Abstract
Biological variance among samples across different cohorts can pose challenges for the long-term validation of developed models. Data-driven normalization methods offer promising tools for mitigating inter-sample biological variance. We applied seven data-driven normalization methods to quantitative metabolome data extracted from rat dried blood spots in the context of the Rice–Vannucci model of hypoxic–ischemic encephalopathy (HIE) in rats. The quality of normalization was assessed through the performance of Orthogonal Partial Least Squares (OPLS) models built on the training datasets; the sensitivity and specificity of these models were calculated by application to validation datasets. PQN, MRN, and VSN demonstrated a higher diagnostic quality of OPLS models than the other methods studied. The OPLS model based on VSN demonstrated superior performance (86% sensitivity and 77% specificity). After VSN, the VIP-identified potential biomarkers notably diverged from those identified using other normalization methods. Glycine consistently emerged as the top marker in six out of seven models, aligning perfectly with our prior research findings. Likewise, alanine exhibited a similar pattern. Notably, VSN uniquely highlighted pathways related to the oxidation of brain fatty acids and purine metabolism. Our findings underscore the widespread utility of VSN in metabolomics, suggesting its potential for use in large-scale and cross-study investigations.
1. Introduction
The biological condition of an organism profoundly impacts the qualitative and quantitative molecular profiles of its tissues and fluids, thereby facilitating the development of personalized medicine and molecular-profile-based diagnostics [1]. However, factors such as external conditions, variations in age–gender compositions, and the relatively small size of cohorts (often just a few dozen samples) can lead to experimental-condition-associated molecular profile variances that overshadow those attributed to individual subjects [2,3,4]. Considering that biological variances are distinct from technical variances associated with sample preparation and analysis conditions, traditional normalization methods utilizing internal standards or quality control samples may prove ineffective. Instead, data-driven approaches such as median/mean normalization, scaling methods, quantile normalization [5], probabilistic quotient normalization (PQN) [6], variance stabilizing normalization (VSN) [7], median ratio normalization (MRN) [8], and trimmed mean m-value normalization (TMM) [9] offer viable alternatives. Based on recent comparative studies conducted by Brix F. et al. [10] and Chua A.E. et al. [11], it is evident that PQN, VSN, and quantile normalization are three commonly employed methods in the field of metabolomics [10,11] . These normalization techniques have been extensively utilized in various research studies to enhance the accuracy and reliability of metabolomic data analysis.
In this study, we evaluate the effectiveness of these normalization methods in correcting validation dataset discrepancies at the level of training datasets, focusing on the molecular markers of the rat hypoxic–ischemic encephalopathy (HIE) model [12]. This study presents several strengths. First, a controlled and easily manipulable model was utilized to induce hypoxic–ischemic injury, aiming to simplify the study of HIE. This model, proposed by Rice J.E. et al. in 1981, remains the most commonly used pre-clinical model for investigating HIE [13]. Over the last four decades, the Rice–Vannucci model has been employed in various laboratories globally. Nonetheless, it exhibits significant inter-subject variability in terms of the severity of brain damage [14]. Second, the study involved conducting three sequential experiments, where each experiment consisted of both a control group and an HIE group. Furthermore, the inclusion of a standard neonatal screening platform, which involved the quantitative analysis of 57 metabolites in blood spots, enhances the translational potential of this research in neonatology. This approach not only brings the study closer to real-world clinical application but also reduces potential technical variability, specifically that stemming from inter-batch effects.
2. Materials and Methods
The dataset utilized in our study was obtained from Shevtsova et al.’s research [12] which comprised three experimental models. From these models, we selected samples from two experiments: “HIE with sampling at different times”, which consisted of intact rats (n = 10), rats with HIE, and sampling at 6 h post-hypoxia (n = 12) as a train dataset; and “Modeling of therapeutic hypothermia”, comprising intact rats (n = 13) and rats with HIE undergoing recovery for 6 h at 37 °C (n = 14) as a test dataset.
The following methods were used for normalization:
- Normalization by total concentration:
Concentrations from the training set were transformed using the following formula:
where represents the original concentration of compound i in sample j and represents the normalized concentration of compound i in sample j.
Concentrations from test set were transformed using the following formula:
where represents the original concentration of compound i in sample j in the test set, is the normalized concentration of compound i in sample j in the test set, and is the normalized concentration of compound i in sample t in the training set.
- 2.
- Autoscaling normalization:
Concentrations from the test dataset were transformed using the following formula:
where represents the original concentration of compound i in sample j of the test set, signifies the normalized concentration of compound i in sample j of the test set, indicates the concentration of compound i in sample t of the training set, and and denote the mean and standard deviation of the concentration of compound i in the test set or training set, respectively.
- 3.
- Quantile normalization:
Quantile normalization is achieved by rearranging and transforming the distribution of values for each species in the samples to match the same distribution across all species [15]. The training dataset underwent normalization using its intrinsic values as the reference for standardization. The normalization process for the test dataset involved a step-by-step procedure. Initially, each sample from the non-normalized test dataset was precisely incorporated into the normalized training dataset. Subsequently, a fresh round of quantile normalization was executed on this merged dataset, which contained both the original test sample and the normalized training dataset. Lastly, the resulting normalized test samples were aggregated to construct the final normalized test dataset.
- 4.
- Probabilistic quotient normalization:
The PQN method hinges on deriving a correction factor, which is uniformly applied to all species within a sample. This correction factor is computed by evaluating the median relative signal intensity of the normalized sample in relation to the signal intensity present in a reference sample or pseudo-sample. This reference sample can either be a quality control sample or the mean/median values of intensity across all samples [6]. The training dataset underwent normalization using median concentration values as the reference. Next, the test dataset was normalized by iteratively adding each sample from the non-normalized test dataset to the normalized training dataset. For each addition, a new PQN was performed with median values as the reference for the newly created dataset (the non-normalized test sample combined with the normalized training dataset). Finally, the normalized test samples were aggregated to construct the normalized test dataset.
- 5.
- Median ratio normalization:
The MRN method shares similarities with PQN, although it distinguishes itself by employing geometric averages of sample concentrations as the reference values for normalization [8]. The training dataset underwent normalization using its own values as the reference point for standardization. Next, the test dataset was normalized by iteratively adding each sample from the non-normalized test dataset to the normalized training dataset. A new median ratio normalization was performed on the combined dataset, containing both the original test sample and the normalized training dataset. Finally, the normalized test samples were aggregated to construct the normalized test dataset.
- 6.
- Trimmed median-m value normalization:
The TMM method involves the utilization of a correction factor, the influence of which is determined by its relative contribution to the overall signal intensity [9,16]. The training dataset underwent normalization using its own values as the reference point for standardization. Next, the test dataset was normalized by iteratively adding each sample from the non-normalized test dataset to the normalized training dataset. A subsequent TMM normalization process was applied to the combined dataset, containing the original test sample with the normalized training data. Finally, the normalized test samples were aggregated to construct the normalized test dataset.
- 7.
- Variance stabilizing normalization.
This approach hinges on determining optimal parameters for glog transformation that effectively reduce signal intensity variation relative to the mean signal intensity. When applied to test samples, these parameters are derived from the pre-existing variation values computed from the training dataset [7,17].
Each normalized training dataset served as the foundation for constructing an Orthogonal Partial Least Squares (OPLS) model, executed through the ropls package [18]) with dependent values (HIE or healthy) acting as pivotal factors. The efficacy of these models was assessed based on the explained variance (R2Y) and predicted variance (Q2Y), alongside sensitivity and specificity metrics derived from validation models applied to the normalized test datasets (Refer to Figure S1 Supplementary). Following this evaluation, blood metabolites exhibiting a variable importance projection (VIP) exceeding 1 were evaluated as potential markers for HIE, with associated metabolite pathways delineated in alignment with prior research [12].
Moreover, the results of principal component analysis (PCA) conducted on raw data sourced from control samples across three distinct experiments [12] were compared with VSN-normalized control samples’ data.
In the context of this study, R 4.3.2 scripts were leveraged within RStudio to execute various normalization techniques. Quantile normalization was administered through the preprocessCore package (https://github.com/bmbolstad/preprocessCore), PQN was implemented using the Rcpm package, MRN was executed utilizing the Ebseq package (https://bioconductor.org/packages/release/bioc/html/EBSeq.html), TMM normalization was performed using the edgeR package [19], and lastly, VSN was executed through the vsn2 package [7].
3. Results
OPLS models constructed using non-normalized data, as well as datasets normalized through total sum of concentrations normalization, probabilistic quotient normalization (PQN), median ratio normalization (MRN), and trimmed mean of m-values (TMM), yielded models of sufficient quality for application in biological studies (with R2Y > 0.5 and Q2Y > 0.4). Notably, the model based on variance stabilizing normalization (VSN) exhibited the highest proportion of prediction-dependent variables and the lowest proportion of described independent variables (Table 1).

Table 1.
The characteristics and diagnostic power of OPLS models constructed using different normalization methods.
Normalization greatly influences the variable importance projection (VIP) distribution. The VIP distribution from the non-normalized dataset-based OPLS model significantly differed from the VIP distributions of the normalized datasets (Figure 1a,b). Total sum normalization, TMM normalization, PQN, and MRN resulted in very similar VIP distributions, with almost identical pairs of distributions between total sum normalization and TMM normalization, and between PQN and MRN (Figure 1a,b). The VIP-determined potential biomarkers after VSN differed dramatically from the other normalization methods (Figure 1a,b).
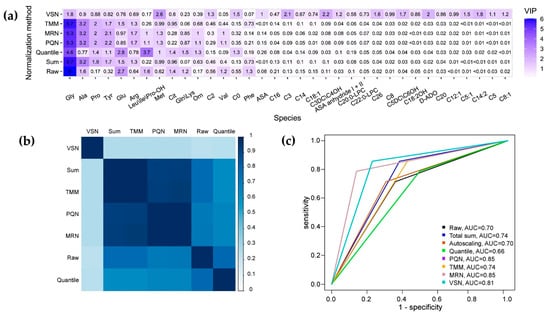
Figure 1.
(a) A heatmap depicting the distribution of VIP from OPLS models based on each normalized dataset. (b) A plot illustrating the similarity in the variable importance projection (VIP) distributions of OPLS models based on different normalization methods. The score of similarity was calculated using the Tanimoto coefficient. (c) The receiver operation curves (ROC), obtained by validation of OPLS models based on normalized datasets.
The total sum of concentrations, PQN, MRN, and TMM-normalized test datasets demonstrated higher accuracy compared to the non-normalized datasets. Furthermore, the PQN and MRN normalized test datasets exhibited the highest accuracy, while the VSN normalized dataset showed the highest sensitivity coupled with good specificity, as depicted in Figure 1c and Table 1.
The combination of OPLS model parameters and the quality of the test set validation indicates that VSN is the most promising method for reducing intra-study diagnostic variability. After VSN, fewer variables contribute to the discriminative components PC1 and PC2. In the non-normalized dataset, PC1 described 63% of the variables and PC2 described 21% (Figure S2a), while in the VSN-normalized dataset, PC1 described 36% and PC2 described 18% (Figure S2c). Additionally, VSN resulted in a more balanced loading of features onto the principal components (Figure S2b,d).
Alanine, arginine, and proline metabolism is enriched across all normalization methods, as depicted in Figure S3 and detailed in Table S1. Notably, total sum normalization, TMM normalization, PQN, and MRN resulted in identical sets of statistically significant enriched pathways (p < 0.1). Moreover, VSN highlighted the oxidation of brain fatty acids and purine metabolism pathways as unique findings, underscoring its distinctive contribution. On the other hand, quantile normalization specifically pinpointed the spermine and spermidine biosynthesis pathway.
4. Discussion
VSN, PQN, and MRN emerge as the most effective methods for reducing intra-study variation. However, OPLS models based on VSN-normalized datasets exhibit exceptionally high R2Y and Q2Y values and an extremely low R2X value. Furthermore, VIP distributions from the PQN and MRN normalized datasets differ significantly from those of the VSN-normalized dataset’s OPLS model. The PQN and MRN methods also yield identical R2X, R2Y, and Q2Y parameters for OPLS models. This similarity may be due to the inherent similarities in the principles of PQN and MRN—they both involve calculating correction factors. However, PQN uses mean or median values, while MRN utilizes the average geometric value of feature intensity or concentration within the cohort [6,8]. On the other hand, VSN operates on the principle of glog transformation, yielding different results.
Total sum normalization and TMM normalization also yield similar parameters and quality in OPLS models. These methods leverage information about total signal or concentration, with total sum normalization utilizing the absolute value of the sum in transformation, while TMM employs information about the ratio of each feature to the total sum [9]. TMM is primarily employed in genomics studies, and there is limited information regarding its use in metabolomic studies.
Quantile normalization of the dataset yields the closest VIP distribution in the OPLS model to the one based on raw data, yet the quality and diagnostic potential decrease after normalization (Table 2). O’Connell et al. demonstrated that quantile normalization, MRN, and TMM have the potential to obscure the biological structure of samples [20]. However, in our study, MRN yielded better results than quantile normalization, TMM normalization, or using non-normalized data. Similarly, in Abbas-Aghababazadeh F. et al.’s (2018) study, MRN improved the differentiation of RNA sequences from various tomato phenotypes compared to non-normalized, TMM-normalized, and quantile-normalized datasets [21].

Table 2.
Summary of results from various normalization methods applied.
VSN, quantile normalization, PQN, and autoscaling were employed in Cook T. et al.’s study for metabolome profile normalization but did not significantly improve sample differentiation between cancer patients (prostate and bladder cancer) and control group patients. This may be attributed to the low number of species analyzed (four species) [22]. Studies by Dressler F.F. et al. and Narasimhan M. et al. indicate that the comparative effectiveness of the VSN and quantile normalization methods depends on the original datasets that were normalized [23,24]. The dataset used for training in our study possessed some imbalance, with 10 samples from the control group and 12 samples from the HIE group. This imbalance may have influenced the normalization process, potentially causing the normalization levels to lean closer to those of the HIE samples rather than the control samples. This imbalance could contribute to the poor specificity observed in validation based on total-sum-normalized, quantile-normalized, and TMM-normalized datasets (Table 2).
Glycine emerges as the predominant marker in six out of seven models and retains significance in the VSN-normalized dataset-based model. This observation aligns with our prior research, which highlighted the correlation between glycine and hypoxic conditions as well as brain ischemia [12]. Similarly, alanine stands out as a crucial marker in five of the seven models, with the associated alanine metabolism pathway consistently represented across all analyses. Notably, the presence of alanine aminotransferase within this pathway underscores its significance as a marker for HIE. Furthermore, the arginine and proline metabolism pathway consistently demonstrates statistically significant enrichment across all normalization methods, signaling its potential relevance to hypoxic injury as indicated in previous studies [25,26]. While the oxidation of branched-chain fatty acids and purine metabolism pathways emerge as uniquely enriched pathways in the VSN-normalized dataset, it is worth noting that oxidative stress has been implicated in hypoxic conditions [27]. Several studies suggest a potential link between purine metabolism and hypoxia, further emphasizing the sophisticated connections that exist between blood metabolites and responses to hypoxic conditions [28,29].
Overall, the comprehensive exploration of these diverse normalization strategies unveiled specific metabolic pathways, shedding light on the interplay between normalization techniques and the identification of enriched pathways in the context of metabolite analysis.
5. Limitations, Future Prospects, and Suggestions
This study is subject to certain limitations that warrant consideration. Firstly, it focused solely on a single task (HIE/health), utilizing only two distinct datasets for analysis. This narrow scope may restrict the generalizability of the findings and the applicability of the normalization methods across diverse biological contexts. Furthermore, the balance of “case/control” samples is a critical factor in ensuring the representativeness and reliability of the data. It is imperative to acknowledge the potential impact of sample proportion imbalance on the quality of normalization procedures. Addressing these limitations and conducting further studies to evaluate the effects of sample proportions on normalization quality are crucial steps toward enhancing the robustness and reliability of data normalization practices in routine research and clinical applications.
Determining the best method for a particular type of biological sample can be a complex process that requires careful consideration of various factors that can influence the experimental outcomes. When it comes to normalization methods in biological sample analysis, several key factors should be taken into account to reach a reliable conclusion: sample features, analytical technique, biological variability, infrastructure and resources, and statistical considerations. In particular, the statistical assumptions underlying different normalization methods should be carefully evaluated. Some methods may introduce bias or distort the data if they are applied incorrectly or if the data do not meet the method’s assumptions. To come to a conclusion about the best method for a particular type of biological sample, it is essential to conduct a thorough evaluation of these factors and perform a meta-analysis to assess the performance of different normalization approaches. Ultimately, the choice of normalization method should be driven by the specific characteristics of the samples and the research objectives to ensure accurate and reliable results.
6. Conclusions
Evaluating the quality of normalization methods in biological sample analysis involves considering several key points. Firstly, the quality of the created model is crucial, with VSN showing promise by generating the best model based on OPLS-model metrics. Secondly, the robustness of the model across datasets is essential for reliability, and methods such as VSN, PQN, and MRN demonstrate the ability to create reliable models across different datasets. Finally, the adequacy in capturing changes in biological marker patterns is a significant factor to consider. VSN has been shown to significantly alter the distribution of biomarker importance, potentially revealing enriched pathways associated with specific conditions like HIE.
In comparison, the PQN and MRN methods maintain a closer alignment in biomarker distribution between themselves and the raw distribution, highlighting their consistency in handling biomarker patterns. In conclusion, VSN emerges as an attractive choice for routine use in reducing between-study biological variation due to its ability to create robust models, significant impact on biomarker importance distribution, and potential for uncovering relevant biological pathways associated with specific conditions like HIE.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/computation12070137/s1. Figure S1: The schematic pipeline illustrates the normalization of training and test datasets, the creation of OPLS models, and their validation; Figure S2: (a) Principal component analysis plot showcasing raw control samples in the principal component space; (b) Loading plot of principal component analysis depicting non-normalized features; (c) Principal component analysis plot illustrating VSN-normalized control samples in the principal component space; (d) Principal component analysis plot based on VSN-normalized features; Figure S3: Barplot showing the mean enrichment of pathways associated with potential biomarkers of HIE; Table S1. Statistically significant enriched pathways observed with various normalization methods.
Author Contributions
Conceptualization, D.S. and N.S.; methodology, A.T.; software, A.T.; validation, N.S. and V.F.; formal analysis, A.T. and D.S.; investigation, A.T. and N.S.; resources, V.F. and D.S.; data curation, V.F. and D.S.; writing—original draft preparation, A.T. and N.S.; writing—review and editing, V.F. and D.S.; visualization, N.S. and A.T.; supervision, V.F. and D.S.; project administration, V.F. and D.S.; funding acquisition, D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation (No. 22-15-00454).
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
List of Used Abbreviations
| HIE—hypoxic–ischemic encephalopathy |
| MRN—median ration normalization |
| OPLS—orthogonal partial least squares |
| PCA—principal component analysis |
| PQN—probabilistic quotient normalization |
| TMM—trimmed mean m-value normalization |
| VSN—variance stabilizing normalization |
| VIP—variable importance projection |
References
- Badrick, T. Biological variation: Understanding why it is so important? Pract. Lab. Med. 2021, 23, e00199. [Google Scholar] [CrossRef] [PubMed]
- Higdon, R.; Kolker, E. Can “normal” protein expression ranges be estimated with high-throughput proteomics? J. Proteome Res. 2015, 14, 2398–2407. [Google Scholar] [CrossRef] [PubMed]
- Chelala, L.; O’Connor, E.E.; Barker, P.B.; Zeffiro, T.A. Meta-analysis of brain metabolite differences in HIV infection. NeuroImage Clin. 2020, 28, 102436. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Siegel, L.; Zhou, J.; Zhu, M.; Tong, T.; Chen, Y.; Chu, H. Estimating the reference interval from a fixed effects meta-analysis. Res. Synth. Methods 2021, 12, 630–640. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.; Lim, M.-S.; Seong, S.J.; Seo, J.J.; Park, S.M.; Lee, H.W.; Yoon, Y.-R. Quantile normalization approach for liquid chromatography—Mass spectrometry-based metabolomic data from healthy human volunteers. Anal. Sci. 2012, 28, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Huber, W.; Von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Brix, F.; Demetrowitsch, T.; Jensen-Kroll, J.; Zacharias, H.U.; Szymczak, S.; Laudes, M.; Schreiber, S.; Schwarz, K. Evaluating the Effect of Data Merging and Postacquisition Normalization on Statistical Analysis of Untargeted High-Resolution Mass Spectrometry Based Urinary Metabolomics Data. Anal. Chem. 2024, 96, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Chua, A.E.; Pfeifer, L.D.; Sekera, E.R.; Hummon, A.B.; Desaire, H. Workflow for Evaluating Normalization Tools for Omics Data Using Supervised and Unsupervised Machine Learning. J. Am. Soc. Mass Spectrom. 2023, 34, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Shevtsova, Y.; Starodubtseva, N.; Tokareva, A.; Goryunov, K.; Sadekova, A.; Vedikhina, I.; Ivanetz, T.; Ionov, O.; Frankevich, V.; Plotnikov, E.; et al. Metabolite Biomarkers for Early Ischemic–Hypoxic Encephalopathy: An Experimental Study Using the NeoBase 2 MSMS Kit in a Rat Model. Int. J. Mol. Sci. 2024, 25, 2035. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.E.; Vannucci, R.C.; Brierley, J.B. The influence of immaturity on hypoxic-ischemic brain damage in the rat. Ann. Neurol. 1981, 9, 131–141. [Google Scholar] [CrossRef]
- Edwards, A.B.; Feindel, K.W.; Cross, J.L.; Anderton, R.S.; Clark, V.W.; Knuckey, N.W.; Meloni, B.P. Modification to the Rice-Vannucci perinatal hypoxic-ischaemic encephalopathy model in the P7 rat improves the reliability of cerebral infarct development after 48 hours. J. Neurosci. Methods 2017, 288, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Bolstad, B.M.; Irizarry, R.A.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.; Hardin, J.; Stoebel, D.M. Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions. Brief. Bioinform. 2018, 19, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Huber, W.; von Heydebreck, A.; Sueltmann, H.; Poustka, A.; Vingron, M. Parameter estimation for the calibration and variance stabilization of microarray data. Stat. Appl. Genet. Mol. Biol. 2003, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, G.C. Variability in donor leukocyte counts confound the use of common RNA sequencing data normalization strategies in transcriptomic biomarker studies performed with whole blood. Sci. Rep. 2023, 13, 15514. [Google Scholar] [CrossRef] [PubMed]
- Abbas-Aghababazadeh, F.; Li, Q.; Fridley, B.L. Comparison of normalization approaches for gene expression studies completed with highthroughput sequencing. PLoS ONE 2018, 13, e0206312. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.; Ma, Y.; Gamagedara, S. Evaluation of statistical techniques to normalize mass spectrometry-based urinary metabolomics data. J. Pharm. Biomed. Anal. 2020, 177, 112854. [Google Scholar] [CrossRef] [PubMed]
- Dressler, F.F.; Brägelmann, J.; Reischl, M.; Perner, S. Normics: Proteomic Normalization by Variance and Data-Inherent Correlation Structure. Mol. Cell. Proteom. 2022, 21, 100269. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, M.; Kannan, S.; Chawade, A.; Bhattacharjee, A.; Govekar, R. Clinical biomarker discovery by SWATH-MS based label-free quantitative proteomics: Impact of criteria for identification of differentiators and data normalization method. J. Transl. Med. 2019, 17, 184. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Wu, D.; Zhang, J.; Pan, Y.; Kan, R.; Gao, J.; Zhou, B. Protective effect and mechanism of procyanidin B2 against hypoxic injury of cardiomyocytes. Heliyon 2023, 9, e21309. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, D.; Chen, D.; Sun, Y.; Feng, X.; Shi, X.; Xu, Y.; Luo, X.; Yu, J.; Li, Y.; et al. Characterizing the effects of hypoxia on the metabolic profiles of mesenchymal stromal cells derived from three tissue sources using chemical isotope labeling liquid chromatography-mass spectrometry. Cell Tissue Res. 2020, 380, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Zhu, P.; Fujino, M.; Zhuang, J.; Guo, H.; Sheikh, I.; Zhao, L.; Li, X.-K. Oxidative stress in hypoxic-ischemic encephalopathy: Molecular mechanisms and therapeutic strategies. Int. J. Mol. Sci. 2016, 17, 2078. [Google Scholar] [CrossRef] [PubMed]
- Denihan, N.M.; Kirwan, J.A.; Walsh, B.H.; Dunn, W.B.; Broadhurst, D.I.; Boylan, G.B.; Murray, D.M. Untargeted metabolomic analysis and pathway discovery in perinatal asphyxia and hypoxic-ischaemic encephalopathy. J. Cereb. Blood Flow Metab. 2019, 39, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Kuligowski, J.; Solberg, R.; Sánchez-Illana, Á.; Pankratov, L.; Parra-Llorca, A.; Quintás, G.; Saugstad, O.D.; Vento, M. Plasma metabolite score correlates with Hypoxia time in a newly born piglet model for asphyxia. Redox Biol. 2017, 12, 1–7. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).