Predicting the Occurrence of Metabolic Syndrome Using Machine Learning Models
Abstract
:1. Introduction
- Waist circumference greater than 102 cm and 88 cm for men and women, respectively.
- Triglyceride levels greater than or equal to 150 mg/dL.
- HDL lower than 40 mg/dL and 50 mg/dL for men and women, respectively.
- Blood pressure, systolic greater than or equal to 130 or diastolic greater than or equal to 85 mmHg.
- Fasting plasma glucose levels greater than or equal to 110 mg/dL.
- Data preprocessing is performed, which involves data cleaning and class balancing using the SMOTE. Thus, we are given the chance to experiment with effective classification models for the accurate identification of the occurrence of metabolic syndrome.
- In terms of feature ranking, three methods, namely information gain, gain ratio, and random forest, are chosen to measure their significance in the MetSyn class.
- In this submission, we experiment with a multitude of supervised ML models to determine the most accurate for classifying an unknown instance into the correct class. Well-known ML metrics with 10-fold cross-validation are used to evaluate the models’ performance. An “ablation experiment” is conducted to measure the role of class balancing in the ML models’ predictive performance. From this perspective, the experiments are executed with and without applying the SMOTE for the models’ training. The performance outcomes illustrate the prevalence of the ensemble model with the stacking technique due to the application of the SMOTE. Moreover, feature importance is measured with and without using the SMOTE.
- A discussion of related works on the prediction of metabolic syndrome using ML techniques and models is presented.
2. Materials and Methods
2.1. Data Collection and Description
2.2. MetSyn Risk Prediction: Methodology
2.2.1. Data Preprocessing
| Algorithm 1: SMOTE |
Steps 2–4 are repeated until the desired proportion S is satisfied. |
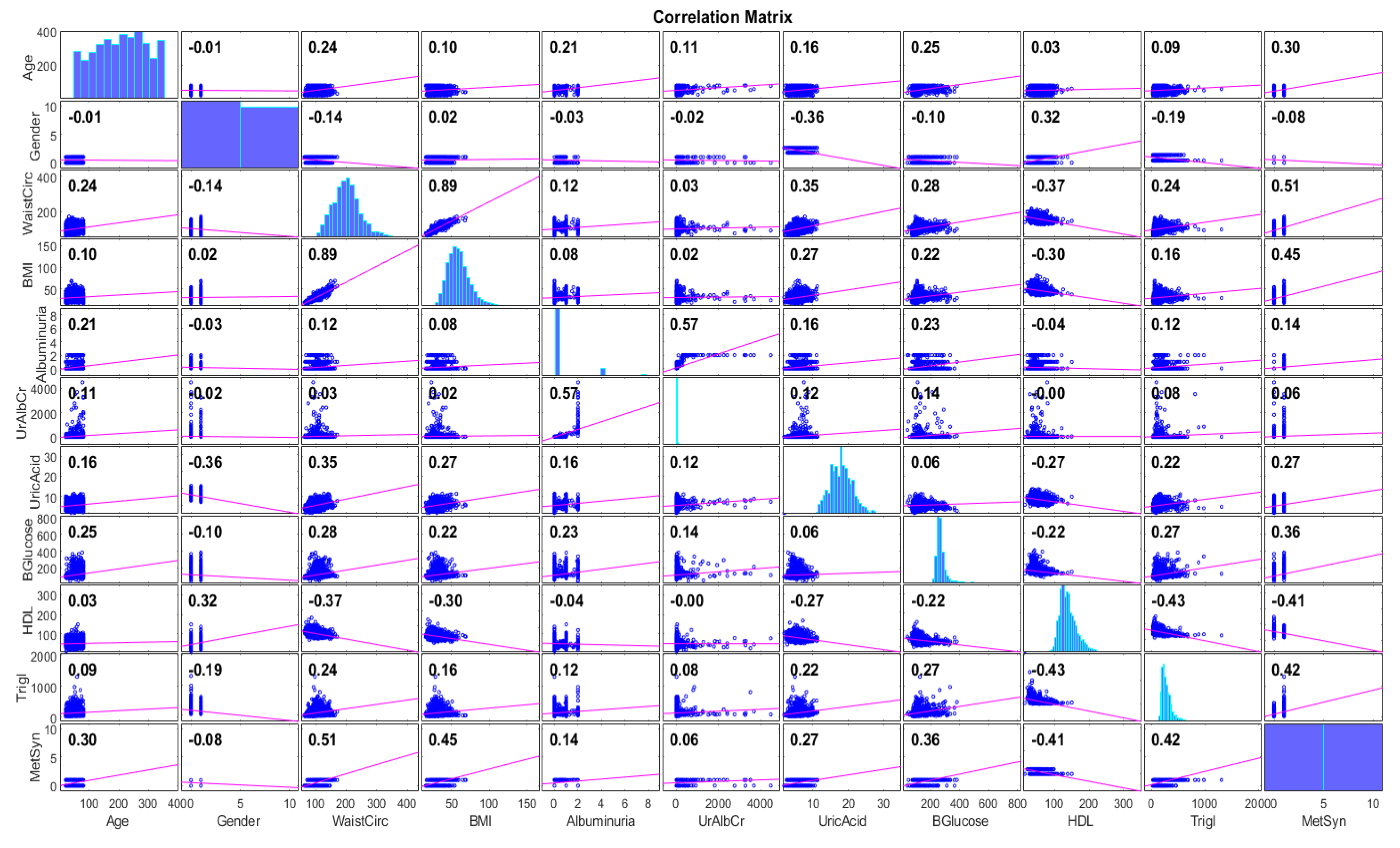
2.2.2. Feature Importance Ranking and Analysis
2.3. Evaluation Models and Metrics
- Accuracy sums up the classification performance by measuring the number of correctly predicted instances out of the total data.
- Precision indicates how many of the instances that were positive for MetSyn actually belong to this class.
- Recall measures the proportion of instances of MetSyn that were correctly considered positive, concerning all positive instances.
- The F1 score is a measure of a model’s accuracy on a given dataset. It is used to evaluate a model’s performance in binary classification problems. It combines the precision and recall values of the model, and it is defined as the harmonic mean of them.
- To evaluate a model’s ability to distinguish MetSyn instances from Non-MetSyn ones, the AUC was utilized. The AUC varied in the range of [0, 1]; the closer to one, the more efficient the ML model.
2.4. Experimental Setup
3. Results
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Conflicts of Interest
References
- Swarup, S.; Goyal, A.; Grigorova, Y.; Zeltser, R. Metabolic syndrome. In StatPearls [Internet]; StatPearls Publishing: St. Petersburg, FL, USA, 2022. [Google Scholar]
- Grundy, S.M. Metabolic Syndrome; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Zafar, U.; Khaliq, S.; Ahmad, H.U.; Manzoor, S.; Lone, K.P. Metabolic syndrome: An update on diagnostic criteria, pathogenesis, and genetic links. Hormones 2018, 17, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Solis, A.L.; Datta Banik, S.; Méndez-González, R.M. Prevalence of metabolic syndrome in Mexico: A systematic review and meta-analysis. Metab. Syndr. Relat. Disord. 2018, 16, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Han, K.; Kim, M.K.; Koh, E.S.; Kim, E.S.; Nam, G.E.; Hong, O.K.; Kim, B.; Kwon, H.S. Combinations of metabolic syndrome components and the risk of type 2 diabetes mellitus: A nationwide cohort study. Diabetes Res. Clin. Pract. 2020, 165, 108237. [Google Scholar] [CrossRef]
- Aboonabi, A.; Meyer, R.R.; Singh, I. The association between metabolic syndrome components and the development of atherosclerosis. J. Hum. Hypertens. 2019, 33, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Salzano, A.; D’Assante, R.; Heaney, L.M.; Monaco, F.; Rengo, G.; Valente, P.; Pasquali, D.; Bossone, E.; Gianfrilli, D.; Lenzi, A.; et al. Klinefelter syndrome, insulin resistance, metabolic syndrome, and diabetes: Review of literature and clinical perspectives. Endocrine 2018, 61, 194–203. [Google Scholar] [CrossRef]
- Paley, C.A.; Johnson, M.I. Abdominal obesity and metabolic syndrome: Exercise as medicine? BMC Sports Sci. Med. Rehabil. 2018, 10, 7. [Google Scholar] [CrossRef]
- Arhire, L.I.; Mihalache, L.; Covasa, M. Irisin: A hope in understanding and managing obesity and metabolic syndrome. Front. Endocrinol. 2019, 10, 524. [Google Scholar] [CrossRef]
- Myers, J.; Kokkinos, P.; Nyelin, E. Physical activity, cardiorespiratory fitness, and the metabolic syndrome. Nutrients 2019, 11, 1652. [Google Scholar] [CrossRef]
- Pilitsi, E.; Farr, O.M.; Polyzos, S.A.; Perakakis, N.; Nolen-Doerr, E.; Papathanasiou, A.E.; Mantzoros, C.S. Pharmacotherapy of obesity: Available medications and drugs under investigation. Metabolism 2019, 92, 170–192. [Google Scholar] [CrossRef]
- Nilsson, P.M.; Tuomilehto, J.; Rydén, L. The metabolic syndrome–What is it and how should it be managed? Eur. J. Prev. Cardiol. 2019, 26, 33–46. [Google Scholar] [CrossRef]
- Finicelli, M.; Squillaro, T.; Di Cristo, F.; Di Salle, A.; Melone, M.A.B.; Galderisi, U.; Peluso, G. Metabolic syndrome, Mediterranean diet, and polyphenols: Evidence and perspectives. J. Cell. Physiol. 2019, 234, 5807–5826. [Google Scholar] [CrossRef]
- Castro-Barquero, S.; Ruiz-León, A.M.; Sierra-Pérez, M.; Estruch, R.; Casas, R. Dietary strategies for metabolic syndrome: A comprehensive review. Nutrients 2020, 12, 2983. [Google Scholar] [CrossRef] [PubMed]
- Takase, H.; Hayashi, K.; Kin, F.; Nakano, S.; Machii, M.; Takayama, S.; Sugiura, T.; Dohi, Y. Dietary salt intake predicts future development of metabolic syndrome in the general population. Hypertens. Res. 2023, 46, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Suliga, E.; Kozieł, D.; Ciesla, E.; Rebak, D.; Głuszek-Osuch, M.; Głuszek, S. Consumption of alcoholic beverages and the prevalence of metabolic syndrome and its components. Nutrients 2019, 11, 2764. [Google Scholar] [CrossRef] [PubMed]
- Morales-Palomo, F.; Ramirez-Jimenez, M.; Ortega, J.F.; Mora-Rodriguez, R. Effectiveness of Aerobic Exercise Programs for Health Promotion in Metabolic Syndrome. Med. Sci. Sports Exerc. 2019, 51, 1876–1883. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Data-driven machine-learning methods for diabetes risk prediction. Sensors 2022, 22, 5304. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, S.; Dritsas, E.; Kocsis, O.; Moustakas, K.; Fakotakis, N. An approach for Personalized Continuous Glucose Prediction with Regression Trees. In Proceedings of the IEEE 2021 6th South-East Europe Design Automation, Computer Engineering, Computer Networks and Social Media Conference (SEEDA-CECNSM), Preveza, Greece, 24–26 September 2021; pp. 1–6. [Google Scholar]
- Dritsas, E.; Trigka, M. Stroke risk prediction with machine learning techniques. Sensors 2022, 22, 4670. [Google Scholar] [CrossRef]
- Dritsas, E.; Alexiou, S.; Moustakas, K. COPD severity prediction in elderly with ML techniques. In Proceedings of the 15th International Conference on PErvasive Technologies Related to Assistive Environments, Corfu, Greece, 29 June–1 July 2022; pp. 185–189. [Google Scholar]
- Dritsas, E.; Trigka, M. Supervised Machine Learning Models to Identify Early-Stage Symptoms of SARS-CoV-2. Sensors 2023, 23, 40. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Machine learning techniques for chronic kidney disease risk prediction. Big Data Cogn. Comput. 2022, 6, 98. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Supervised machine learning models for liver disease risk prediction. Computers 2023, 12, 19. [Google Scholar] [CrossRef]
- Dritsas, E.; Trigka, M. Efficient data-driven machine learning models for cardiovascular diseases risk prediction. Sensors 2023, 23, 1161. [Google Scholar] [CrossRef]
- Dritsas, E.; Alexiou, S.; Moustakas, K. Efficient data-driven machine learning models for hypertension risk prediction. In Proceedings of the IEEE 2022 International Conference on INnovations in Intelligent SysTems and Applications (INISTA), Biarritz, France, 8–12 August 2022; pp. 1–6. [Google Scholar]
- Dritsas, E.; Trigka, M. Lung Cancer Risk Prediction with Machine Learning Models. Big Data Cogn. Comput. 2022, 6, 139. [Google Scholar] [CrossRef]
- Metabolic Syndrome Dataset. Available online: https://data.world/informatics-edu/metabolic-syndrome-prediction (accessed on 26 February 2023).
- Zhang, J.; Zhu, W.; Qiu, L.; Huang, L.; Fang, L. Sex- and age-specific optimal anthropometric indices as screening tools for metabolic syndrome in Chinese adults. Int. J. Endocrinol. 2018, 2018, 1067603. [Google Scholar] [CrossRef]
- Jeong, S.; Cho, S.-i.; Kong, S.Y. Effect of income level on stroke incidence and the mediated effect of simultaneous diagnosis of metabolic syndrome diseases; a nationwide cohort study in South Korea. Diabetol. Metab. Syndr. 2022, 14, 110. [Google Scholar] [CrossRef]
- Suliga, E.; Cieśla, E.; Rębak, D.; Kozieł, D.; Głuszek, S. Relationship between sitting time, physical activity, and metabolic syndrome among adults depending on body mass index (BMI). Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 7633. [Google Scholar] [CrossRef] [PubMed]
- Rashidbeygi, E.; Safabakhsh, M.; Mohammed, S.H.; Alizadeh, S. Metabolic syndrome and its components are related to a higher risk for albuminuria and proteinuria: Evidence from a meta-analysis on 10,603,067 subjects from 57 studies. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 830–843. [Google Scholar] [CrossRef]
- Mohtashami, A. Effects of bread with Nigella sativa on blood glucose, blood pressure and anthropometric indices in patients with metabolic syndrome. Clin. Nutr. Res. 2019, 8, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Mocciaro, G.; D’amore, S.; Jenkins, B.; Kay, R.; Murgia, A.; Herrera-Marcos, L.V.; Neun, S.; Sowton, A.P.; Hall, Z.; Palma-Duran, S.A.; et al. Lipidomic approaches to study HDL metabolism in patients with central obesity diagnosed with metabolic syndrome. Int. J. Mol. Sci. 2022, 23, 6786. [Google Scholar] [CrossRef]
- Chen, J.; Huang, H.; Cohn, A.G.; Zhang, D.; Zhou, M. Machine learning-based classification of rock discontinuity trace: SMOTE oversampling integrated with GBT ensemble learning. Int. J. Min. Sci. Technol. 2022, 32, 309–322. [Google Scholar] [CrossRef]
- Zhang, R.; Nie, F.; Li, X.; Wei, X. Feature selection with multi-view data: A survey. Inf. Fusion 2019, 50, 158–167. [Google Scholar] [CrossRef]
- Berrar, D. Bayes’ theorem and naive Bayes classifier. Encycl. Bioinform. Comput. Biol. ABC Bioinform. 2018, 403. [Google Scholar]
- Nusinovici, S.; Tham, Y.C.; Yan, M.Y.C.; Ting, D.S.W.; Li, J.; Sabanayagam, C.; Wong, T.Y.; Cheng, C.Y. Logistic regression was as good as machine learning for predicting major chronic diseases. J. Clin. Epidemiol. 2020, 122, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Posonia, A.M.; Vigneshwari, S.; Rani, D.J. Machine Learning based Diabetes Prediction using Decision Tree J48. In Proceedings of the IEEE 2020 3rd International Conference on Intelligent Sustainable Systems (ICISS), Thoothukudi, India, 3–5 December 2020; pp. 498–502. [Google Scholar]
- Pisner, D.A.; Schnyer, D.M. Support vector machine. In Machine learning; Elsevier: Amsterdam, The Netherlands, 2020; pp. 101–121. [Google Scholar]
- González, S.; García, S.; Del Ser, J.; Rokach, L.; Herrera, F. A practical tutorial on bagging and boosting based ensembles for machine learning: Algorithms, software tools, performance study, practical perspectives and opportunities. Inf. Fusion 2020, 64, 205–237. [Google Scholar] [CrossRef]
- Palimkar, P.; Shaw, R.N.; Ghosh, A. Machine learning technique to prognosis diabetes disease: Random forest classifier approach. In Advanced Computing and Intelligent Technologies; Springer: Berlin/Heidelberg, Germany, 2022; pp. 219–244. [Google Scholar]
- Husna, N.A.; Bustamam, A.; Yanuar, A.; Sarwinda, D. The drug design for diabetes mellitus type II using rotation forest ensemble classifier. Procedia Comput. Sci. 2021, 179, 161–168. [Google Scholar] [CrossRef]
- Dogan, A.; Birant, D. A weighted majority voting ensemble approach for classification. In Proceedings of the IEEE 2019 4th International Conference on Computer Science and Engineering (UBMK), Samsun, Turkey, 11–15 September 2019; pp. 1–6. [Google Scholar]
- Pavlyshenko, B. Using stacking approaches for machine learning models. In Proceedings of the 2018 IEEE Second International Conference on Data Stream Mining & Processing (DSMP), Lviv, Ukraine, 21–25 August 2018; pp. 255–258. [Google Scholar]
- Masih, N.; Naz, H.; Ahuja, S. Multilayer perceptron based deep neural network for early detection of coronary heart disease. Health Technol. 2021, 11, 127–138. [Google Scholar] [CrossRef]
- Cunningham, P.; Delany, S.J. k-Nearest neighbour classifiers—A Tutorial. ACM Comput. Surv. (CSUR) 2021, 54, 1–25. [Google Scholar] [CrossRef]
- Weka. Available online: https://www.weka.io/ (accessed on 26 February 2023).
- Gutiérrez-Esparza, G.O.; Infante Vázquez, O.; Vallejo, M.; Hernández-Torruco, J. Prediction of metabolic syndrome in a Mexican population applying machine learning algorithms. Symmetry 2020, 12, 581. [Google Scholar] [CrossRef]
- Karimi-Alavijeh, F.; Jalili, S.; Sadeghi, M. Predicting metabolic syndrome using decision tree and support vector machine methods. ARYA Atheroscler. 2016, 12, 146. [Google Scholar]
- Tavares, L.D.; Manoel, A.; Donato, T.H.R.; Cesena, F.; Minanni, C.A.; Kashiwagi, N.M.; da Silva, L.P.; Amaro, E., Jr.; Szlejf, C. Prediction of metabolic syndrome: A machine learning approach to help primary prevention. Diabetes Res. Clin. Pract. 2022, 191, 110047. [Google Scholar] [CrossRef]
- Yu, C.S.; Lin, Y.J.; Lin, C.H.; Wang, S.T.; Lin, S.Y.; Lin, S.H.; Wu, J.L.; Chang, S.S. Predicting metabolic syndrome with machine learning models using a decision tree algorithm: Retrospective cohort study. JMIR Med. Inform. 2020, 8, e17110. [Google Scholar] [CrossRef]
- Choe, E.K.; Rhee, H.; Lee, S.; Shin, E.; Oh, S.W.; Lee, J.E.; Choi, S.H. Metabolic syndrome prediction using machine learning models with genetic and clinical information from a nonobese healthy population. Genom. Inform. 2018, 16, e31. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Schraplau, A.; Da Cruz, H.F.; Sachs, J.P.; Mayer, F.; Böttinger, E. A machine learning approach for non-invasive diagnosis of metabolic syndrome. In Proceedings of the 2019 IEEE 19th International Conference on Bioinformatics and Bioengineering (BIBE), Athens, Greece, 28–30 October 2019; pp. 933–940. [Google Scholar]
- Cheong, K.C.; Ghazali, S.M.; Hock, L.K.; Subenthiran, S.; Huey, T.C.; Kuay, L.K.; Mustapha, F.I.; Yusoff, A.F.; Mustafa, A.N. The discriminative ability of waist circumference, body mass index and waist-to-hip ratio in identifying metabolic syndrome: Variations by age, sex and race. Diabetes Metab. Syndr. Clin. Res. Rev. 2015, 9, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Esfahani, F.; Alafchi, B.; Cheraghi, Z.; Doosti-Irani, A.; Mirmiran, P.; Khalili, D.; Azizi, F. Using machine learning techniques to predict factors contributing to the incidence of metabolic syndrome in tehran: Cohort study. JMIR Public Health Surveill. 2021, 7, e27304. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, H.; Choi, J.R.; Koh, S.B. Development and validation of prediction model for risk reduction of metabolic syndrome by body weight control: A prospective population-based study. Sci. Rep. 2020, 10, 10006. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yu, B.; OUYang, P.; Li, X.; Lai, X.; Zhang, G.; Zhang, H. Machine learning-aided risk prediction for metabolic syndrome based on 3 years study. Sci. Rep. 2022, 12, 2248. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, S.K.; Kim, J.Y.; Cho, N.; Shin, C. Sasang constitutional types for the risk prediction of metabolic syndrome: A 14-year longitudinal prospective cohort study. BMC Complement. Altern. Med. 2017, 17, 438. [Google Scholar] [CrossRef]
- Li, G.; Esangbedo, I.C.; Xu, L.; Fu, J.; Li, L.; Feng, D.; Han, L.; Xiao, X.; Li, M.; Mi, J.; et al. Childhood retinol-binding protein 4 (RBP4) levels predicting the 10-year risk of insulin resistance and metabolic syndrome: The BCAMS study. Cardiovasc. Diabetol. 2018, 17, 69. [Google Scholar] [CrossRef]
- Zou, T.T.; Zhou, Y.J.; Zhou, X.D.; Liu, W.Y.; Van Poucke, S.; Wu, W.J.; Zheng, J.N.; Gu, X.M.; Zhang, D.C.; Zheng, M.H.; et al. MetS risk score: A clear scoring model to predict a 3-year risk for metabolic syndrome. Horm. Metab. Res. 2018, 50, 683–689. [Google Scholar] [CrossRef]
- Molnar, C.; König, G.; Herbinger, J.; Freiesleben, T.; Dandl, S.; Scholbeck, C.A.; Casalicchio, G.; Grosse-Wentrup, M.; Bischl, B. General pitfalls of model-agnostic interpretation methods for machine learning models. In Proceedings of the xxAI-Beyond Explainable AI: International Workshop, Held in Conjunction with ICML 2020, Vienna, Austria, 18 July 2020; Revised and Extended Papers. Springer: Berlin/Heidelberg, Germany, 2022; pp. 39–68. [Google Scholar]
- Devassy, B.M.; George, S. Dimensionality reduction and visualisation of hyperspectral ink data using t-SNE. Forensic Sci. Int. 2020, 311, 110194. [Google Scholar] [CrossRef]
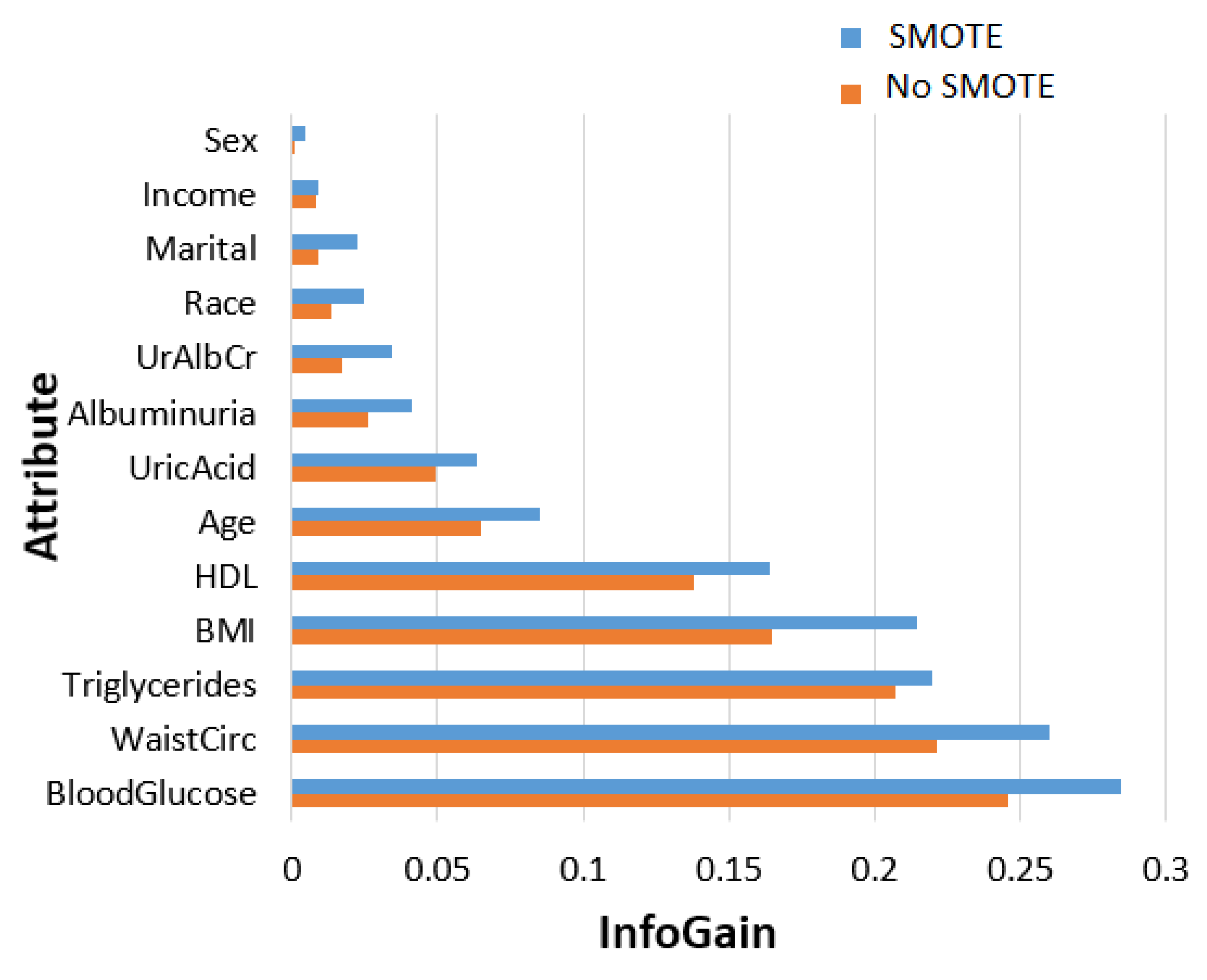
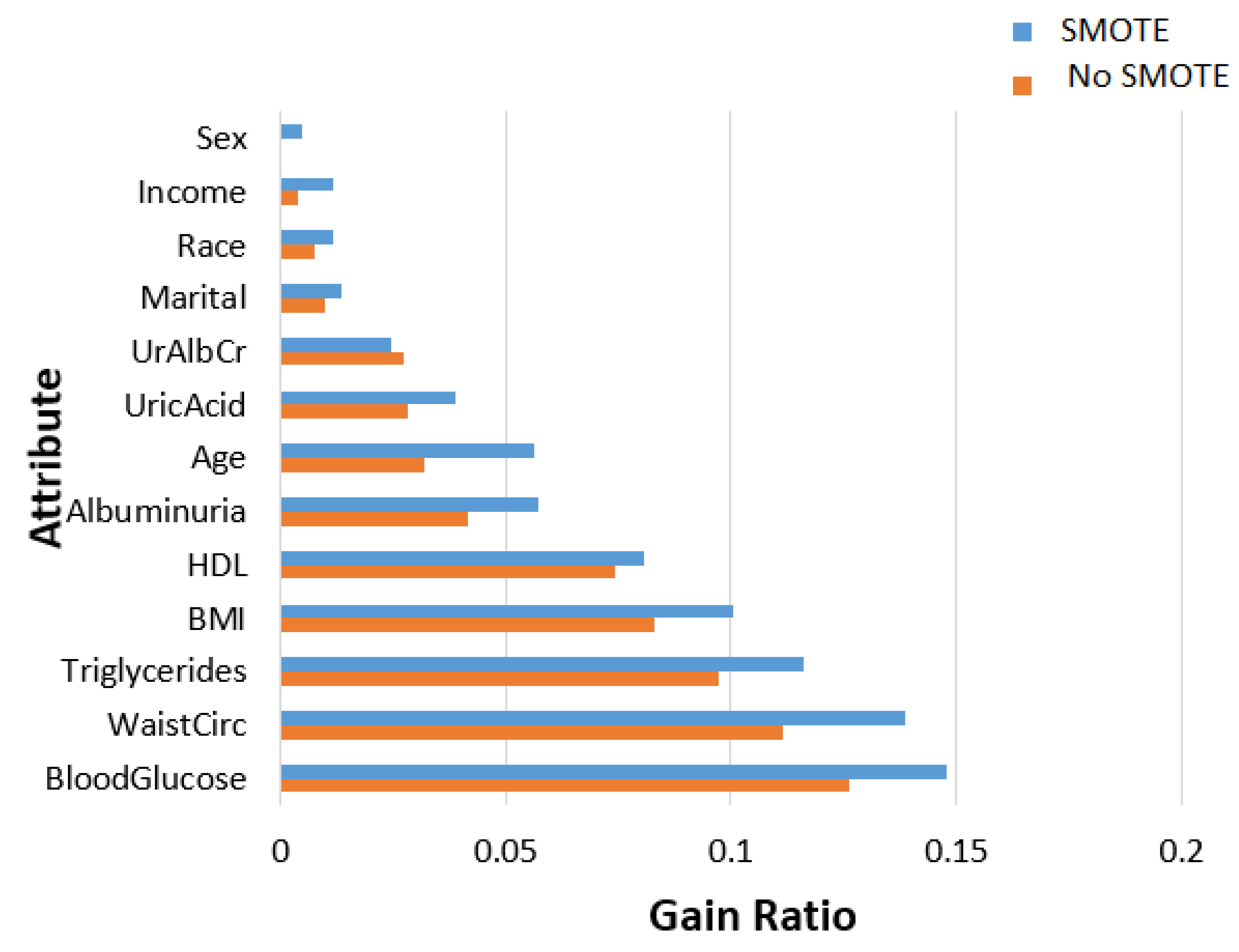
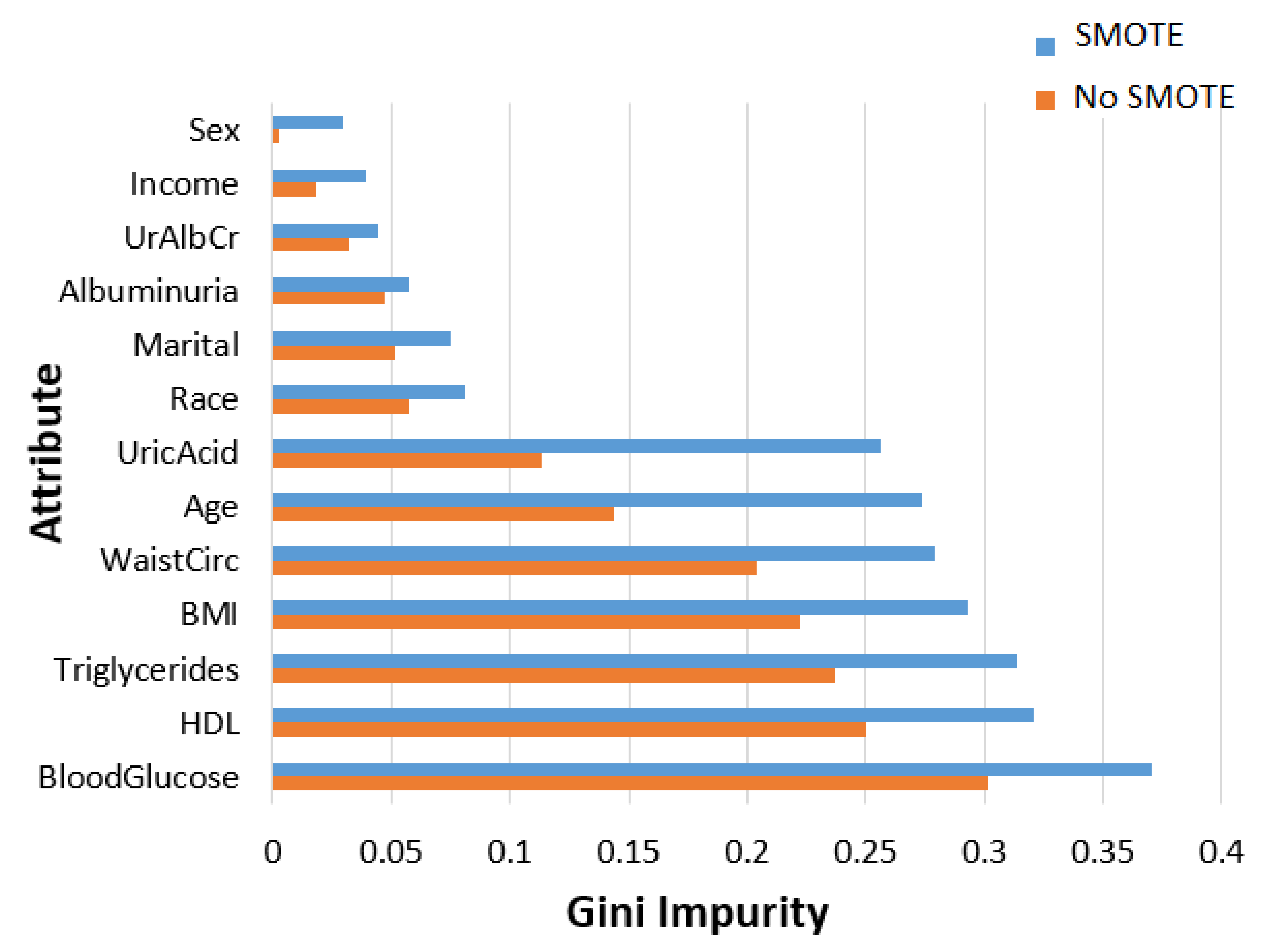
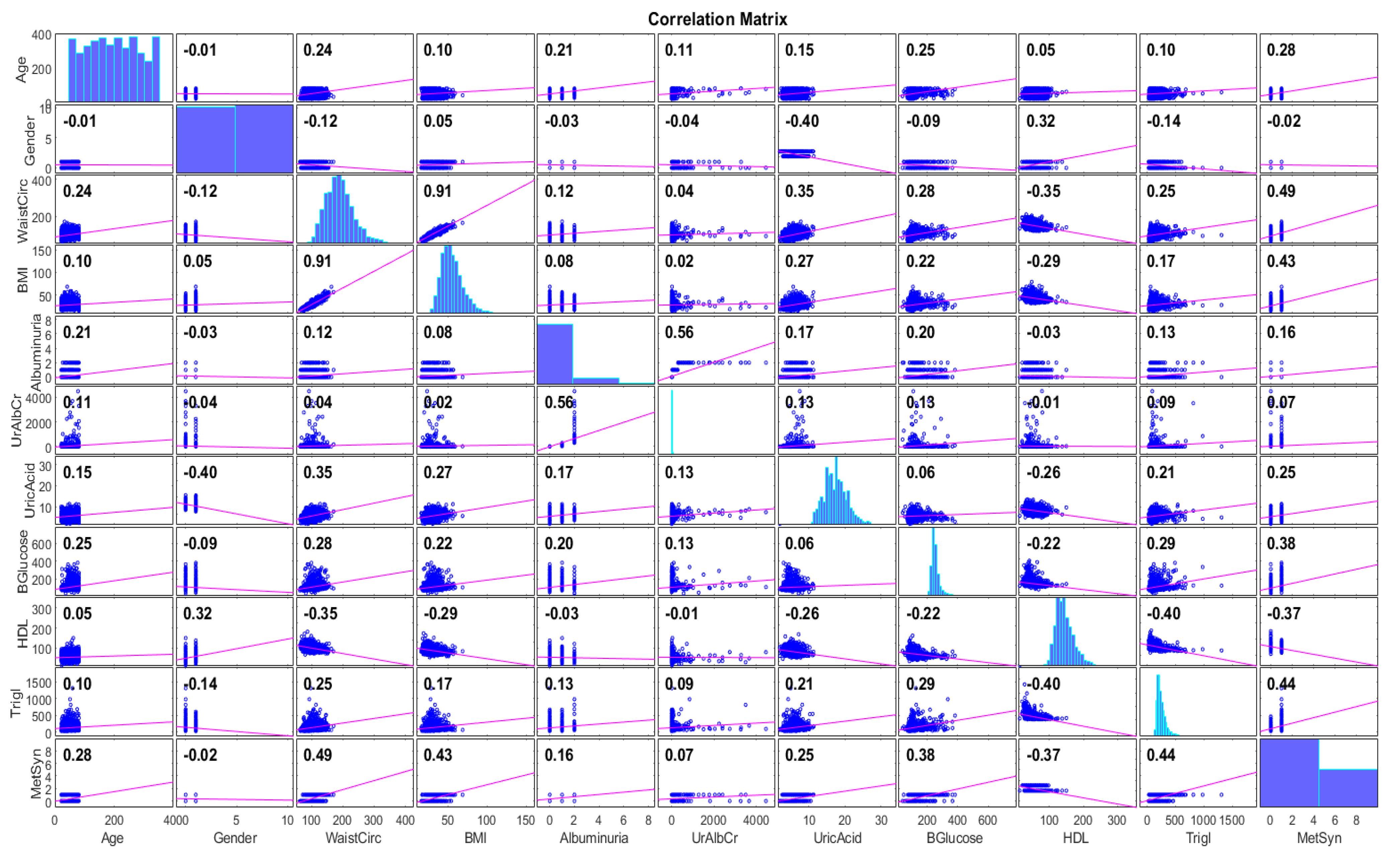

| Attribute | Description | ||
|---|---|---|---|
| Min | Max | Mean ± std Dev | |
| age | 20 | 80 | 49.2 ± 17.4 |
| income | 300 | 9000 | 4147.2 ± 2984.6 |
| waistcir | 63.1 | 170.5 | 98.5 ± 16.3 |
| BMI | 15.7 | 68.7 | 28.7 ± 6.58 |
| albuminuria | 0 | 2 | 0.15 ± 0.41 |
| UrAlbCr | 1.4 | 4462.8 | 42.3 ± 241.4 |
| uric acid | 1.8 | 11.3 | 5.5 ± 1.4 |
| blood glucose | 39 | 382 | 108 ± 33.6 |
| HDL | 14 | 150 | 53.5 ± 15 |
| triglycerides | 26 | 1311 | 126.9 ± 89.8 |
| Model | Parameters |
|---|---|
| LR | use Conjugate Gradient Descent: True |
| J48 | reduced Error Pruning: False save lnstance Data: True use MDL Correction: True subtree Raising: True binary Splits: True collapse Tree: True |
| MLP | learning rate = 0.1 training time = 200 |
| k-NN | k = 3 Search Algorithm: Linear NN Search with Euclidean cross-validate: True |
| NB | use Kernel Estimator: False use Supervised Discretization: True |
| SVM | kernel type: linear |
| RF | break Ties Radomly: True store out of Bag Predictions: True |
| XGBoost | batch Size: 100 num Decimal Places: 2 |
| RotF | classifier: RF number of Groups: True projection Filter: Principal Components |
| Stacking | classifiers: RF and J48 meta Classifier: LR |
| Voting | classifiers: RF and J48 combination Rule: average of probabilities |
| Bagging | classifiers: RF print Classifiers: True store out of Bag Predictions: True |
| Accuracy (%) | Precision | Recall | F1 Score | AUC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No SMOTE | SMOTE | No SMOTE | SMOTE | No SMOTE | SMOTE | No SMOTE | SMOTE | No SMOTE | SMOTE | |
| NB | 84.57 | 84.87 | 0.849 | 0.854 | 0.846 | 0.858 | 0.847 | 0.856 | 0.926 | 0.930 |
| LR | 83.27 | 83.56 | 0.831 | 0.835 | 0.833 | 0.837 | 0.832 | 0.836 | 0.891 | 0.912 |
| SVM | 82.83 | 82.96 | 0.826 | 0.829 | 0.828 | 0.833 | 0.827 | 0.831 | 0.796 | 0.806 |
| MLP | 84.87 | 84.98 | 0.847 | 0.851 | 0.849 | 0.853 | 0.848 | 0.852 | 0.901 | 0.919 |
| 3-NN | 75.91 | 75.96 | 0.754 | 0.757 | 0.759 | 0.763 | 0.756 | 0.760 | 0.779 | 0.789 |
| J-48 | 86.56 | 86.61 | 0.865 | 0.869 | 0.865 | 0.868 | 0.865 | 0.868 | 0.895 | 0.918 |
| RF | 88.93 | 89.15 | 0.890 | 0.894 | 0.890 | 0.894 | 0.890 | 0.894 | 0.958 | 0.962 |
| RotF | 85.51 | 86.71 | 0.854 | 0.868 | 0.854 | 0.867 | 0.854 | 0.867 | 0.937 | 0.944 |
| Stacking | 88.95 | 89.35 | 0.889 | 0.898 | 0.889 | 0.898 | 0.889 | 0.898 | 0.960 | 0.965 |
| Bagging | 88.80 | 89.10 | 0.888 | 0.891 | 0.892 | 0.893 | 0.890 | 0.892 | 0.958 | 0.963 |
| Voting | 87.36 | 87.49 | 0.873 | 0.885 | 0.878 | 0.887 | 0.875 | 0.886 | 0.956 | 0.957 |
| XGBoost | 87.50 | 88.15 | 0.821 | 0.873 | 0.832 | 0.889 | 0.826 | 0.881 | 0.949 | 0.961 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trigka, M.; Dritsas, E. Predicting the Occurrence of Metabolic Syndrome Using Machine Learning Models. Computation 2023, 11, 170. https://doi.org/10.3390/computation11090170
Trigka M, Dritsas E. Predicting the Occurrence of Metabolic Syndrome Using Machine Learning Models. Computation. 2023; 11(9):170. https://doi.org/10.3390/computation11090170
Chicago/Turabian StyleTrigka, Maria, and Elias Dritsas. 2023. "Predicting the Occurrence of Metabolic Syndrome Using Machine Learning Models" Computation 11, no. 9: 170. https://doi.org/10.3390/computation11090170
APA StyleTrigka, M., & Dritsas, E. (2023). Predicting the Occurrence of Metabolic Syndrome Using Machine Learning Models. Computation, 11(9), 170. https://doi.org/10.3390/computation11090170








