Computationally Efficient Outlier Detection for High-Dimensional Data Using the MDP Algorithm
Abstract
1. Introduction
2. High-Dimensional Multivariate Outlier Detection with the MDP
- Step 1
- Set the number of observations to , repeating Steps 2–5 m times.
- Step 2
- Randomly select a small sample of size 2 and compute the mean vector and diagonal covariance matrix . Use these values to compute the Mahalanobis distances of all observations as follows:
- Step 3
- The h smallest are assigned a weight 1, while the others are assigned a weight of 0.
- Step 4
- Recompute and using h observations with weight 1 and recompute the Mahalanobis distances.
- Step 5
- Repeat Steps 3–4 15 times until the weights no further change.
- Step 6
- Compute the Mahalanobis distances using the and of the h observations obtained from Steps 2–5 as follows:
- Step 7
- Refine the Mahalanobis distances:
- Step 8
- Compute , where denotes the correlation matrix of h observations: .
- Step 9
- Compute the following quantity:where . Then, retain the observations , for which , which is the quantile of the standard normal distribution.
- Step 10
- Using the observations from Step 9, update the Mahalanobis distances, termed this time for convenience, and compute the updated trace of the correlation matrix . Then, update .
- Step 11
- Compute the following quantities:
- Step 12
- Compute the following quantity:
- Step 13
- The observations for which the inequality holds true are possible or candidate outliers.
Breaking Down the R Code
RMDP <- function(y, alpha = 0.05, itertime = 100) {
## y is the data
## alpha is the significance level
## itertime is the number of iterations
y <- as.matrix(y) ## makes sure y is a matrix
n <- nrow(y) ## sample size
p <- ncol(y) ## dimensionality
h <- round(n/2) + 1
init_h <- 2
delta <- alpha/2
bestdet <- 0
jvec <- array(0, dim = c(n, 1))
#####
for (A in 1:itertime) {
id <- sample(n, init_h)
ny <- y[id, ]
mu_t <- apply(ny, 2, mean)
var_t <- apply(ny, 2, var)
dist <- apply( ( t((t(y) - mu_t) / var_t^0.5) )^2, 1, sum)
crit <- 10
l <- 0
while (crit != 0 & l <= 15) {
l <- l + 1
ivec <- array(0, dim = c(n, 1))
dist_perm <- order(dist)
ivec[ dist_perm[1:h] ] <- 1
crit <- sum( abs(ivec - jvec) )
jvec <- ivec
newy <- y[dist_perm[1:h], ]
mu_t <- apply(newy, 2, mean)
var_t <- apply(newy, 2, var)
dist <- apply( ( t( (t(y) - mu_t ) / var_t^0.5) )^2, 1, sum )
}
tempdet <- prod(var_t)
if (bestdet == 0 | tempdet < bestdet) {
bestdet <- tempdet
final_vec <- jvec
}
}
#####
submcd <- seq(1, n)[final_vec != 0]
mu_t <- apply( y[submcd, ], 2, mean )
var_t <- apply( y[submcd, ], 2, var )
dist <- apply( ( t( (t(y) - mu_t)/var_t^0.5) )^2, 1, sum )
dist <- dist * p / median(dist)
ER <- cor( y[submcd, ] )
ER <- ER %*% ER
tr2_h <- sum( diag(ER) )
tr2 <- tr2_h - p^2 / h
cpn_0 <- 1 + (tr2_h) / p^1.5
w0 <- (dist - p) / sqrt( 2 * tr2 * cpn_0 ) < qnorm(1 - delta)
nw <- sum(w0)
sub <- seq(1, n)[w0]
mu_t <- apply( y[sub,], 2, mean )
var_t <- apply( y[sub,], 2, var )
ER <- cor( y[sub, ] )
ER <- ER %*% ER
tr2_h <- sum( diag(ER) )
tr2 <- tr2_h - p^2 / nw
dist <- apply( ( t( (t(y) - mu_t)/var_t^0.5) )^2, 1, sum )
scale <- 1 + 1/sqrt( 2 * pi) * exp( - qnorm(1 - delta)^2 / 2 ) /
(1 - delta) * sqrt( 2 * tr2) / p
dist <- dist / scale
cpn_1 <- 1 + (tr2_h) / p^1.5
w1 <- (dist - p) / sqrt(2 * tr2 * cpn_1 ) < qnorm(1 - alpha)
list(decision = w1)
}
rmdp <- function(y, alpha = 0.05, itertime = 100) {
dm <- dim(y)
n <- dm[1]
p <- dm[2]
h <- round(n/2) + 1
init_h <- 2
delta <- alpha/2
runtime <- proc.time()
id <- replicate( itertime, sample.int(n, 2) ) - 1
final_vec <- as.vector( .Call(“Rfast_rmdp”, PACKAGE = “Rfast”, y, h, id, itertime) )
submcd <- seq(1, n)[final_vec != 0]
mu_t <- Rfast::colmeans(y[submcd, ])
var_t <- Rfast::colVars(y[submcd, ])
sama <- ( t(y) - mu_t )^2 / var_t
disa <- Rfast::colsums(sama)
disa <- disa * p / Rfast::Median(disa)
b <- Rfast::hd.eigen(y[submcd, ], center = TRUE, scale = TRUE)$values
tr2_h <- sum(b^2)
tr2 <- tr2_h - p^2/h
cpn_0 <- 1 + (tr2_h) / p^1.5
w0 <- (disa - p) / sqrt(2 * tr2 * cpn_0) < qnorm(1 - delta)
nw <- sum(w0)
sub <- seq(1, n)[w0]
mu_t <- Rfast::colmeans(y[sub, ])
var_t <- Rfast::colVars(y[sub, ])
sama <- (t(y) - mu_t)^2/var_t
disa <- Rfast::colsums(sama)
b <- Rfast::hd.eigen(y[sub, ], center = TRUE, scale = TRUE)$values
tr2_h <- sum(b^2)
tr2 <- tr2_h - p^2/nw
scal <- 1 + exp( -qnorm(1 - delta)^2/2 ) / (1 - delta) * sqrt(tr2) / p / sqrt(pi)
disa <- disa/scal
cpn_1 <- 1 + (tr2_h) / p^1.5
dis <- (disa - p) / sqrt(2 * tr2 * cpn_1)
wei <- dis < qnorm(1 - alpha)
runtime <- proc.time() - runtime
list(runtime = runtime, dis = dis, wei = wei)
}
-
Translation to C++: The chunk of code in the function RMDP that was transferred to C++ is indicated by five hashtags in the RMDP() function (in rmdp(), this is indicated by the command Call (“Rfast_rmdp”, PACKAGE = “Rfast”, y, h, id, itertime)). This part contains a for and a while loop and it is known that these loops are much faster in C++ than in R. Translation is essential due to the fact that R is an interpreted language; that is, it “interprets” the source code line by line. R translates it into an efficient intermediate representation or object code and immediately executes that. On the contrary, C++ is a compiled language meaning that it is generally translated into a machine language that can be understood directly by the system, making the generated program highly efficient. Further, the execution of the for loop in parallel (in C++) reduces the computational cost by a factor of 2, as seen by the time experiments in the next section.
-
Use of more efficient R functions:
-
The functions used to compute the column-wise means and variances, apply(y, 2, mean) and apply(y, 2, var), are substituted by the functions colmeans() and colVars(), both of which are available in Rfast.
-
The built-in R function median() is substituted with Rfast’s command Median(). These functions are, to the best of our knowledge, the most efficient ones contained in a contributed R package.
-
The chunk of code,
mu_t <- apply( y[submcd, ], 2, mean ) var_t <- apply( y[submcd, ], 2, var ) dist <- apply( ( t( (t(y) - mu_t)/var_t^0.5) )^2, 1, sum ),
was written in a much more efficient manner as follows:mu_t <- Rfast::colmeans(y[sub, ]) var_t <- Rfast::colVars(y[sub, ]) sama <- (t(y) - mu_t)^2/var_t disa <- Rfast::colsums(sama).
This is an example of efficiently written R code. The third line above (object sama) is written in this way due to R’s execution style, which is similar to C++’s execution style. R does not apply row-wise operations using a vector in a matrix, but rather column-wise operations. This means that if one wants to subtract a vector from each row of a matrix, they need to transpose the matrix and subtract the vector from each column. -
We further examine the case of the hd.eigen() function that is used twice in the code. The data must be normalized prior to the application of the eigendecomposition. Thus, the functions colmeans() and colVars() are present within the hd.eigen() function. However, for the case of , their call requires only 0.03 s; hence, no significant computational reduction is gained. Another alternative would be to translate the whole function rmdp() into C++, but we believe that the computational reduction will not be dramatic and perhaps would save only a few seconds in the difficult cases examined in the following sections.
-
-
Linear algebra tricks: Steps 8 and 10 of the high-dimensional MDP algorithm require computation of the trace of the square of the correlation matrix . Notice that this computation occurs twice, one time at every step. It is well known from linear algebra [6] that for a given matrix , its eigendecomposition is given by , where is an orthogonal matrix with p eigenvectors and is a diagonal matrix that contains the p eigenvalues, and also . Further, . This trick allows one to avoid computing the correlation matrix, multiplying it by itself, and then, computing the sum of its diagonal elements.R’s built-in function prcomp() performs the eigendecomposition of a matrix pretty fast and in a memory-efficient manner. The drawback of prcomp() is that it computes p eigenvectors and p eigenvalues when, in fact, most n eigenvalues are non-zero since the rank of the correlation matrix in this case is at most n. Hence, instead of the function prcomp(), the function hd.eigen() available in Rfast is applied. This function is an optimized function for the case of that returns the first n eigenvectors and eigenvalues. Thus, hd.eigen() is not only faster than prcomp() in the high-dimensional settings, but requires less memory. The function takes the original data () and standardizes them (), performing the eigendcomposition of the matrix . The sum of the squared eigenvalues divided by equals .
3. Experiments
4. Application in Bioinformatics
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rousseeuw, P.J.; Hubert, M. Robust statistics for outlier detection. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011, 1, 73–79. [Google Scholar] [CrossRef]
- Omar, S.; Ngadi, A.; Jebur, H.H. Machine Learning Techniques for Anomaly Detection: An Overview. Int. J. Comput. Appl. 2013, 79, 33–41. [Google Scholar] [CrossRef]
- Ro, K.; Zou, C.; Wang, Z.; Yin, G. Outlier detection for high-dimensional data. Biometrika 2015, 102, 589–599. [Google Scholar] [CrossRef]
- Rousseeuw, P.J.; Driessen, K.V. A fast algorithm for the minimum covariance determinant estimator. Technometrics 1999, 41, 212–223. [Google Scholar] [CrossRef]
- Papadakis, M.; Tsagris, M.; Dimitriadis, M.; Fafalios, S.; Tsamardinos, I.; Fasiolo, M.; Borboudakis, G.; Burkardt, J.; Zou, C.; Lakiotaki, K.; et al. Rfast: A Collection of Efficient and Extremely Fast R Functions; R Package Version 2.1.0; 2023. Available online: https://cloud.r-project.org/web/packages/Rfast/Rfast.pdf (accessed on 3 September 2024).
- Strang, G. Introduction to Linear Algebra, 6th ed.; Wellesley-Cambridge Press: Wellesley, MA, USA, 2023. [Google Scholar]
- Lakiotaki, K.; Vorniotakis, N.; Tsagris, M.; Georgakopoulos, G.; Tsamardinos, I. BioDataome: A collection of uniformly preprocessed and automatically annotated datasets for data-driven biology. Database 2018, 2018, bay011. [Google Scholar] [CrossRef] [PubMed]
- Fayomi, A.; Pantazis, Y.; Tsagris, M.; Wood, A.T. Cauchy robust principal component analysis with applications to high-dimensional data sets. Stat. Comput. 2024, 34, 26. [Google Scholar] [CrossRef]
| Dimensionality | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Function | p = 100 | p = 200 | p = 500 | p = 1000 | p = 2000 | p = 5000 | p = 10,000 | p = 20,000 | |
| n = 50 | RMDP | 8.550 | 16.074 | 37.604 | 68.324 | 128.710 | 645.438 | 3114.512 | 20,738.400 |
| rmdp | 0.104 | 0.208 | 0.416 | 0.696 | 1.138 | 5.818 | 20.996 | 22.260 | |
| rmdp (parallel) | 0.014 | 0.028 | 0.058 | 0.284 | 0.668 | 2.578 | 5.498 | 11.384 | |
| Ratio | 82.212 | 77.279 | 90.394 | 98.167 | 113.102 | 110.938 | 148.338 | 931.644 | |
| Ratio (parallel) | 610.714 | 574.071 | 648.345 | 240.578 | 192.680 | 250.364 | 566.481 | 1821.715 | |
| n = 100 | RMDP | 23.283 | 59.890 | 106.085 | 260.060 | 707.158 | 3068.273 | 25,523.890 | |
| rmdp | 0.500 | 1.163 | 2.163 | 9.053 | 13.542 | 21.240 | 43.950 | ||
| rmdp (parallel) | 0.076 | 0.430 | 1.106 | 3.216 | 6.996 | 11.848 | 23.578 | ||
| Ratio | 46.565 | 51.518 | 49.057 | 28.728 | 52.220 | 144.457 | 580.748 | ||
| Ratio (parallel) | 306.355 | 139.279 | 95.917 | 80.864 | 101.080 | 258.970 | 1082.53 | ||
| n = 500 | RMDP | 178.020 | 359.840 | 1199.200 | 3747.340 | 26,115.310 | |||
| rmdp | 22.220 | 40.660 | 106.230 | 177.460 | 470.520 | ||||
| rmdp (parallel) | 10.138 | 19.450 | 51.780 | 116.834 | 224.634 | ||||
| Ratio | 8.012 | 8.850 | 11.289 | 21.117 | 55.503 | ||||
| Ratio (parallel) | 17.560 | 18.500 | 23.160 | 32.074 | 116.257 | ||||
| Dimensionality | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Function | p = 100 | p = 200 | p = 500 | p = 1000 | p = 2000 | p = 5000 | p = 10,000 | p = 20,000 | |
| n = 50 | RMDP | 23.087 | 38.887 | 81.663 | 153.907 | 270.630 | 719.727 | 4354.165 | 22,314.542 |
| rmdp | 0.337 | 0.430 | 0.887 | 1.653 | 3.463 | 7.617 | 25.700 | 46.151 | |
| rmdp (parallel) | 0.038 | 0.046 | 0.110 | 0.636 | 1.336 | 5.624 | 11.238 | 23.768 | |
| Ratio | 68.574 | 90.434 | 92.102 | 93.089 | 78.141 | 94.494 | 169.423 | 483.522 | |
| Ratio (parallel) | 607.552 | 845.370 | 742.391 | 241.992 | 202.567 | 127.974 | 387.450 | 938.848 | |
| n = 100 | RMDP | 40.023 | 83.737 | 162.663 | 322.120 | 963.857 | 3825.913 | 20,216.713 | |
| rmdp | 0.820 | 1.610 | 3.050 | 11.810 | 26.453 | 44.380 | 81.181 | ||
| rmdp (parallel) | 0.130 | 0.818 | 1.996 | 6.568 | 15.062 | 36.194 | 54.246 | ||
| Ratio | 48.809 | 52.010 | 53.332 | 27.275 | 36.436 | 86.208 | 249.159 | ||
| Ratio (parallel) | 307.869 | 102.368 | 81.495 | 49.044 | 63.993 | 105.706 | 372.686 | ||
| n = 500 | RMDP | 737.460 | 1337.820 | 2844.760 | 6902.070 | 27,725.263 | |||
| rmdp | 86.610 | 120.250 | 212.600 | 444.250 | 801.275 | ||||
| rmdp (parallel) | 24.426 | 42.734 | 102.318 | 219.148 | 410.798 | ||||
| Ratio | 8.515 | 11.125 | 13.381 | 15.536 | 34.602 | ||||
| Ratio (parallel) | 30.192 | 31.306 | 27.803 | 31.495 | 67.491 | ||||
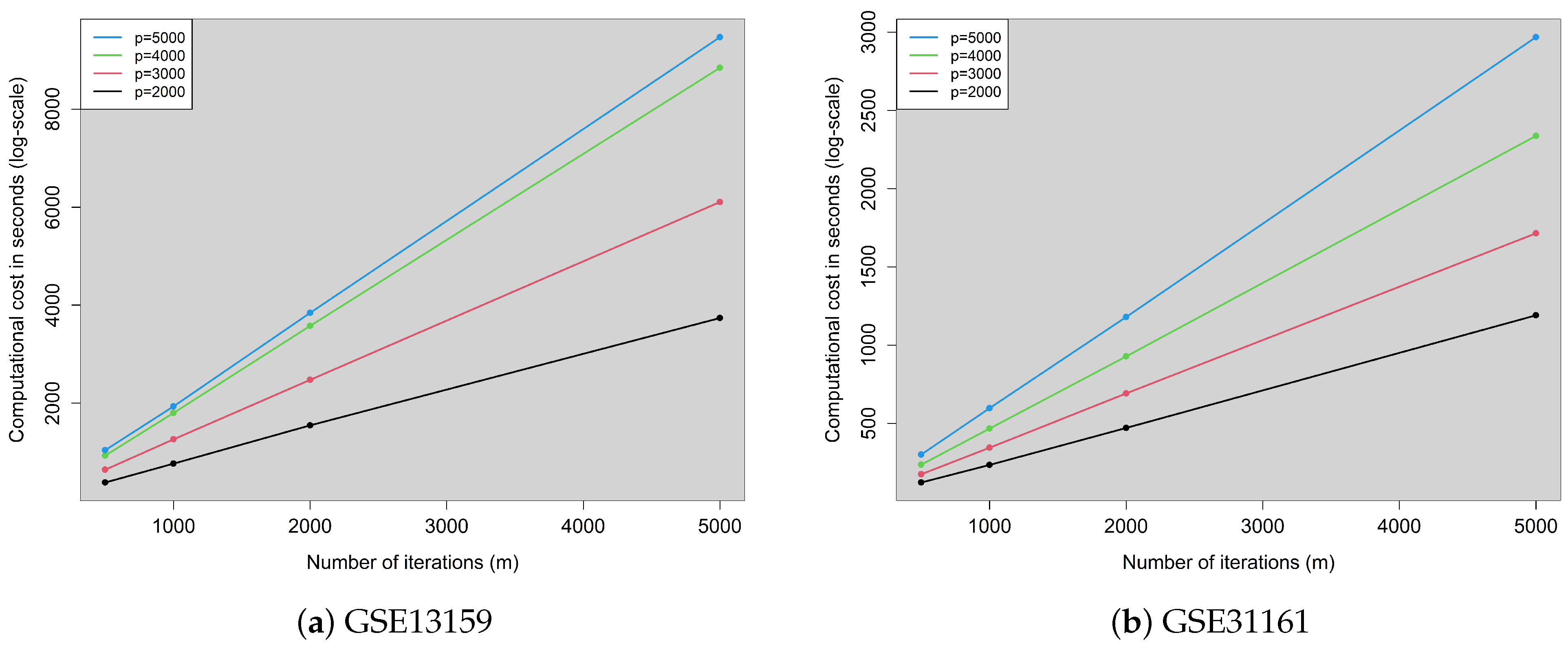
| GSE13159 | ||||||||
| m = 500 | m = 1000 | m = 2000 | m = 5000 | m = 500 | m = 1000 | m = 2000 | m = 5000 | |
| p = 2000 | 190.04 | 382.91 | 772.72 | 1869.28 | 99.71% | 99.89% | 99.79% | 99.89% |
| p = 3000 | 321.00 | 630.74 | 1237.70 | 3053.17 | 99.93% | 99.80% | 99.90% | 99.79% |
| p = 4000 | 463.85 | 899.88 | 1788.67 | 4423.02 | 99.95% | 99.98% | 100.00% | 99.96% |
| p = 5000 | 520.19 | 967.27 | 1921.61 | 4735.42 | 99.98% | 100.00% | 100.00% | 99.96% |
| GSE13159 | ||||||||
| p = 2000 | 60.77 | 116.79 | 235.03 | 594.82 | 99.14% | 99.49% | 98.97% | 99.33% |
| p = 3000 | 87.00 | 172.05 | 345.24 | 856.76 | 99.02% | 98.72% | 98.46% | 98.45% |
| p = 4000 | 117.79 | 232.86 | 463.42 | 1168.23 | 99.17% | 99.23% | 99.28% | 99.14% |
| p = 5000 | 149.99 | 298.34 | 589.61 | 1483.72 | 97.92% | 98.85% | 98.65% | 98.60% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsagris, M.; Papadakis, M.; Alenazi, A.; Alzeley, O. Computationally Efficient Outlier Detection for High-Dimensional Data Using the MDP Algorithm. Computation 2024, 12, 185. https://doi.org/10.3390/computation12090185
Tsagris M, Papadakis M, Alenazi A, Alzeley O. Computationally Efficient Outlier Detection for High-Dimensional Data Using the MDP Algorithm. Computation. 2024; 12(9):185. https://doi.org/10.3390/computation12090185
Chicago/Turabian StyleTsagris, Michail, Manos Papadakis, Abdulaziz Alenazi, and Omar Alzeley. 2024. "Computationally Efficient Outlier Detection for High-Dimensional Data Using the MDP Algorithm" Computation 12, no. 9: 185. https://doi.org/10.3390/computation12090185
APA StyleTsagris, M., Papadakis, M., Alenazi, A., & Alzeley, O. (2024). Computationally Efficient Outlier Detection for High-Dimensional Data Using the MDP Algorithm. Computation, 12(9), 185. https://doi.org/10.3390/computation12090185






