Abstract
The DNA virus responsible for monkeypox, transmitted from animals to humans, exhibits two distinct genetic lineages in central and eastern Africa. Beyond the zoonotic transmission involving direct contact with the infected animals’ bodily fluids and blood, the spread of monkeypox can also occur through skin lesions and respiratory secretions among humans. Both monkeypox and chickenpox involve skin lesions and can also be transmitted through respiratory secretions, but they are caused by different viruses. The key difference is that monkeypox is caused by an orthopox-virus, while chickenpox is caused by the varicella-zoster virus. In this study, the utilization of a patch-based vision transformer (ViT) model for the identification of monkeypox and chickenpox disease from human skin color images marks a significant advancement in medical diagnostics. Employing a transfer learning approach, the research investigates the ViT model’s capability to discern subtle patterns which are indicative of monkeypox and chickenpox. The dataset was enriched through carefully selected image augmentation techniques, enhancing the model’s ability to generalize across diverse scenarios. During the evaluation phase, the patch-based ViT model demonstrated substantial proficiency, achieving an accuracy, precision, recall, and F1 rating of 93%. This positive outcome underscores the practicality of employing sophisticated deep learning architectures, specifically vision transformers, in the realm of medical image analysis. Through the integration of transfer learning and image augmentation, not only is the model’s responsiveness to monkeypox- and chickenpox-related features enhanced, but concerns regarding data scarcity are also effectively addressed. The model outperformed the state-of-the-art studies and the CNN-based pre-trained models in terms of accuracy.
1. Introduction
After the third wave of COVID-19, which started in January 2022, the condition of the pandemic got progressively less severe in the first half of the year 2022. Sadly, a new threat appeared in just a few weeks and quickly spread around the world, with the risk of becoming a pandemic. This sickness, called human monkeypox, although not a new one [1], was first found in 1970, and over the next ten years, more and more cases were found. Notably, this is not the first time that human monkeypox has spread. The 2003 Midwest monkeypox outbreak and the 2017–2019 Nigeria monkeypox outbreak are evidence of this [2]. There have also been rare cases of the disease in places like the UK, Singapore, and different parts of the US [3]. On the other hand, over the past nine months, the 2022 monkeypox outbreak has spread to more than 100 countries and regions [4]. Although this virus is comparatively less contagious due to its mode of transmission [5], the imperative for the development of a cost-effective and expeditious detection system remains paramount, given its continued spread. Understanding the genetic diversity and transmission patterns of the monkeypox virus is crucial for public health efforts, outbreak control, and vaccine development.
- Genetic Diversity: The monkeypox virus has genetic variety, similarly to other viruses. Mutations that occur during viral replication and recombination activities give rise to this variety. Various strains of MPXV with different genetic compositions have been identified through genomic investigations. These variations may have an effect on host range, transmissibility, and pathogenicity. Through genome sequencing and analysis, researchers are able to follow the evolution of the virus, gaining insight into its epidemiology.
- Transmission Patterns: Non-human primates, especially African rodents, are the main reservoir hosts for monkeypox infections. Direct contact with diseased animals or their body fluids, as well as contact with contaminated objects or surfaces, can result in human diseases. Although it happens less frequently, human-to-human transmission can happen when skin lesions or respiratory droplets come into contact with one another. Human behavior, healthcare practices, vaccine coverage, and population density are some of the factors that affect the spread of the disease.
- Globalization and Travel: Globalization and increased travel facilitate the spread of infectious diseases, including monkeypox. The importation of infected animals or humans can introduce the virus to new regions. Surveillance systems at ports of entry help detect and contain imported cases, preventing local transmission.
Belonging to the Poxyviridae family [6], this virus finds its natural hosts among mammalian species, including squirrels, rats, and various primates. The disease caused by this virus exhibits an infectious course, lasting from two to four weeks, typically manifesting its initial symptoms approximately five to twenty-one days after exposure. As of now, the known symptoms include fever, muscle and joint pain, chills, swollen lymph nodes, and the appearance of blistering spots. [7]. These rashes usually show up in three days, primarily appearing on the face, hands, and bottoms of the feet. There is also potential for these rashes to extend to other areas, such as the mouth, eyes, and genital region. Subsequently, the disease progresses to a phase characterized by skin eruptions, which evolve through four distinct stages. At first, lesions have flat bases and are called macules. Later, they get raised, harden, and are then called papules. After that, these papules fill with pus and turn into pustules, which then turn into solid crusts [8].
The duration of monkeypox symptoms typically spans a period of 2 to 4 weeks, and it is noteworthy that severe cases can manifest. A study from the World Health Organization (WHO) says that the latest case fatality rate is somewhere between 3% and 6%. Monkeypox usually takes between 6 and 13 days to incubate, but it is important to know that it can take anywhere from 5 to 21 days. The spread happens over two separate time periods. During the first few weeks after the attack, patients often had back pain, fever, swollen lymph nodes, severe headaches, muscle aches, and a general lack of energy. The next phase usually starts one to three days after the fever starts, and this is when the familiar skin sores show up. These skin lesions show up on the face in about 95% of the cases, on the palms and soles of the feet in about 75% of the cases, on the inside of the mouth about 70% of the time, on the external sexual organs in about 30% of the cases, and on the conjunctivae, including the eyeball, in about 20% of the cases [9]. Transmitting the virus mostly happens through close touch between people or through bedding and clothes that have been contaminated [9]. According to [10], it is anticipated that more cases will be detected. However, it is important to note that the availability of polymerase chain reactions (PCR) and other biochemical tests is currently limited in terms of sufficient quantities, as indicated by [11].
Multiplex polymerase chain reaction (PCR) testing is the most common way to detect human monkeypox. However, the accuracy of the results obtained through this test can be compromised, often yielding inconclusive outcomes due to the virus’s transient presence in the bloodstream, as highlighted by [12]. This method of diagnosis also needs extra details, like the current stage of the rashes, the patient’s age, and the exact times when the fever and rash started. Furthermore, PCR tests are not widely utilized because they require a lot of resources, which causes them to be unavailable in most rural or remote places. In light of these challenges, there is a compelling case for the development of an alternative diagnostic system which operates independently of these metrics and leverages real-time data while utilizing readily accessible devices. Such an approach holds the potential to offer a near-perfect diagnostic solution for monkeypox, significantly enhancing both its effectiveness and efficiency.
Utilizing artificial intelligence (AI) and its various parts has been used in healthcare for a long time [12,13]. When it comes to healthcare, employing deep neural networks, especially for computer vision tasks, opens up a whole new world of possibilities. This method can harness the huge amount of healthcare data that is available to train convolutional neural networks (CNNs). These networks can then use current devices to solve new healthcare challenges [14]. A similar deep learning model based on patches has been proposed to identify monkeypox and chickenpox using skin images. This model utilizes RGB images of skin lesions captured using the cameras commonly found on smartphones.
2. Literature Review
The first recorded instance of monkeypox affecting humans was documented in 1970, marking the inception of human monkeypox studies in the scientific literature [15,16]. Over recent years, the research on human monkeypox has gained momentum, prompted by the alarming global spread of monkeypox infections. In fact, some researchers [17,18] have explicitly noted the pressing need for further investigation in this area.
Despite the historical presence of human monkeypox cases, the application of computer vision for early disease diagnosis is a relatively recent development. Currently, there is a dearth of comprehensive studies on this subject. Ahsan et al. [19], researchers collected image data of monkeypox-infected cases from Google called “Monkeypox2022” and conducted an in-depth analysis using advanced deep learning techniques. Specifically, they harnessed a modified VGG16 network for this purpose. Their model had great performance measures; its accuracy, sensitivity, recall, and f1-score all reached an amazing 97%. Ali et al. [11] involved the creation of a dedicated database of human monkeypox images, subsequently subjecting them to classification. In their classification efforts, the researchers employed four distinct deep learning networks, namely VGG16, ResNet50, InceptionV3, and Ensemble.
A lot of experts have used deep learning to figure out how to diagnose the monkeypox (Mpox) virus. In a different study, Abdelhamid et al. [20] used the AI-Biruni Earth Radius Optimization method, along with GoogLeNet, to pull out features for their Mpox diagnosis. They got a maximum accuracy rate of 98.8% by using different deep learning methods. The f1-score, sensitivity, and recall reached 62.5%, 99.8%, and 76%, respectively.
To make it easier for people to get medical help, a mobile app was made that can diagnose Mpox from pictures of skin lesions [21]. The creation process used Java and Android technologies, which led to an excellent maximum accuracy rate of 91.11%. The sensitivity score was 85%, the memory score was 94%, and the f1-score was 89%. A study by Islam et al. [22] used deep learning methods and a dataset with pictures of measles, mumps, chickenpox, smallpox, cowpox, and typhus. They utilized seven different classifiers, and the results were 83% for accuracy, 85% for sensitivity, 94% for recall, and 89% for the f1-score. Finally, Sitaula et al. [23] used eight different deep learning models that had already been trained to tell the difference between four groups and identify a case of mumps. They got an f1-score of 85%, an accuracy rate of 87.13%, a sensitivity rate of 85%.
Alakus et al. [24] used wart DNA segments and deep learning models to tell the difference between warts and monkeypox in a distinctive way. This classification process involved three stages and achieved an impressive maximum accuracy of 96.08%. Given the potential for monkeypox to emerge as a significant global health concern, efficient resource utilization is imperative. Disease diagnosis is one of the many areas in which artificial intelligence (AI) is essential. This work advances our knowledge of and ability to treat monkeypox by using a variety of transfer learning models to classify images of the illness. The objectives of this research are as follows:
- The implementation of augmentation techniques was considered essential to ensure the model proper and consistent training with balanced class representation.
- A state-of-the-art vision transformer model was employed, utilizing a transfer learning approach to detect instances of monkeypox from skin images.
- An empirical exploration and adjustment of hyperparameters related to the proposed model and its training process were carried out to optimize performance.
- The proposed model’s performance was systematically compared with that of other deep learning models and relevant studies. This comparative analysis aimed to derive insights into the significance of the proposed model within the broader research context.
3. Proposed Methodology
This section discusses the proposed methodology with the description of the dataset. The augmentation technique and the splitting of the model is also part of this section. Lastly, the model architecture is discussed with proper working of the model. The complete architecture of the proposed work is presented in Figure 1.
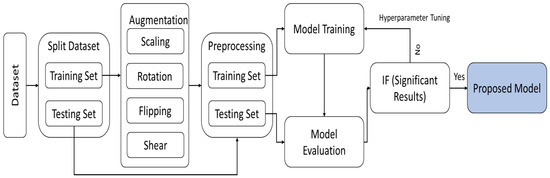
Figure 1.
Illustrating the comprehensive design and process flow of the proposed method.
3.1. Dataset Description
The experiment for classifying monkeypox was conducted using the Monkeypox Skin Image Dataset (MSID). This dataset, sourced from Kaggle (https://www.kaggle.com/datasets/dipuiucse/monkeypoxskinimagedataset, (accessed on 1 December 2023)), comprises images of human skin, representing four distinct skin diseases: monkeypox, chickenpox, measles, and uninfected skin, as presented in Figure 2. Within the dataset, a total of 279 instances of monkeypox were identified, along with 107, 91, and 293 instances of chickenpox, measles, and uninfected skin, respectively. In total, the dataset comprises 770 images. Originally, the images in the dataset were in PNG format with a resolution size of 224 × 224 pixels in RGB.
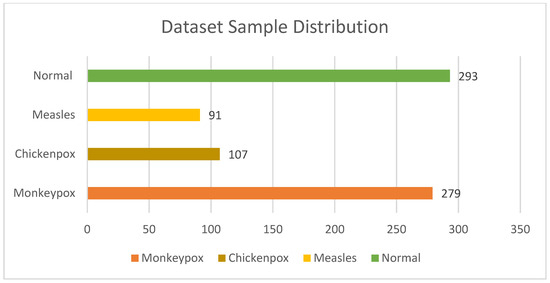
Figure 2.
Distribution of samples in datasets.
3.2. Data Pre-Processing
This section discusses the preprocessing of the splitting of the dataset before diving into the training and testing of the proposed model for monkeypox identification. Initially, the distribution of the samples across different classes was very diverse, and few classes have a very limited number of samples as shown in Table 1. By analyzing this behavior of the MSID dataset, 30 images of each class were split in a test set in order to evaluate the model on unseen samples. Furthermore, the rest of the dataset (training set) contains very few numbers of samples for some classes, like measles, that are not sufficient for the proper training of the model.

Table 1.
The distribution of samples in different subsets.
In order to expand the number of dataset samples for each class, data augmentation is performed on the training samples. Additionally, data augmentation keeps the model from overfitting, and helps to make it more resilient. This popular method is used to expand the number of samples that are automatically generated by using various image transformation methods, including cropping, translation, rotation, shearing, mirroring, and vertical and horizontal flipping. In order to partially balance the dataset samples for each class, four data augmentation techniques, brightness, rotation, zooming, and shear, are applied to the dataset in this study, as presented in Figure 3. The images in the monkeypox, chickenpox, measles, and uninfected classes are augmented by factors of 2, 5, 7, and 2, respectively. Since the measles class has the fewest images, it has undergone the most augmentation. Table 1 also summarizes the total number of samples for each class in the training set before and after the augmentation.
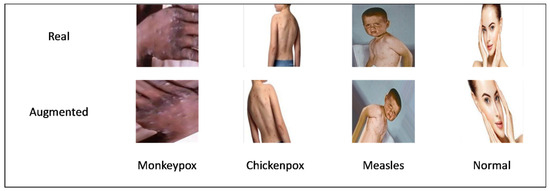
Figure 3.
Overview of samples before and after augmentation.
Lastly, the labels of the training and testing sets were encoded into 1, 2, 3, and 4 for monkeypox, chickenpox, measles, and normal classes.
3.3. Proposed Architecture
This section presents the ViT framework, emphasizing its key concepts, organization, self-attention mechanism, multi-headed self-attention, and the mathematical foundations that informed its development. In 2020, the ViT—a deep neural network architecture—was first introduced, and was specifically designed for image recognition tasks [25]. It expands the transformer architecture, which was initially developed for natural language processing through the use of the innovative notion of viewing images as sequences of tokens, which are frequently represented by image patches. ViT uses the transformer design’s capabilities to handle these token sequences efficiently. Notably, ViT’s transformer design has proven to be broadly applicable and effective, as shown by its successful application to a range of tasks, such as object identification, image restoration and identification [26,27,28].
Important steps in the ViT architecture include tokenization and input image embedding. To move the image to a higher-dimensional space, it is first divided into a grid of non-overlapping patches, flattened, and then linearly converted and normalized. The ViT model supports comprehensive learning by extracting both global and local information from the image through tokenization and embedding.
Despite having the ability to handle sequences, the transformer design does not specifically account for the location of each token inside the sequence. The ViT architecture uses pre-defined positional embeddings to overcome this restriction. These embeddings, which are extra vectors, encode the sequence positions of each token before being transmitted into the transformer layers. Through this integration, the model is able to deduce spatial information from the input image, comprehending the relative positions of the tokens.
The multi-head self-attention (MSA) mechanism is the central component of the ViT architecture. The model may focus on many areas of the image simultaneously because of this feature. The discrete “heads” that comprise MSA compute attention independently. These attention heads can focus on different parts of the image, creating a variety of representations that are then integrated in order to create the final image representation. ViT records complex interactions between input items by continually monitoring several sections. However, because it requires additional processing to aggregate the results from all heads and to pay greater attention to the heads, this upgrade increases computational costs and complexity. The mathematical expression for MSA is as follows:
MSA(Q,K,V) = Concat (H1, H2, …, Hn)
Equation (1) defines Q, K, and V as the query, key, and value matrices. The H1, H2, …, Hn denote the outputs of several attention heads. In neural networks, notably in transformers, multi-head attention employs multiple sets of attention weights (attention heads) to grasp various facets of relationships within the input data. Each output corresponds to the i-th attention head. The self-attention mechanism is crucial in transformers, forming the cornerstone for explicitly modeling interactions and relationships across all sequences in prediction tasks. Unlike CNNs, the self-attention layer aggregates insights and features from the whole input sequence in order to gather both local and global knowledge. Self-attention stands out from CNNs because of this characteristic, which encourages a more thorough analysis and representation of the data.
The attention mechanism computes the dot product between the query and key vectors, normalizes the attention scores using SoftMax activation function, and adjusts the value vectors to produce better output representation. Cordonnier et al.’s study [29] investigated the connection between convolution processes and self-attention. Their findings show that when self-attention is given a wide range of factors, it develops into a very flexible and adaptable mechanism that can extract both local and global features. This proves that self-attention is a more adaptable and versatile approach than conventional convolutional neural networks.
Figure 4 displays the abstract level ViT network diagram, which is based on the following key elements of the ViT model:
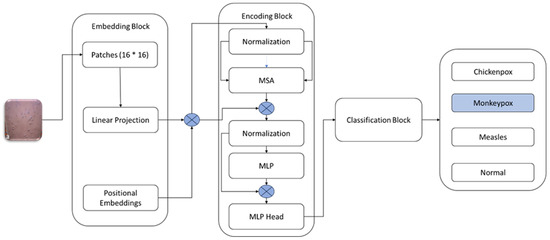
Figure 4.
Architecture of Vision Transformer Model [25].
Patch Embedding: Within the ViT framework, the initial image input undergoes a process wherein it is subdivided into non-overlapping patches of fixed sizes. Subsequently, each patch undergoes linear projection, facilitated by a learned linear transformation matrix, which operates to transform the 2D spatial characteristics of the image into a sequential arrangement of embeddings.
Positional Embedding: Given the inherent lack of spatial understanding within the transformer architecture, positional information becomes crucial. To address this, positional embeddings are introduced. These embeddings are incorporated into the patch embeddings, offering insights into the spatial positions of each patch within the overall image structure.
Transformer Encoder: The positional embeddings (E_POS) pass through an encoder transformer. This encoder consists of several layers, each using feedforward neural networks and self-attention processes. Each patch is able to consider other patches, due to the self-attention process, capturing the image’s overall associations. This is followed by feedforward neural networks, processing the attended representations further. As a result, the encoder generates contextualized embeddings that capture local and global visual information for every patch.
Classification Head: Finally, the transformer encoder yields final contextualized embeddings that provide the basis for other tasks, most notably image classification. Various approaches can be employed for processing these embeddings in classification tasks. One widely used technique is calculating the mean of all embeddings or the embedding of a particular token (e.g., a classification token). After processing, the data is put through one or more fully connected layers, creating class predictions as a result.
4. Experimental Results and Discussion
This section provides a thorough analysis of the assessment metrics used to determine the efficacy of the suggested approach. It also explores the system and software prerequisites that are necessary for model evaluation and training. Extensive details pertaining to the diverse hyper-parameters and their associated values are meticulously outlined. Additionally, a comprehensive analysis of the outcomes attained using the suggested approach is methodically provided in this section.
4.1. Evaluation Measures
Assessment metrics are quantitative measurements that are crucial for evaluating a deep learning model’s efficacy. They are essential in the evaluation of how well different models or algorithms perform on a given task, determining the performance of a model or algorithm in solving a particular issue, and identifying possible areas for improvement. The assessment metrics utilized in this study include recall/sensitivity, ROC curve, accuracy, f1-score, precision, and confusion matrix. Together, these measures offer a thorough evaluation of the model’s performance and offer insightful information about its advantages and potential improvement areas.
Accuracy: The accuracy metric calculates the ratio of correctly categorized cases to total samples, which assesses the overall correctness of the model’s predictions. However, depending only on accuracy might not be sufficient enough to provide a thorough assessment in instances where different types of errors carry different degrees of relevance, or in cases where datasets are imbalanced.
Accuracy = (TP + TN)/(TP + TN + FP + FN)
Precision: The precision of a model refers to its ability to accurately identify positive samples from the set of real positives. The ratio of genuine positives to the total of true positives and false positives is measured by this metric. To put it simply, accuracy tells us how well the model works when it makes a favorable prediction.
Precision = TP/(TP + FP)
Recall: Recall, sometimes referred to as sensitivity or the true positive rate, is used to assess how well the model distinguishes positive samples from the real positives pool. The ratio of true positives to the total of true positives and false negatives is used to calculate this measure. Recall essentially provides an evaluation of how comprehensive the model’s positive predictions are.
Recall = TP/(TP + FN)
F1-Score: The f1-score is a complete statistic that balances precision and recall. It is computed as the harmonic mean of these two measurements. This is especially useful when the distribution of errors between the classes is not equal, or when the importance of the two categories of errors is the same. The f1-score is a consolidated evaluation of the precision and recall capabilities of the model, using a range from 0 to 1. It performs best at 1.
f1-score = (2 × (Precision × Recall))/(Precision + Recall)
4.2. Environmental Setup
Different experiments, including the training and testing of the model, were carried out in the Colab environment. The model was trained and evaluated using TensorFlow and Keras, employing the Python programming language. The experiments made use of a NVIDIA Tesla T4 GPU with 15 GB of RAM on the free version of Google Colab (https://colab.research.google.com/, (accessed on 3 December 2023)).
4.3. Hyper-Parameter Settings
To achieve optimal performance in model training for monkeypox classification, a comprehensive process of empirical experimentation was undertaken to fine-tune various hyperparameters. These critical factors include batch size, choice of optimizers, learning rate, epochs, embedding size, patch size, and the selection of an appropriate loss function. Through systematic iteration and testing, the aim was to identify the combination of hyperparameter values that yields the best results in classifying monkeypox. This iterative optimization process is crucial in ensuring that the model achieves the desired level of accuracy and robustness in distinguishing monkeypox cases effectively. The details of the parameters are given in Table 2.

Table 2.
Hyperparameters settings.
4.4. Results Analysis and Discussion
In the proposed study, a vision transformer model is used with transfer learning technique for the classification of skin-related diseases, including monkeypox. The dataset was originally based on 770 samples, and some of the classes have very insufficient samples for the proper training of the model. Firstly, 30 samples were separated to form the test set. Furthermore, the augmentation was performed on the rest of the samples in the dataset. Finally, all the original samples, except for the test samples and the augmented samples, collectively made the training set. The training of the vision transformer model was completed using the training set. The 10% samples of the training set were used as the validation set during the training of the model.
The model showed an accuracy of 0.992% and 0.967% for training and validation during the training of the model, as presented in Figure 5. The model also showed a loss of 0.038% and 0.097% during the training and validation, respectively, as shown in Figure 5.
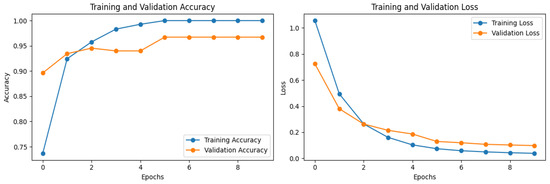
Figure 5.
The accuracy and loss of the model during training.
Overfitting was likely avoided in this study due to the consistency between the training and validation accuracies and losses. The model exhibited a training accuracy of 0.992% and a validation accuracy of 0.967%, indicating that it performed well not only on the training data, but also on unseen validation data. Similarly, the training and validation losses were low at 0.038% and 0.097%, respectively, suggesting that the model generalized well to new data without overfitting to the training set. Furthermore, augmentation samples contribute significantly to achieving both high scores and a well-generalized model. By diversifying the training data through augmentation techniques, the model becomes exposed to a broader range of variations and scenarios, thus enhancing its ability to generalize to unseen data. This helps prevent overfitting by ensuring that the model learns robust features and patterns that are applicable across different instances of the data. As a result, augmentation plays a crucial role in improving the performance and robustness of machine learning models.
A confusion matrix technique was used for the assessment of the trained model on the unseen test data. Confusion matrix is an evaluation measure that shows the predicted label of the model on the x-axis and the actual label of the samples on the y-axis. Furthermore, it calculates the count of accurate matches when the predicted label is truly matched with the actual label. The confusion matrix of the trained ViT model is presented in Figure 6. The rest of the evaluation measures, including accuracy, precision, recall, and f1-score, were also calculated as described in equations 2–5, using the confusion matrix. A detailed report of the model for disease classification on unseen samples is given in Table 3. In Table 3, “Macro AVG” refers to the unweighted average of metrics calculated independently for each class. In other words, it treats all classes equally, regardless of their frequency or importance, and computes the average of their individual performance metrics. This provides a balanced assessment across all classes. On the other hand, “Weighted AVG” considers the class imbalance by computing the average of the metrics weighted by the number of samples in each class. This means that classes with more instances have a greater influence on the overall average, compared to classes with fewer instances. “Weighted AVG” is particularly useful when dealing with imbalanced datasets, as it gives more weight to the performance of classes that are more representative of the overall distribution.
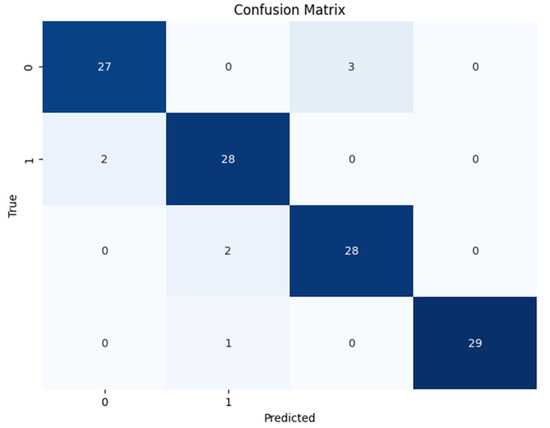
Figure 6.
Confusion matrix of proposed model on the test set.

Table 3.
The classification report of the model on test set.
4.5. Comparative and Ablation Analysis
This part assessed the suggested model’s performance by contrasting it with the pretrained models and the researchers’ proposed model. For this, numerous pretrained models including the VGG16, ResNet50, and DenseNet-121 were trained on the training data and evaluated on the test data. For the training of the pretrained model, an augmented trained set was used. Furthermore, a similar setting for the model was used as for the proposed model, allowing for a fair comparison of the model.
By following the training of the selected pretrained models, the test set used for the assessment of the models (see Figure 7, Figure 8 and Figure 9). During the assessment of the models, the ResNet-50 model showed the highest accuracy score, but this was not higher than the proposed ViT model, as shown in Table 4. The comparative performance of the model reveals the significance of the ViT model for the identification of monkeypox, among other skin diseases.
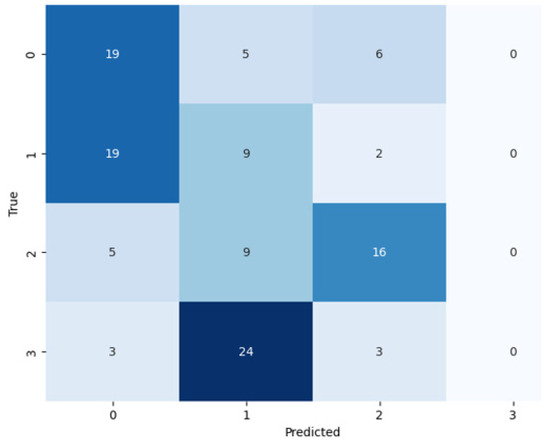
Figure 7.
Confusion matrix of ResNet-50 on the test set.
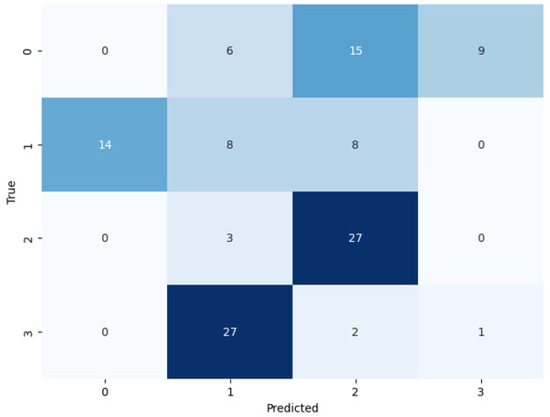
Figure 8.
Confusion matrix of VGG-16 on the test set.
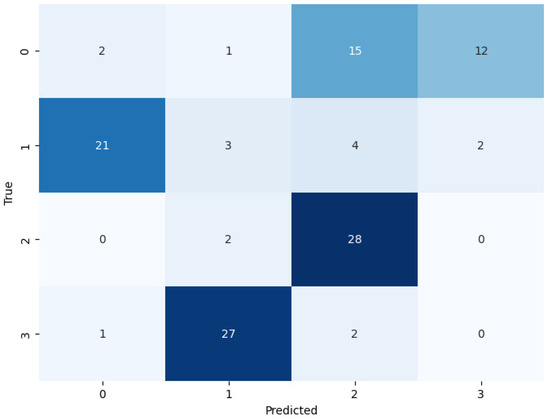
Figure 9.
Confusion matrix of DenseNet-201 on the test set.

Table 4.
The comparative analysis of the proposed model.
Furthermore, the performance of the proposed model was evaluated by comparing it with existing studies. The comparison with the existing studies also reveals that the proposed model outperforms all the existing studies, as shown in Table 4. The main reason for the superior performance of the transformer model is the use of the global feature extraction technique and patch-based learning.
5. Conclusions
As we come to the end of our research, the full review of our monkeypox and chickenpox detection model shows some interesting results. Through careful ablation and comparison studies, we carefully looked at how our model worked internally to discover its strengths and weaknesses. We carefully looked at how each part of the model affected its success in the area of ablation studies. Through this process, we were able to pinpoint the important factors, understanding how they interact with each other and affect our ability to recognize things. Putting our patch-wise ViT model up against other methods in a comparative study also gave us useful standards. Our model regularly did better than competitors in accuracy tests, with a score of 93%, which supports the reliability of the method. These results were passively added to our model, which shows both how well it works, and where it could be improved and explored further in future studies. In every case, our model did better than the others, showing that it could be a useful tool for diagnosing monkeypox and chickenpox disease using images of human skin. Through the combination of advanced deep learning techniques and healthcare problems, this journey is a monumental step towards the improvement of medical diagnoses.
Author Contributions
Conceptualization, M.A.A. and C.D.; methodology, M.A.A. and H.A.R.; validation, H.J.C., S.A. and M.A.A.; investigation, M.A.A., S.A., H.J.C. and C.D.; data curation, M.A.A.; writing—original draft preparation, H.A.R.; writing—review and editing, M.A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Monkeypox Skin Images Dataset (MSID) (https://www.kaggle.com/datasets/dipuiucse/monkeypoxskinimagedataset, (accessed on 1 December 2023)), The final prepared dataset will be provided on request.
Acknowledgments
The authors would like to thank all colleagues from Satya Wacana Christian University, Indonesia, and all involved in this research. This research is supported by the Vice-Rector of Research, Innovation and Entrepreneurship at Satya Wacana Christian University. We also acknowledge the use of ChatGPT (https://chat.openai.com/, (accessed on 1 December 2023)) for English correction across all manuscript sections.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Rizk, J.; Lippi, G.; Henry, B.; Forthal, D.; Rizk, R. Prevention and treatment of monkeypox. Drugs 2022, 82, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Yinka-Ogunleye, A.; Aruna, O.; Dalhat, M.; Ogoina, D.; McCollum, A.; Disu, Y.; Mamadu, I.; Akinpelu, A.; Ahmad, A.; Burga, J.; et al. Outbreak of human monkeypox in Nigeria in 2017–18: A clinical and epidemiological report. Lancet Infect. Dis. 2019, 19, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Zachary, K.C.; Shenoy, E.S. Transmission following exposure in healthcare facilities in nonendemic settings: Low risk but limited literature. Infect. Control. Hosp. Epidemiol. 2022, 43, 920–924. [Google Scholar] [CrossRef] [PubMed]
- Chadha, J.; Khullar, L.; Gulati, P.; Chhibber, S.; Harjai, K. Insights into the monkeypox virus: Making of another pandemic within the pandemic? Environ. Microbiol. 2022, 24, 4547–4560. [Google Scholar] [CrossRef] [PubMed]
- Uwishema, O.; Adekunbi, O.; Peñamante, C.A.; Bekele, B.K.; Khoury, C.; Mhanna, M.; Nicholas, A.; Adanur, I.; Dost, B.; Onyeaka, H. The burden of monkeypox virus amidst the Covid-19 pandemic in Africa: A double battle for Africa. Ann. Med. Surg. 2022, 80, 104197. [Google Scholar] [CrossRef] [PubMed]
- Nayak, T.; Chadaga, K.; Sampathila, N.; Mayrose, H.; Gokulkrishnan, N.; Prabhu, S.; Umakanth, S. Deep learning based detection of monkeypox virus using skin lesion images. Med. Nov. Technol. Devices 2023, 18, 100243. [Google Scholar] [CrossRef] [PubMed]
- De Baetselier, I.; Van Dijck, C.; Kenyon, C.; Coppens, J.; Michiels, J.; de Block, T.; Smet, H.; Coppens, S.; Vanroye, F.; Bugert, J.J.; et al. Retrospective detection of asymptomatic monkeypox virus infections among male sexual health clinic attendees in Belgium. Nat. Med. 2022, 28, 2288–2292. [Google Scholar] [CrossRef]
- Altindis, M.; Puca, E.; Shapo, L. Diagnosis of monkeypox virus—An overview. Travel. Med. Infect. Dis. 2022, 50, 102459. [Google Scholar] [CrossRef]
- Multi-Country Monkeypox Outbreak in Non-Endemic Countries: Update. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON388 (accessed on 23 November 2023).
- Zumla, A.; Valdoleiros, S.R.; Haider, N.; Asogun, D.; Ntoumi, F.; Petersen, E.; Kock, R. Monkeypox outbreaks outside endemic regions: Scientific and social priorities. Lancet Infect. Dis. 2022, 22, 929–931. [Google Scholar] [CrossRef]
- Ali, S.N.; Ahmed, M.T.; Paul, J.; Jahan, T.; Sani, S.M.; Noor, N.; Hasan, T. Monkeypox Skin Lesion Detection Using Deep Learning Models: A Feasibility Study. arXiv 2022, arXiv:2207.03342. Available online: https://arxiv.org/abs/2207.03342v1 (accessed on 23 November 2023).
- Paniz-Mondolfi, A.; Guerra, S.; Munoz, M.; Luna, N.; Hernandez, M.M.; Patino, L.H.; Reidy, J.; Banu, R.; Shrestha, P.; Liggayu, B.; et al. Evaluation and validation of an RT-PCR assay for specific detection of monkeypox virus (MPXV). J. Med. Virol. 2022, 95, e28247. [Google Scholar] [CrossRef]
- Chadaga, K.; Prabhu, S.; Sampathila, N.; Nireshwalya, S.; Katta, S.S.; Tan, R.S.; Acharya, U.R. Application of artificial intelligence techniques for monkeypox: A systematic review. Diagnostics 2023, 13, 824. [Google Scholar] [CrossRef]
- Norgeot, B.; Glicksberg, B.S.; Butte, A.J. A call for deep-learning healthcare. Nat. Med. 2023, 25, 14–15. [Google Scholar] [CrossRef]
- Bulletin of the World Health Organization. Available online: https://www.who.int/publications/journals/bulletin (accessed on 23 November 2023).
- Heymann, D.L.; Szczeniowski, M.; Esteves, K. Re-emergence of monkeypox in Africa: A review of the past six years. Br. Med. Bull. 1998, 54, 693–702. [Google Scholar] [CrossRef]
- Bragazzi, N.L.; Kong, J.D.; Mahroum, N.; Tsigalou, C.; Khamisy-Farah, R.; Converti, M.; Wu, J. Epidemiological trends and clinical features of the ongoing monkeypox epidemic: A preliminary pooled data analysis and literature review. J. Med. Virol. 2023, 95, e27931. [Google Scholar] [CrossRef]
- Wilson, M.E.; Hughes, J.M.; McCollum, A.M.; Damon, I.K. Human Monkeypox. Clin. Infect. Dis. 2014, 58, 260–267. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Uddin, M.R.; Farjana, M.; Sakib, A.N.; Al Momin, K.; Luna, S.A. Image Data collection and implementation of deep learning-based model in detecting Monkeypox disease using modified VGG16. arXiv 2022, arXiv:2206.0186. Available online: https://arxiv.org/abs/2206.01862v1 (accessed on 23 November 2023).
- Abdelhamid, A.A.; El-Kenawy, E.S.M.; Khodadadi, N.; Mirjalili, S.; Khafaga, D.S.; Alharbi, A.H.; Ibrahim, A.; Eid, M.M.; Saber, M. Classification of monkeypox images based on transfer learning and the Al-Biruni Earth Radius Optimization algorithm. Mathematics 2022, 10, 3614. [Google Scholar] [CrossRef]
- Sahin, V.H.; Oztel, I.; Yolcu Oztel, G. Human monkeypox classification from skin lesion images with deep pre-trained network using mobile application. J. Med. Syst. 2022, 46, 79. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.A.; Islam, T.; Chowdhury, F.U.H.; Islam, B.R. Can Artificial Intelligence Detect Monkeypox from Digital Skin Images? bioRxiv 2022. [Google Scholar] [CrossRef]
- Sitaula, C.; Shahi, T.B. Monkeypox Virus Detection Using Pre-trained Deep Learning-based Approaches. J. Med. Syst. 2022, 46, 78. [Google Scholar] [CrossRef] [PubMed]
- Alakus, T.B.; Baykara, M. Comparison of Monkeypox and wart DNA sequences with deep learning model. Appl. Sci. 2022, 12, 10216. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the ICLR 2021—9th International Conference on Learning Representations, Virtual Event, 3–7 May 2021; Available online: https://arxiv.org/abs/2010.11929v2 (accessed on 23 November 2023).
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2020; Available online: https://link.springer.com/chapter/10.1007/978-3-030-58452-8_13 (accessed on 23 November 2023).
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the NAACL HLT 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies—Proceedings of the Conference, Minneapolis, Minnesota, 2–7 June 2019; Volume 1, pp. 4171–4186. [Google Scholar]
- Arshed, M.A.; Mumtaz, S.; Ibrahim, M.; Dewi, C.; Tanveer, M.; Ahmed, S. Multiclass AI-Generated Deepfake Face Detection Using Patch-Wise Deep Learning Model. Computers 2024, 13, 31. [Google Scholar] [CrossRef]
- Cordonnier, J.B.; Loukas, A.; Jaggi, M. On the relationship between self-attention and convolutional layeRS. In Proceedings of the 8th International Conference on Learning Representations, ICLR 2020, Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Uysal, F. Detection of Monkeypox Disease from Human Skin Images with a Hybrid Deep Learning Model. Diagnostics 2023, 13, 1772. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).