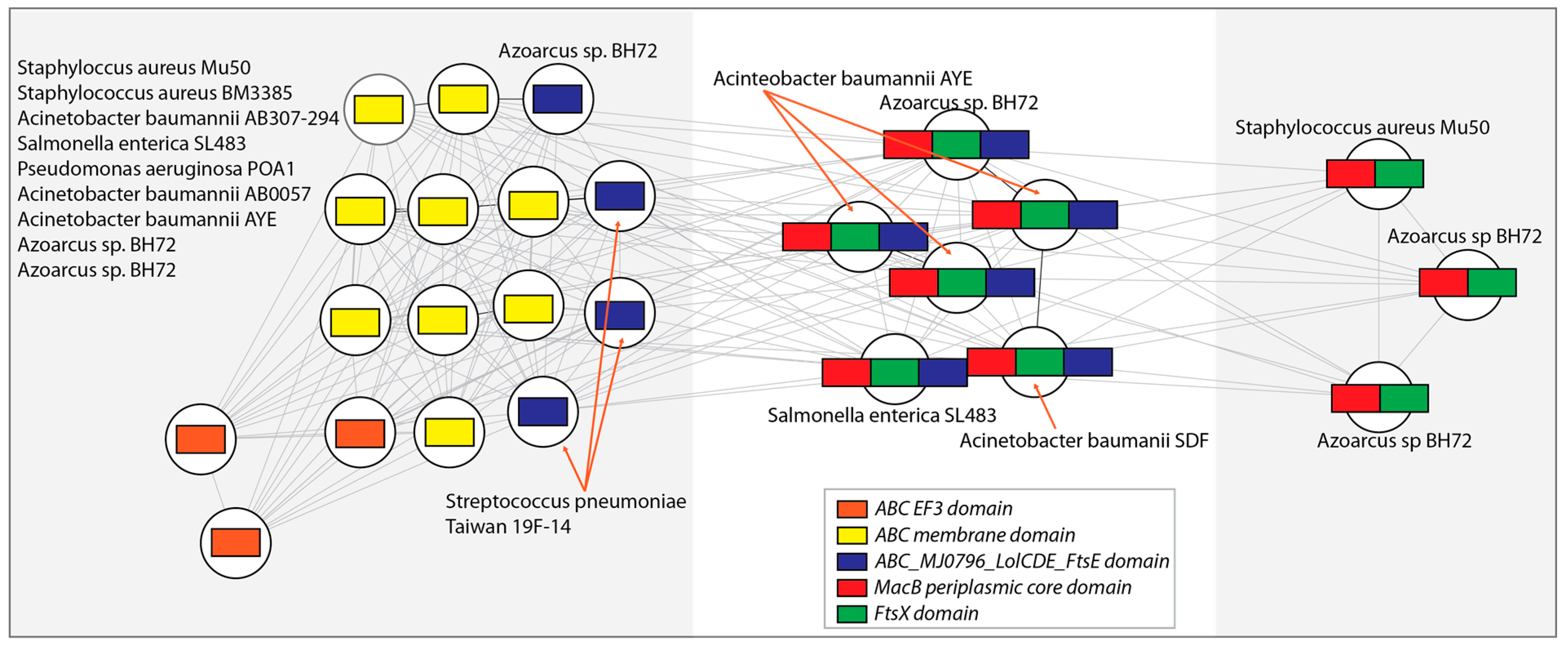
3.4. Case 2: Antibiotic Resistance Genes
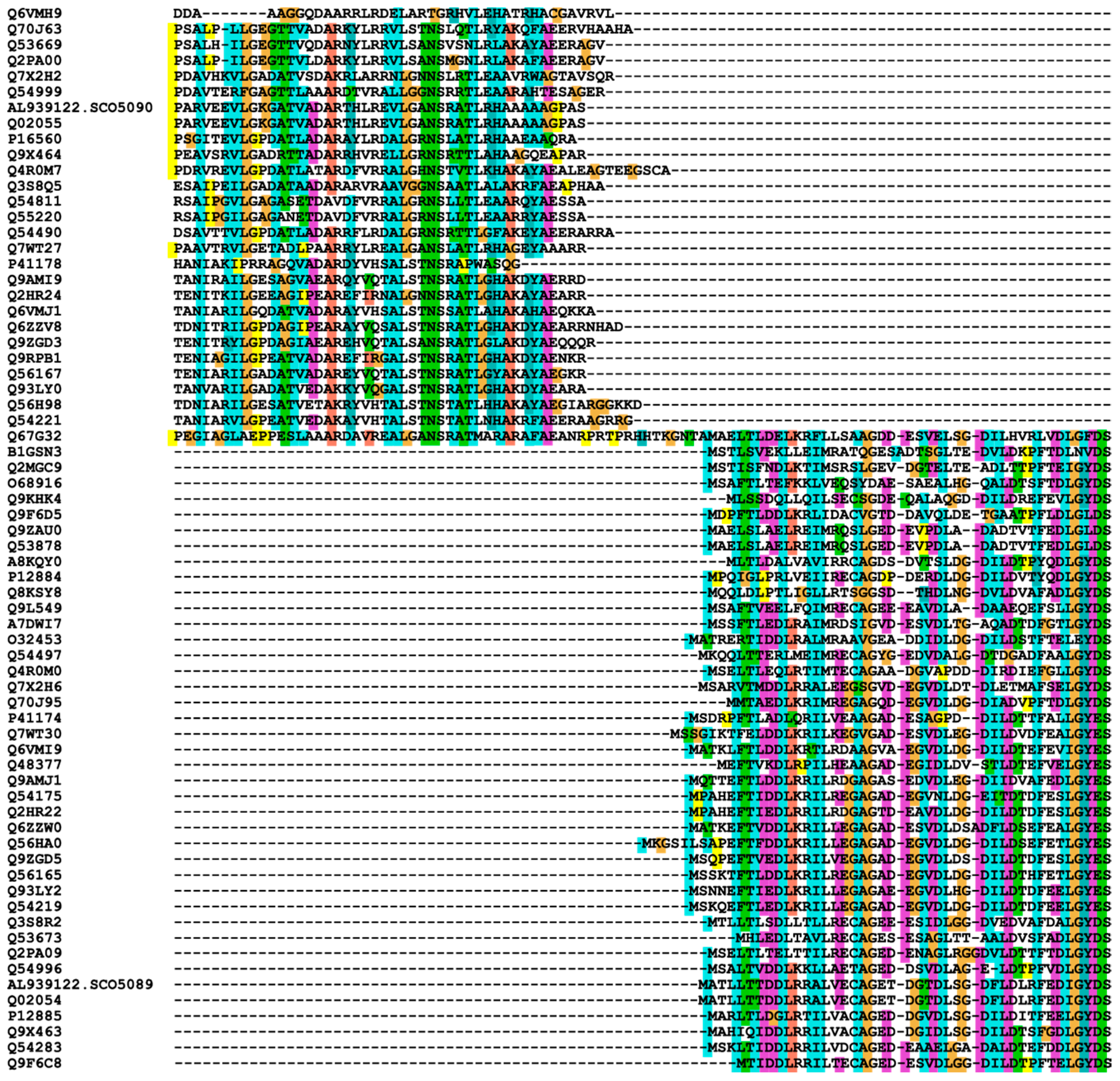
3.5. Sequence Alignments
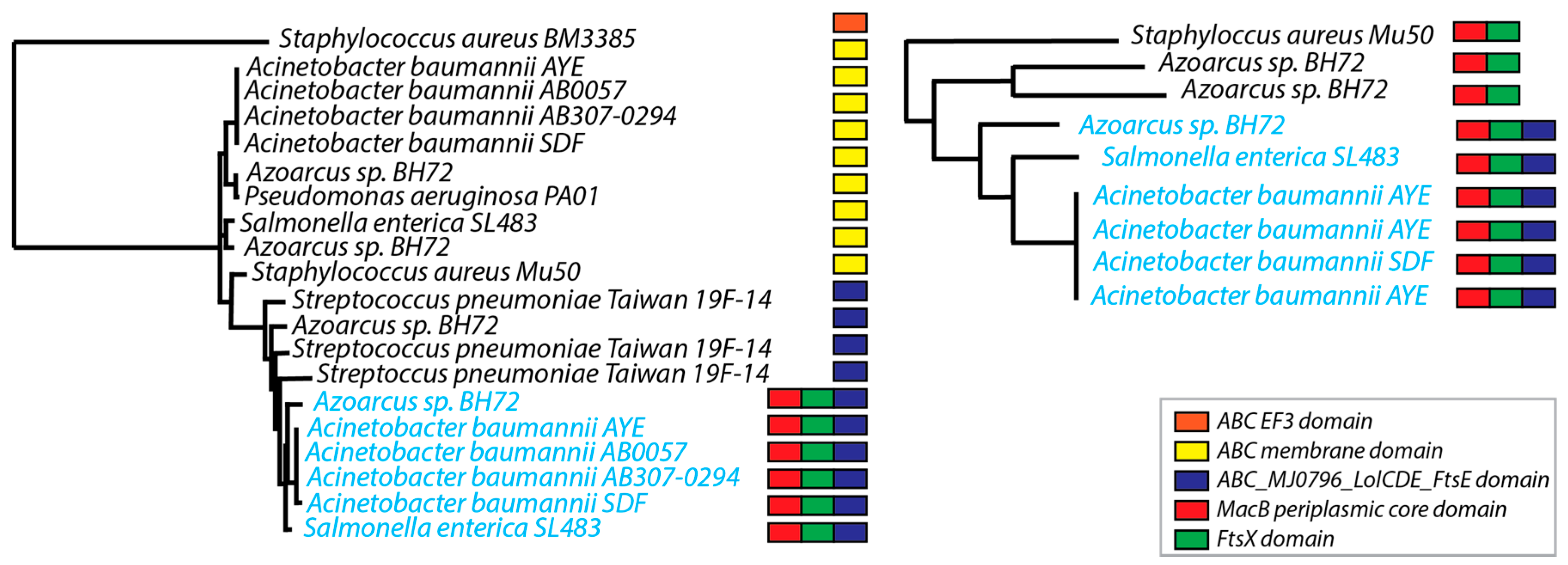
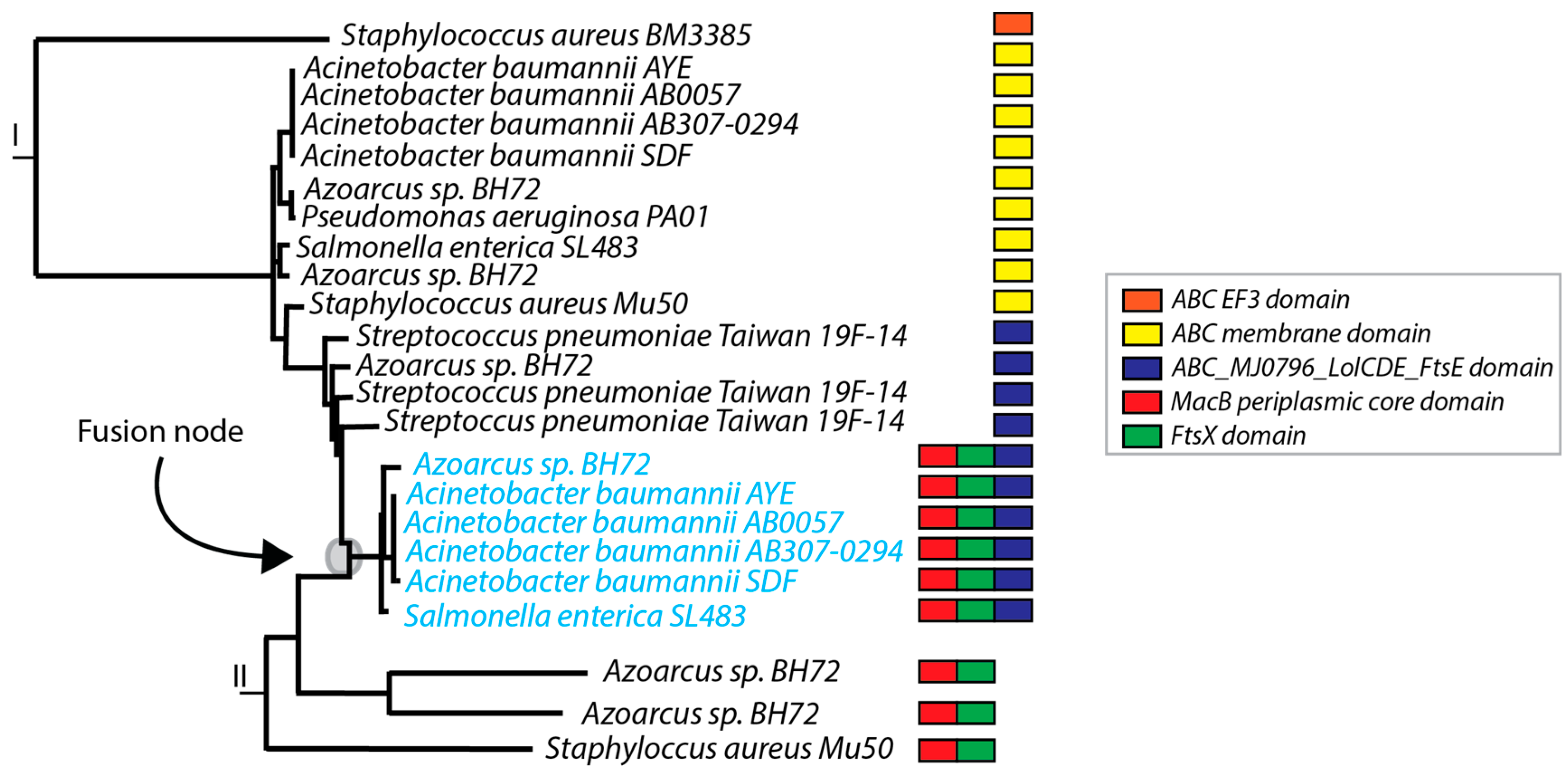
3.6. Phylogenetic Trees
3.7. N-Rooted Graph
4. Conclusions
Author Contributions
Conflicts of Interest
References
- Dagan, T. Phylogenomic networks. Trends Microbiol. 2011, 19, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Darwin, C. On the Origin of Species 1859 Chapter IV “Character of Natural Selection” in the sub-section on “Divergence of Character.”; John Murray: London, UK, 1859. [Google Scholar]
- Huson, D.H.; Rupp, R.; Scornavacca, C. Phylogenetic Networks-Concepts, Algorithms and Applications; Cambridge University Press: Cambridge, UK, 2011; Volume 1. [Google Scholar]
- Bapteste, E.; van Iersel, L.; Janke, A.; Kelchner, S.; Kelk, S.; McInerney, J.O.; Morrison, D.A.; Nakhleh, L.; Steel, M.; Stougie, L.; et al. Networks: Expanding evolutionary thinking. Cell 2013, 29, 439–441. [Google Scholar]
- Myers, C.L.; Robson, D.; Wible, A.; Hibbs, M.A.; Chiriac, C.; Thessfeld, C.L.; Dolinski, K.; Troyanskaya, O.G. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005, 6. [Google Scholar] [CrossRef]
- Larremore, D.B.; Clauset, A.; Buckee, C.O. A Network Approach to Analyzing Highly Recombinant Malaria Parasite Genes. PloS Comput. Biol. 2013, 9. [Google Scholar] [CrossRef]
- Jordan, W.C.; Turnquist, M.A. A stochastic, dynamic network model for railroad car distribution. Transp. Sci. 1983, 17, 123–145. [Google Scholar] [CrossRef]
- Viswanath, B.; Mislove, A.; Cha, M.; Gummadi, K.P. On the evolution of user interactions in Facebook. In Proceedings of the 2nd ACM Workshop on Online Social Networks (WOSN’09), Barcelona, Spain, 17 August 2009; pp. 37–42.
- Dagan, T.; Martin, W. Getting a better picture of microbial evolution en route to a network of genomes. Philos. Trans. Royal Soc. Boil. Sci. 2009, 364, 2187–2196. [Google Scholar] [CrossRef]
- Halary, S.; Leigh, J.W.; Cheaib, B.; Lopez, P.; Bapteste, E. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. USA 2010, 107, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Haggerty, L.; Jachiet, P.A.; Hanage, W.P.; Fitzpatrick, D.A.; Lopez, F.; O’Connell, M.J.; Pisani, D.; Wilkinson, M.; Bapteste, E.; McInerney, J.O. A pluralistic account of homology: Adapting the models to the data. Mol. Boil. Evol. 2013, 22. [Google Scholar] [CrossRef]
- Alvarez-Ponce, D.; Lopez, P.; Bapteste, E.; McInerney, J.O. Gene similarity networks provide tools for understanding eukaryotic origins and evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 1594–1603. [Google Scholar] [CrossRef]
- Halary, S.; Mc Inerney, J.O.; Lopez, P.; Bapteste, E. EGN: A wizard for construction of gene and genome similarity networks. BMC Evol. Biol. 2013, 13. [Google Scholar] [CrossRef]
- Fondi, M.; Fani, R. The horizontal flow of plasmid resistome: Clues from inter-generic similarity networks. Wiley 2010, 12, 3228–3242. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, M.P.; Nagulapalli, K.; Palakal, M.J. Cliques for the identification of gene signatures for colorectal cancer across population. BMC Syst. Biol. 2012, 6. [Google Scholar] [CrossRef] [PubMed]
- Long, M.; Betrán, E.; Thornton, K.; Wang, W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003, 4, 865–875. [Google Scholar] [CrossRef] [PubMed]
- Durrens, P.; Nikolski, M.; Sherman, D. Fusion and Fission of Genes Define a Metric between Fungal Genomes. PloS Comput. Biol. 2008, 4. [Google Scholar] [CrossRef]
- Kummerfeld, S.K.; Teichmann, S.A. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005, 21, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Long, M.Y. A new function evolved from gene fusion. Genome Res. 2000, 10, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Snel, B.; Bork, P.; Huynen, M. Genome evolution—Gene fusion versus gene fission. Trends Genet. 2000, 16, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Marcotte, E.M.; Pellegrini, M.; Ng, H.L.; Rice, D.W.; Yeates, T.O.; Eisenberg, D. Detecting protein function and protein-protein interactions from genome sequences. Science 1999, 285, 751–753. [Google Scholar] [CrossRef] [PubMed]
- Civjan, N. Natural Products in Chemical Biology; Wiley: Hoboken, NJ, USA, 2012; pp. 143–189. [Google Scholar]
- Ridley, C.P.; Lee, H.Y.; Khosla, C. Evolution of polyketide synthases in bacteria. Proc. Natl. Acad. Sci. USA 2007, 105, 4595–4600. [Google Scholar] [CrossRef]
- Wright, G.D.; Poinar, H. Antibiotic resistance is ancient: Implications for drug discovery. Trends Microbiol. 2012, 20, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Galerunti, R.A. The Antibiotic Paradox: How Miracle Drugs Are Destroying the Miracle. Health Values 1994, 18, 60–61. [Google Scholar]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yi, G.S. PKMiner: A database for exploring type II polyketide synthases. BMC Microbiol. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; de Pascale, G.; Ejim, L.; et al. The Comprehensive Antibiotic Resistance Database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Jachiet, P.A.; Pogorelcnik, R.; Berry, A.; Lopez, P.; Bapteste, E. MosaicFinder: Identification of fused gene families in sequence similarity networks. Bioinformatics 2013, 29, 837–844. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39D, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Gouy, M.; Gautier, C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Computer Appl. Biosci. 1996, 12, 543–548. [Google Scholar]
- Ames, B.D.; Korman, T.P.; Zhang, W.; Smith, P.; Vu, T.; Tang, Y.; Tsai, S.C. Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclisation specificity of aromatic polyketides. Proc. Natl. Acad. Sci. USA 2008, 105, 5349–5354. [Google Scholar] [CrossRef] [PubMed]
- Britannica Encyclopedia (2014) “Homology”. Encyclopaedia Britannica Online, Encyclopædia Britannica Inc. 2014. Available online: http://www.britannica.com/EBchecked/topic/270557/homology (accessed on 10 April 2014).
- Brown, J.R. Ancient horizontal gene transfer. Nat. Rev. Genet. 2003, 4, 121–132. [Google Scholar] [CrossRef] [PubMed]
- McInerney, J.O.; Cotton, J.A.; Pisani, D. The prokaryotic tree of life: past, present ... and future? Trends Ecol. Evol. 2008, 23, 276–281. [Google Scholar] [CrossRef] [PubMed]
- Bapteste, E.; O’Malley, M.A.; Beiko, R.G.; Ereshefsky, M.; Gogarten, J.P.; Franklin-Hall, L.; Lapointe, F.J.; Dupre, J.; Dagan, T.; Boucher, Y.; et al. Prokaryotic evolution and the tree of life are two different things. Biol. Direct. 2009, 4. [Google Scholar] [CrossRef]
- Bapteste, E.; Lopez, P.; Bouchard, F.; Baquero, F.; McInerney, J.O.; Burian, R.M. Evolutionary analyses of non-genealogical bonds produced by introgressive descent. Proc. Natl. Acad. Sci. USA 2012, 109, 18266–18272. [Google Scholar] [CrossRef] [PubMed]
- Mitelman, F.; Johansson, B.; Mertens, F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat. Genet. 2004, 36, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Long, M. A New Function Evolved from Gene Fusion. Genome Res. 2000, 10, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Oh, S.H.; Coleman, D.A.; Hoyer, L.L. ALS51, a newly discovered gene in the Candida albicans ALS family, created by intergenic recombination: Analysis of the gene and protein, and implications for evolution of microbial gene families. FEMS Immunol. Med. Microbiol. 2011, 61, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Micci, F.; Thorsen, J.; Panagopoulos, I.; Nyquist, K.B.; Zeller, B.; Tierens, A.; Heim, S. High-throughput sequencing identifies an NFIA/CBFA2T3 fusion gene in acute erythroid leukemia with t(1;16)(p31;q24). Leukemia 2013, 27, 980–982. [Google Scholar] [CrossRef] [PubMed]
- McInerney, J.O.; Pisani, D.; Bapteste, E.; O’Connell, M.J. The public goods hypothesis for the evolution of life on Earth. Biol. Direct. 2011, 6. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coleman, O.; Hogan, R.; McGoldrick, N.; Rudden, N.; McInerney, J.O. Evolution by Pervasive Gene Fusion in Antibiotic Resistance and Antibiotic Synthesizing Genes. Computation 2015, 3, 114-127. https://doi.org/10.3390/computation3020114
Coleman O, Hogan R, McGoldrick N, Rudden N, McInerney JO. Evolution by Pervasive Gene Fusion in Antibiotic Resistance and Antibiotic Synthesizing Genes. Computation. 2015; 3(2):114-127. https://doi.org/10.3390/computation3020114
Chicago/Turabian StyleColeman, Orla, Ruth Hogan, Nicole McGoldrick, Niamh Rudden, and James O. McInerney. 2015. "Evolution by Pervasive Gene Fusion in Antibiotic Resistance and Antibiotic Synthesizing Genes" Computation 3, no. 2: 114-127. https://doi.org/10.3390/computation3020114
APA StyleColeman, O., Hogan, R., McGoldrick, N., Rudden, N., & McInerney, J. O. (2015). Evolution by Pervasive Gene Fusion in Antibiotic Resistance and Antibiotic Synthesizing Genes. Computation, 3(2), 114-127. https://doi.org/10.3390/computation3020114




