Quantum Mechanics/Molecular Mechanics Simulations for Chiral-Selective Aminoacylation: Unraveling the Nature of Life
Abstract
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Perutz, M.F.; Rossmann, M.G.; Cullis, A.F.; Muirhead, H.; Will, G.; North, A.C.T. Structure of hæmoglobin: A three-dimensional Fourier synthesis at 5.5-Å. resolution, obtained by X-ray analysis. Nature 1960, 185, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Kendrew, J.C.; Dickerson, R.E.; Strandberg, B.E.; Hart, R.G.; Davies, D.R.; Phillips, D.C.; Shore, V.C. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 Å. resolution. Nature 1950, 185, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Hodgkin, D.C. The X-ray analysis of complicated molecules. Science 1965, 150, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Blundell, T.L. The first resolution revolution in protein structure analysis: X-ray diffraction of polypeptide conformations and globular protein folds in 1950s and 1960s. Prog. Biophys. Mol. Biol. 2021, 167, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Henkel, A.; Oberthür, D. A snapshot love story: What serial crystallography has done and will do for us. Acta. Crystallogr. D Struct. Biol. 2024, 80, 563–579. [Google Scholar] [CrossRef]
- Aue, W.P.; Bartholdi, E.; Ernst, R.R. Two-dimensional spectroscopy. Application to nuclear magnetic resonance. J. Chem. Phys. 1976, 64, 2229–2246. [Google Scholar] [CrossRef]
- Wagner, G.; Wüthrich, K. Dynamic model of globular protein conformations based on NMR studies in solution. Nature 1978, 275, 247–248. [Google Scholar] [CrossRef]
- Yao, S.; Keizer, D.W.; Babon, J.J.; Separovic, F. NMR measurement of biomolecular translational and rotational motion for evaluating changes of protein oligomeric state in solution. Eur. Biophys. J. 2022, 51, 193–204. [Google Scholar] [CrossRef]
- Han, B.; Yang, J.; Zhang, Z. Selective methods promote protein solid-state NMR. J. Phys. Chem. Lett. 2024, 15, 11300–11311. [Google Scholar] [CrossRef]
- Shen, P.S. The 2017 Nobel Prize in Chemistry: Cryo-EM comes of age. Anal. Bioanal. Chem. 2018, 410, 2053–2057. [Google Scholar] [CrossRef] [PubMed]
- Cabral, A.; Cabral, J.E.; McNulty, R. Cryo-EM for small molecules. Curr. Protoc. 2022, 2, e632. [Google Scholar] [CrossRef] [PubMed]
- Mazal, H.; Wieser, F.F.; Sandoghdar, V. Insights into protein structure using cryogenic light microscopy. Biochem. Soc. Trans. 2023, 51, 2041–2059. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. Origin of life: The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Crick, F.H.C. On Degenerate Templates and the Adapter Hypothesis. 1955. Available online: https://collections.nlm.nih.gov/catalog/nlm:nlmuid-101584582X73-doc (accessed on 25 November 2024).
- Tamura, K. Origins and early evolution of the tRNA molecule. Life 2015, 5, 1687–1699. [Google Scholar] [CrossRef]
- Giegé, R.; Eriani, G. The tRNA identity landscape for aminoacylation and beyond. Nucleic Acids Res. 2023, 51, 1528–1570. [Google Scholar] [CrossRef]
- Tennakoon, R.; Cui, H. Aminoacyl-tRNA synthetases. Curr. Biol. 2024, 34, R884–R888. [Google Scholar] [CrossRef]
- Kim, S.H.; Suddath, F.L.; Quigley, G.J.; McPherson, A.; Sussman, J.L.; Wang, A.H.; Seeman, N.C.; Rich, A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 1974, 185, 435–440. [Google Scholar] [CrossRef]
- Robertus, J.D.; Ladner, J.E.; Finch, J.T.; Rhodes, D.; Brown, R.S.; Clark, B.F.; Klug, A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 1974, 250, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Francklyn, C.; Schimmel, P. Aminoacylation of RNA minihelices with alanine. Nature 1989, 337, 478–481. [Google Scholar] [CrossRef]
- Frugier, M.; Florentz, C.; Giegé, R. Efficient aminoacylation of resected RNA helices by class II aspartyl-tRNA synthetase dependent on a single nucleotide. EMBO J. 1994, 13, 2219–2226. [Google Scholar] [CrossRef]
- Martinis, S.A.; Schimmel, P. Small RNA oligonucleotide substrates for specific aminoacylations. In tRNA: Structure, Biosynthesis, and Function; ASM Press: Washington, DC, USA, 1997; pp. 349–370. [Google Scholar]
- Musier-Forsyth, K.; Schimmel, P. Atomic determinants for aminoacylation of RNA minihelices and relationship to genetic code. Acc. Chem. Res. 1999, 32, 368–375. [Google Scholar] [CrossRef]
- Schimmel, P.; Giegé, R.; Moras, D.; Yokoyama, S. An operational RNA code for amino-acids and possible relationship to genetic-code. Proc. Natl. Acad. Sci. USA 1993, 90, 8763–8768. [Google Scholar] [CrossRef]
- Lei, L.; Burton, Z.F. The 3 31 nucleotide minihelix tRNA evolution theorem and the origin of life. Life 2023, 13, 2224. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.Q.; Hu, H.; Douglas, J.; Carter, C.W., Jr. Primordial aminoacyl-tRNA synthetases preferred minihelices to full-length tRNA. Nucleic Acids Res. 2024, 52, 7096–7111. [Google Scholar] [CrossRef]
- Schimmel, P.; Ribas de Pouplana, L. Transfer RNA: From minihelix to genetic code. Cell 1995, 81, 983–986. [Google Scholar] [CrossRef]
- Sarzynska, J.; Popenda, M.; Antczak, M.; Szachniuk, M. RNA tertiary structure prediction using RNAComposer in CASP15. Proteins 2023, 91, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P. Aminoacyl tRNA synthetases: General scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987, 56, 125–158. [Google Scholar] [CrossRef]
- Lux, M.C.; Standke, L.C.; Tan, D.S. Targeting adenylate-forming enzymes with designed sulfonyladenosine inhibitors. J. Antibiot. 2019, 72, 325–349. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Atencio, A. Separation of chiral molecules: A way to homochirality. Orig. Life Evol. Biosph. 2012, 42, 55–73. [Google Scholar] [CrossRef]
- Tamura, K. Perspectives on the origin of biological homochirality on Earth. J. Mol. Evol. 2019, 87, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Hegstrom, R.A. Parity violation and chiral symmetry breaking of a racemic mixture. Biosystems 1987, 20, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Bonner, W.A. Parity violation and the evolution of biomolecular homochirality. Chirality 2000, 12, 114–126. [Google Scholar] [CrossRef]
- Fukue, T.; Tamura, M.; Kandori, R.; Kusakabe, N.; Hough, J.H.; Bailey, J.; Whittet, D.C.; Lucas, P.W.; Nakajima, Y.; Hashimoto, J. Extended high circular polarization in the Orion massive star forming region: Implications for the origin of homochirality in the solar system. Orig. Life Evol. Biosph. 2010, 40, 335–346. [Google Scholar] [CrossRef]
- Soai, K.; Shibata, T.; Morioka, H.; Choji, K. Asymmetric autocatalysis and amplification of enantiomeric excess of a chiral molecule. Nature 1995, 378, 767–768. [Google Scholar] [CrossRef]
- Bada, J.L.; Miller, S.L. Racemization and the origin of optically active organic compounds in living organisms. Biosystems 1987, 20, 21–26. [Google Scholar] [CrossRef]
- Tamura, K.; Schimmel, P. Chiral-selective aminoacylation of an RNA minihelix. Science 2004, 305, 1253. [Google Scholar] [CrossRef]
- Karplus, M.; McCammon, J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef]
- Ando, T.; Takahashi, S.; Tamura, K. Principles of chemical geometry underlying chiral selectivity in RNA minihelix aminoacylation. Nucleic Acids Res. 2018, 46, 11144–11152. [Google Scholar] [CrossRef]
- Krepl, M.; Zgarbová, M.; Stadlbauer, P.; Otyepka, M.; Banáš, P.; Koča, J.; Cheatham, T.E., 3rd; Jurečka, P.; Šponer, J. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: Quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theory Comput. 2012, 8, 2506–2520. [Google Scholar] [CrossRef]
- Zgarbová, M.; Luque, F.J.; Šponer, J.; Cheatham, T.E., 3rd; Otyepka, M.; Jurečka, P. Toward improved description of DNA backbone: Revisiting epsilon and zeta torsion force field parameters. J. Chem. Theory Comput. 2013, 9, 2339–2354. [Google Scholar] [CrossRef] [PubMed]
- Zgarbová, M.; Šponer, J.; Otyepka, M.; Cheatham, T.E., 3rd; Galindo-Murillo, R.; Jurečka, P. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015, 11, 5723–5736. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.; Marchán, I.; Svozil, D.; Sponer, J.; Cheatham, T.E., 3rd; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef]
- Zgarbová, M.; Otyepka, M.; Šponer, J.; Mládek, A.; Banáš, P.; Cheatham, T.E., 3rd; Jurečka, P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011, 7, 2886–2902. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Ando, T.; Tamura, K. Mechanism of chiral-selective aminoacylation of an RNA minihelix explored by QM/MM free-energy simulations. Life 2023, 13, 722. [Google Scholar] [CrossRef]
- Kubař, T.; Elstner, M.; Cui, Q. Hybrid quantum mechanical/molecular mechanical methods for studying energy transduction in biomolecular machines. Annu. Rev. Biophys. 2023, 52, 525–551. [Google Scholar] [CrossRef]
- Schlick, T. Molecular Modeling and Simulation: An Interdisciplinary Guide, 2nd ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- Brunk, E.; Rothlisberger, U. Mixed quantum mechanical/molecular mechanical molecular dynamics simulations of biological systems in ground and electronically excited states. Chem. Rev. 2015, 115, 6217–6263. [Google Scholar] [CrossRef]
- Kar, R.K. Benefits of hybrid QM/MM over traditional classical mechanics in pharmaceutical systems. Drug Discov. Today 2023, 28, 103374. [Google Scholar] [CrossRef]
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- Gaus, M.; Cui, Q.; Elstner, M. DFTB3: Extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB). J. Chem. Theory Comput. 2012, 7, 931–948. [Google Scholar] [CrossRef] [PubMed]
- Service, R.F. AI tools set off an explosion of designer proteins. Science 2024, 386, 260–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Watson, J.L.; Lisanza, S.L. Protein design using structure-prediction networks: AlphaFold and RoseTTAFold as protein structure foundation models. Cold Spring Harb. Perspect. Biol. 2024, 16, a041472. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Q.; Nasif, K.F.A.; Xie, Y.; Deng, B.; Niu, S.; Pouriyeh, S.; Dai, Z.; Chen, J.; Xie, C.Y. AI-driven deep learning techniques in protein structure prediction. Int. J. Mol. Sci. 2024, 25, 8426. [Google Scholar] [CrossRef]
- Cui, Q.; Pal, T.; Xie, L. Biomolecular QM/MM simulations: What are some of the “burning issues”? J. Phys. Chem. B 2021, 125, 689–702. [Google Scholar] [CrossRef]



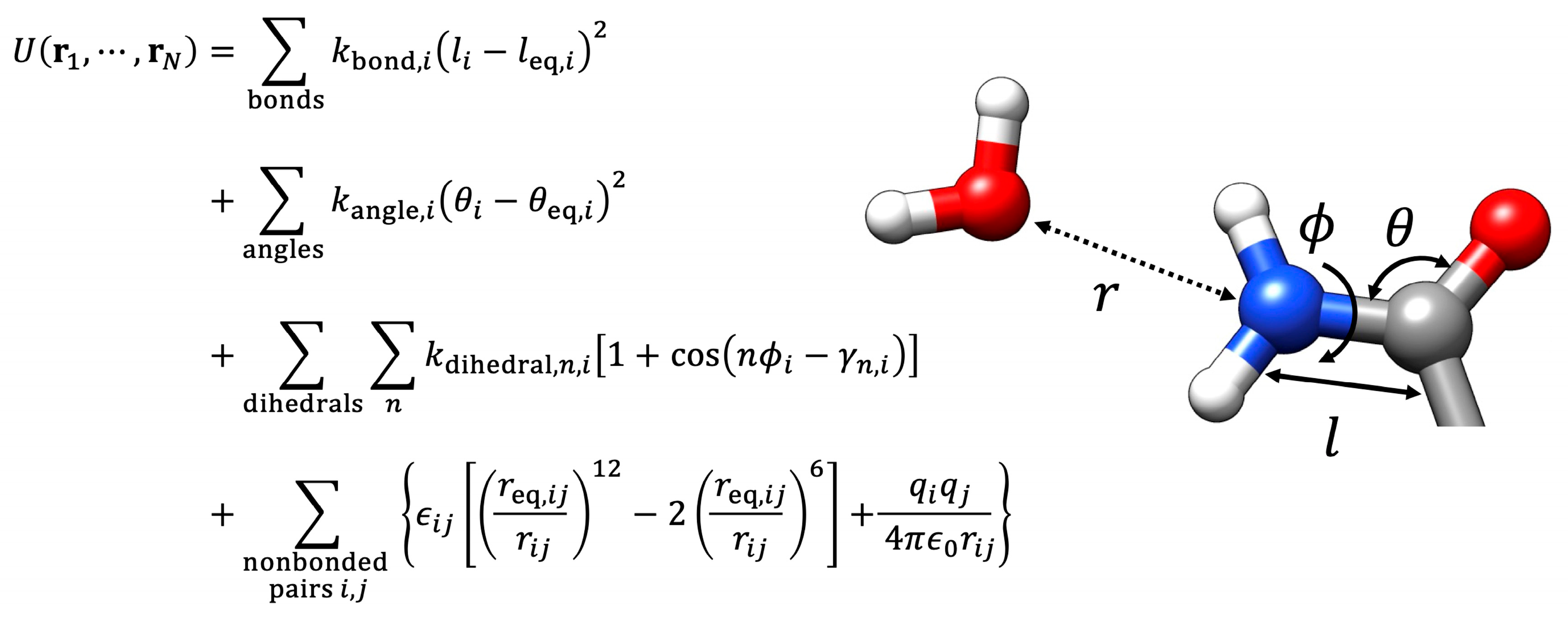
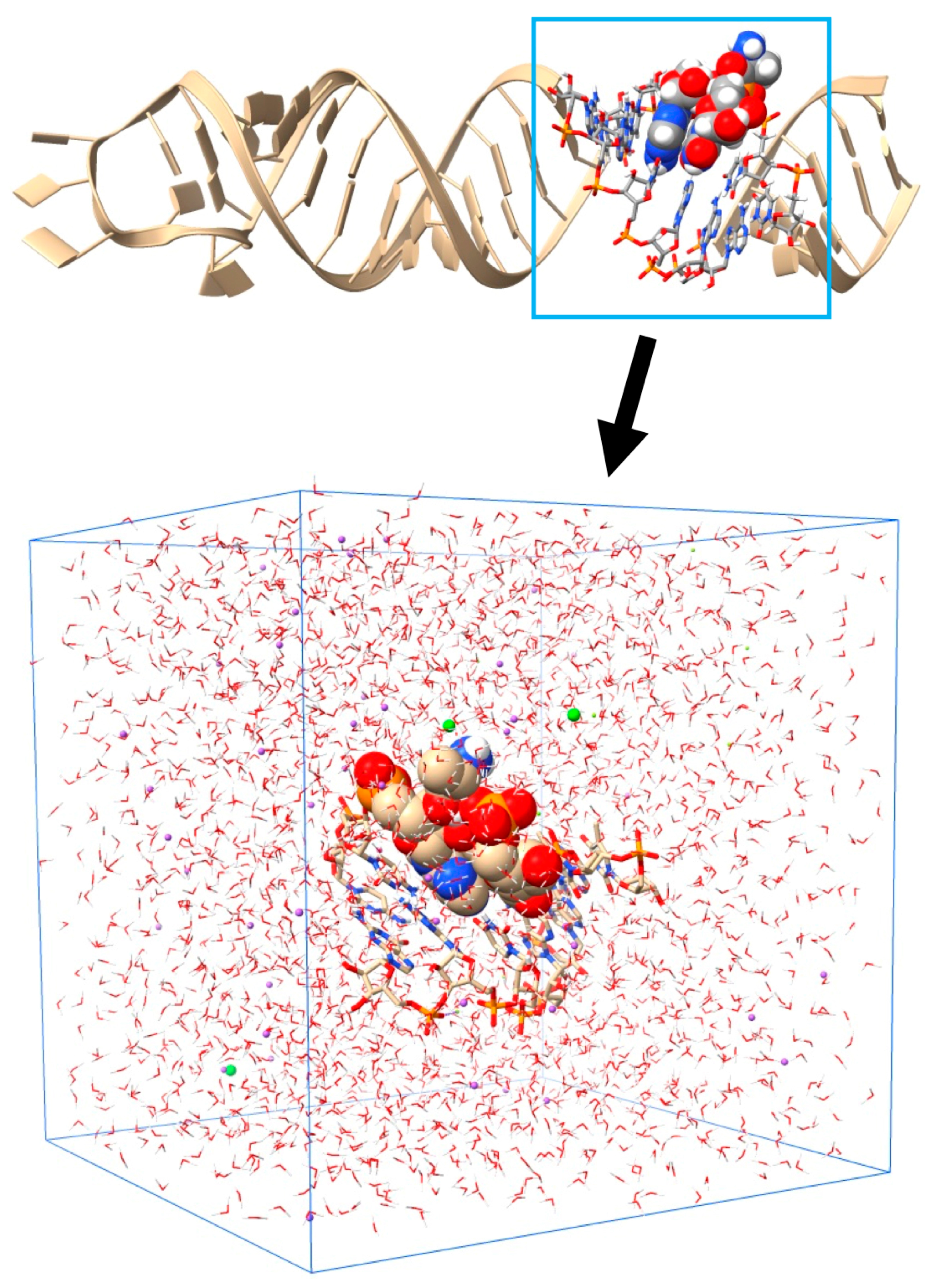

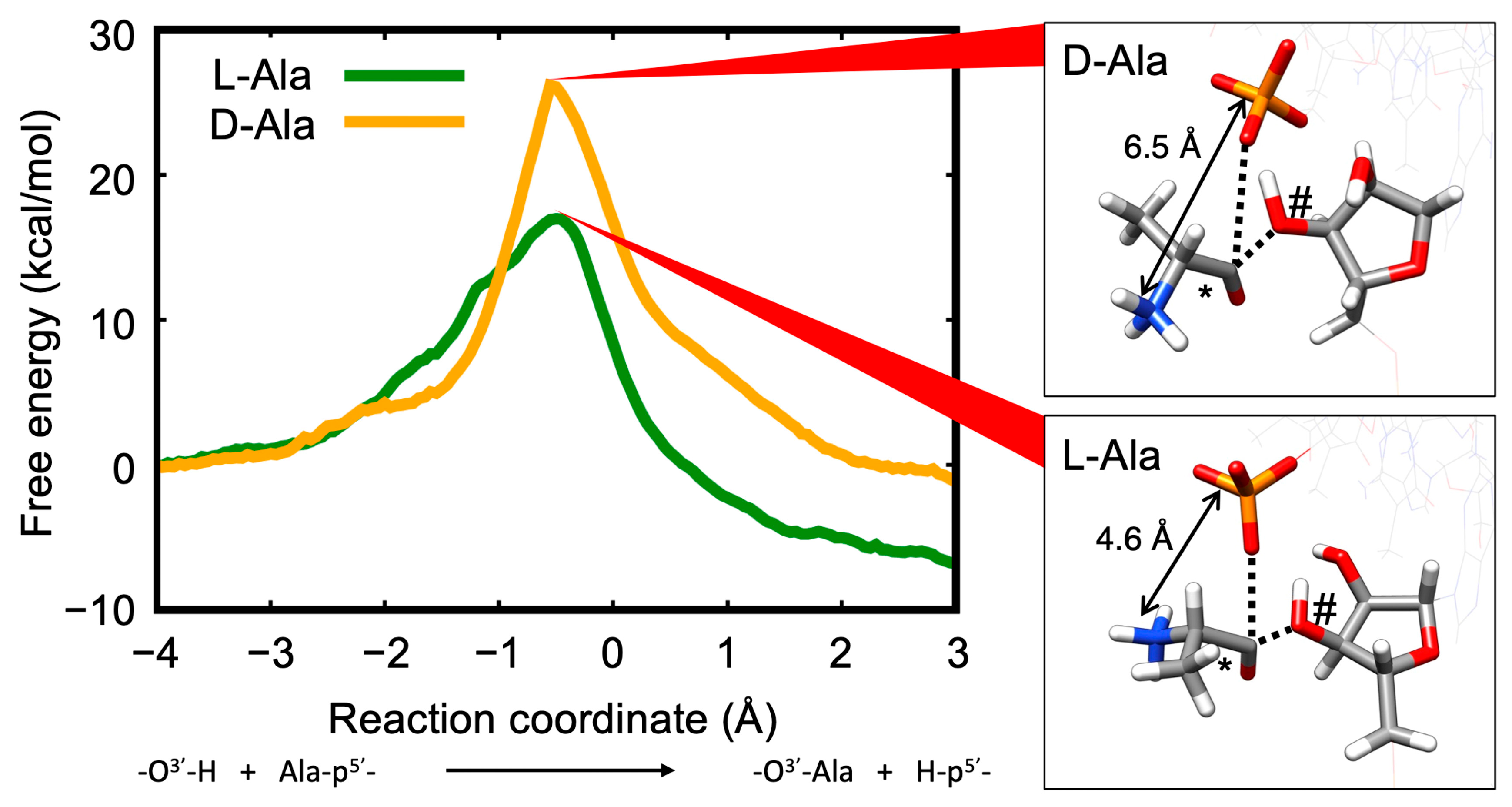
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ando, T.; Tamura, K. Quantum Mechanics/Molecular Mechanics Simulations for Chiral-Selective Aminoacylation: Unraveling the Nature of Life. Computation 2024, 12, 238. https://doi.org/10.3390/computation12120238
Ando T, Tamura K. Quantum Mechanics/Molecular Mechanics Simulations for Chiral-Selective Aminoacylation: Unraveling the Nature of Life. Computation. 2024; 12(12):238. https://doi.org/10.3390/computation12120238
Chicago/Turabian StyleAndo, Tadashi, and Koji Tamura. 2024. "Quantum Mechanics/Molecular Mechanics Simulations for Chiral-Selective Aminoacylation: Unraveling the Nature of Life" Computation 12, no. 12: 238. https://doi.org/10.3390/computation12120238
APA StyleAndo, T., & Tamura, K. (2024). Quantum Mechanics/Molecular Mechanics Simulations for Chiral-Selective Aminoacylation: Unraveling the Nature of Life. Computation, 12(12), 238. https://doi.org/10.3390/computation12120238





