Two-Stage Input-Space Image Augmentation and Interpretable Technique for Accurate and Explainable Skin Cancer Diagnosis
Abstract
:1. Introduction
- The integration of geometric and GAN-based augmentation for skin cancer detection;
- In this study, we provide an explainable AI using SHAP to explain how the model makes decisions or predictions.
2. Related Works
3. Materials and Methods
3.1. Dataset
3.2. The Proposed Skin Cancer Detection Method
- Rotation: images can be rotated by a certain angle, either clockwise or counterclockwise.
- Translation: images can be shifted in various directions, both horizontally and vertically.
- Scaling: images can be resized to become larger or smaller.
- Shearing: images can undergo linear distortions, such as changing the angles.
- Flipping: images can be flipped horizontally or vertically.
- Cropping: parts of the image can be cut out to create variations.
- Perspective Distortion: images can undergo perspective distortions to change the viewpoint.
3.3. Design of Experiments
3.4. Performance Metrics
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bozkurt, F. Skin lesion classification on dermatoscopic images using effective data augmentation and pre-trained deep learning approach. Multimed. Tools Appl. 2023, 82, 18985–19003. [Google Scholar] [CrossRef]
- Kalpana, B.; Reshmy, A.; Senthil Pandi, S.; Dhanasekaran, S. OESV-KRF: Optimal ensemble support vector kernel random forest based early detection and classification of skin diseases. Biomed. Signal Process. Control 2023, 85, 104779. [Google Scholar] [CrossRef]
- Girdhar, N.; Sinha, A.; Gupta, S. DenseNet-II: An improved deep convolutional neural network for melanoma cancer detection. Soft Comput. 2023, 27, 13285–13304. [Google Scholar] [CrossRef]
- Gomathi, E.; Jayasheela, M.; Thamarai, M.; Geetha, M. Skin cancer detection using dual optimization based deep learning network. Biomed. Signal Process. Control 2023, 84, 104968. [Google Scholar] [CrossRef]
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Med. Image Anal. 2022, 75, 102305. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.S.; Tembhurne, J.V.; Diwan, T. A multi-class skin Cancer classification using deep convolutional neural networks. Multimed. Tools Appl. 2020, 79, 28477–28498. [Google Scholar] [CrossRef]
- Alsahafi, Y.S.; Kassem, M.A.; Hosny, K.M. Skin-Net: A novel deep residual network for skin lesions classification using multilevel feature extraction and cross-channel correlation with detection of outlier. J. Big Data 2023, 10, 105. [Google Scholar] [CrossRef]
- Mumuni, A.; Mumuni, F. Data augmentation: A comprehensive survey of modern approaches. Array 2022, 16, 100258. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Zhang, Z.; Liu, J.; Feng, Y.; Wee, L.; Dekker, A.; Chen, Q.; Traverso, A. GAN-based one dimensional medical data augmentation. Soft Comput. 2023, 27, 10481–10491. [Google Scholar] [CrossRef]
- Abayomi-Alli, O.O.; Damaševičius, R.; Misra, S.; Maskeliūnas, R.; Abayomi-Alli, A. Malignant skin melanoma detection using image augmentation by oversampling in nonlinear lower-dimensional embedding manifold. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 2600–2614. [Google Scholar] [CrossRef]
- Leguen-deVarona, I.; Madera, J.; Martínez-López, Y.; Hernández-Nieto, J.C. SMOTE-Cov: A New Oversampling Method Based on the Covariance Matrix. In Data Analysis and Optimization for Engineering and Computing Problems: Proceedings of the 3rd EAI International Conference on Computer Science and Engineering and Health Services, Mexico City, Mexico, 28–29 November 2019; Vasant, P., Litvinchev, I., Marmolejo-Saucedo, J.A., Rodriguez-Aguilar, R., Martinez-Rios, F., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 207–215. [Google Scholar]
- Douzas, G.; Bacao, F.; Last, F. Improving imbalanced learning through a heuristic oversampling method based on k-means and SMOTE. Inf. Sci. 2018, 465, 1–20. [Google Scholar] [CrossRef]
- Chang, C.C.; Li, Y.Z.; Wu, H.C.; Tseng, M.H. Melanoma Detection Using XGB Classifier Combined with Feature Extraction and K-Means SMOTE Techniques. Diagnostics 2022, 12, 1747. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Naeem, A.; Malik, H.; Tanveer, J.; Naqvi, R.A.; Lee, S.W. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers 2023, 15, 2179. [Google Scholar] [CrossRef] [PubMed]
- Batista, G.E.A.P.A.; Bazzan, A.L.C.; Monard, M.C. Balancing Training Data for Automated Annotation of Keywords: A Case Study. WOB 2003, 3, 10–18. [Google Scholar]
- Alam, T.M.; Shaukat, K.; Khan, W.A.; Hameed, I.A.; Almuqren, L.A.; Raza, M.A.; Aslam, M.; Luo, S. An Efficient Deep Learning-Based Skin Cancer Classifier for an Imbalanced Dataset. Diagnostics 2022, 12, 2115. [Google Scholar] [CrossRef] [PubMed]
- Sae-Lim, W.; Wettayaprasit, W.; Aiyarak, P. Convolutional Neural Networks Using MobileNet for Skin Lesion Classification. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chonburi, Thailand, 10–12 July 2019; pp. 242–247. [Google Scholar] [CrossRef]
- Alsaidi, M.; Jan, M.T.; Altaher, A.; Zhuang, H.; Zhu, X. Tackling the class imbalanced dermoscopic image classification using data augmentation and GAN. Multimed. Tools Appl. 2023. [CrossRef]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef]
- Ali, I.S.; Mohamed, M.F.; Mahdy, Y.B. Data Augmentation for Skin Lesion using Self-Attention based Progressive Generative Adversarial Network. arXiv 2019, arXiv:1910.11960. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Nets. In Proceedings of the Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- Shamsolmoali, P.; Zareapoor, M.; Shen, L.; Sadka, A.H.; Yang, J. Imbalanced data learning by minority class augmentation using capsule adversarial networks. Neurocomputing 2021, 459, 481–493. [Google Scholar] [CrossRef]
- Shahin, M.; Chen, F.F.; Hosseinzadeh, A.; Khodadadi Koodiani, H.; Shahin, A.; Ali Nafi, O. A smartphone-based application for an early skin disease prognosis: Towards a lean healthcare system via computer-based vision. Adv. Eng. Inform. 2023, 57, 102036. [Google Scholar] [CrossRef]
- Bhandari, M.; Shahi, T.B.; Neupane, A. Evaluating Retinal Disease Diagnosis with an Interpretable Lightweight CNN Model Resistant to Adversarial Attacks. J. Imaging 2023, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Shan, P.; Chen, J.; Fu, C.; Cao, L.; Tie, M.; Sham, C.W. Automatic skin lesion classification using a novel densely connected convolutional network integrated with an attention module. J. Ambient. Intell. Humaniz. Comput. 2023, 14, 8943–8956. [Google Scholar] [CrossRef]
- Alwakid, G.; Gouda, W.; Humayun, M.; Jhanjhi, N.Z. Diagnosing Melanomas in Dermoscopy Images Using Deep Learning. Diagnostics 2023, 13, 1815. [Google Scholar] [CrossRef] [PubMed]
- Ameri, A. A Deep Learning Approach to Skin Cancer Detection in Dermoscopy Images. J. Biomed. Phys. Eng. 2020, 10, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.S.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M.K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Sevli, O. A deep convolutional neural network-based pigmented skin lesion classification application and experts evaluation. Neural Comput. Appl. 2021, 33, 12039–12050. [Google Scholar] [CrossRef]
- Fraiwan, M.; Faouri, E. On the Automatic Detection and Classification of Skin Cancer Using Deep Transfer Learning. Sensors 2022, 22, 4963. [Google Scholar] [CrossRef]
- Balambigai, S.; Elavarasi, K.; Abarna, M.; Abinaya, R.; Vignesh, N.A. Detection and optimization of skin cancer using deep learning. J. Phys. Conf. Ser. 2022, 2318, 012040. [Google Scholar] [CrossRef]
- Shaheen, H.; Singhn, M.P. Multiclass skin cancer classification using particle swarm optimization and convolutional neural network with information security. J. Electron. Imaging 2022, 32, 042102. [Google Scholar] [CrossRef]
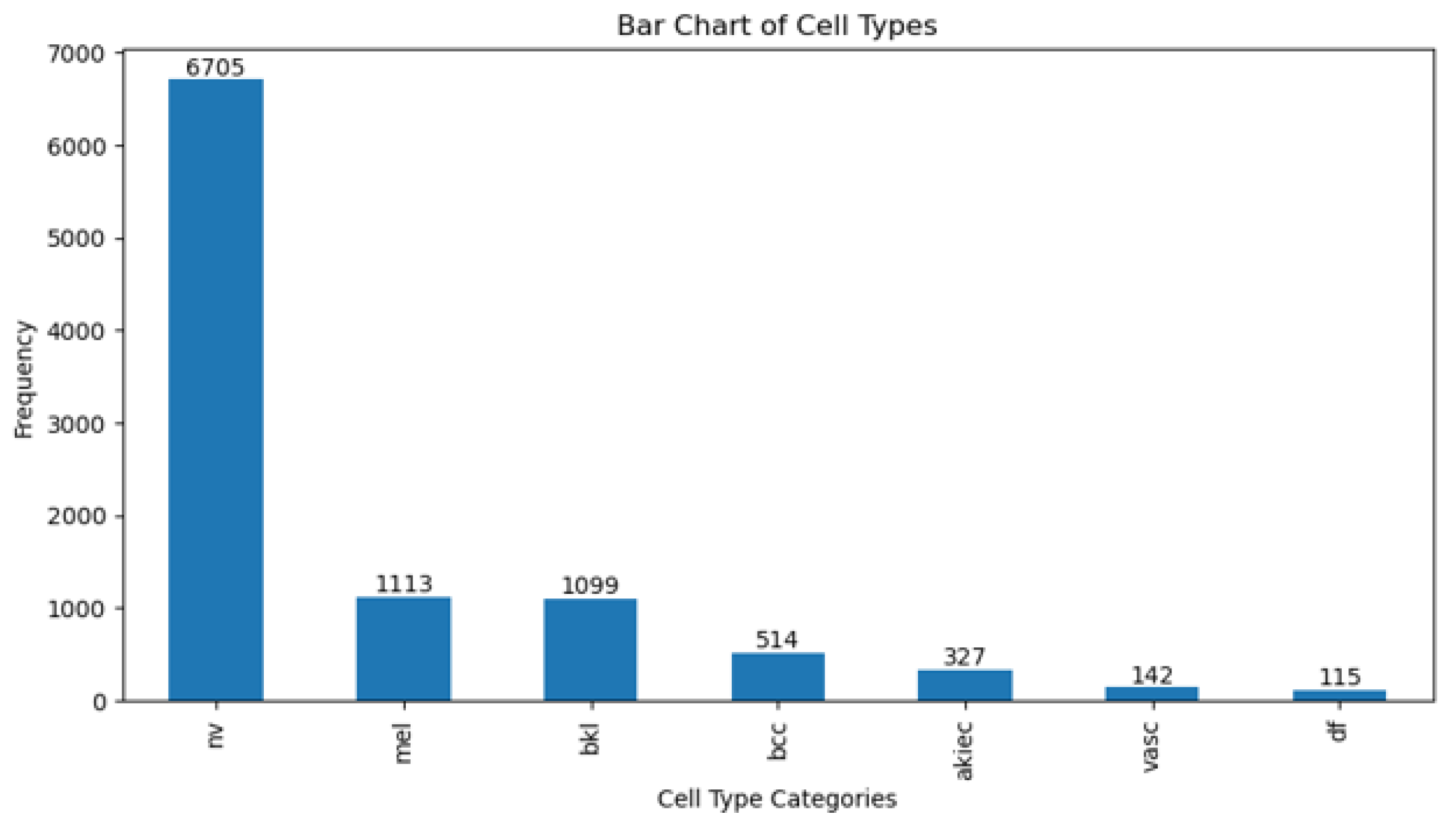

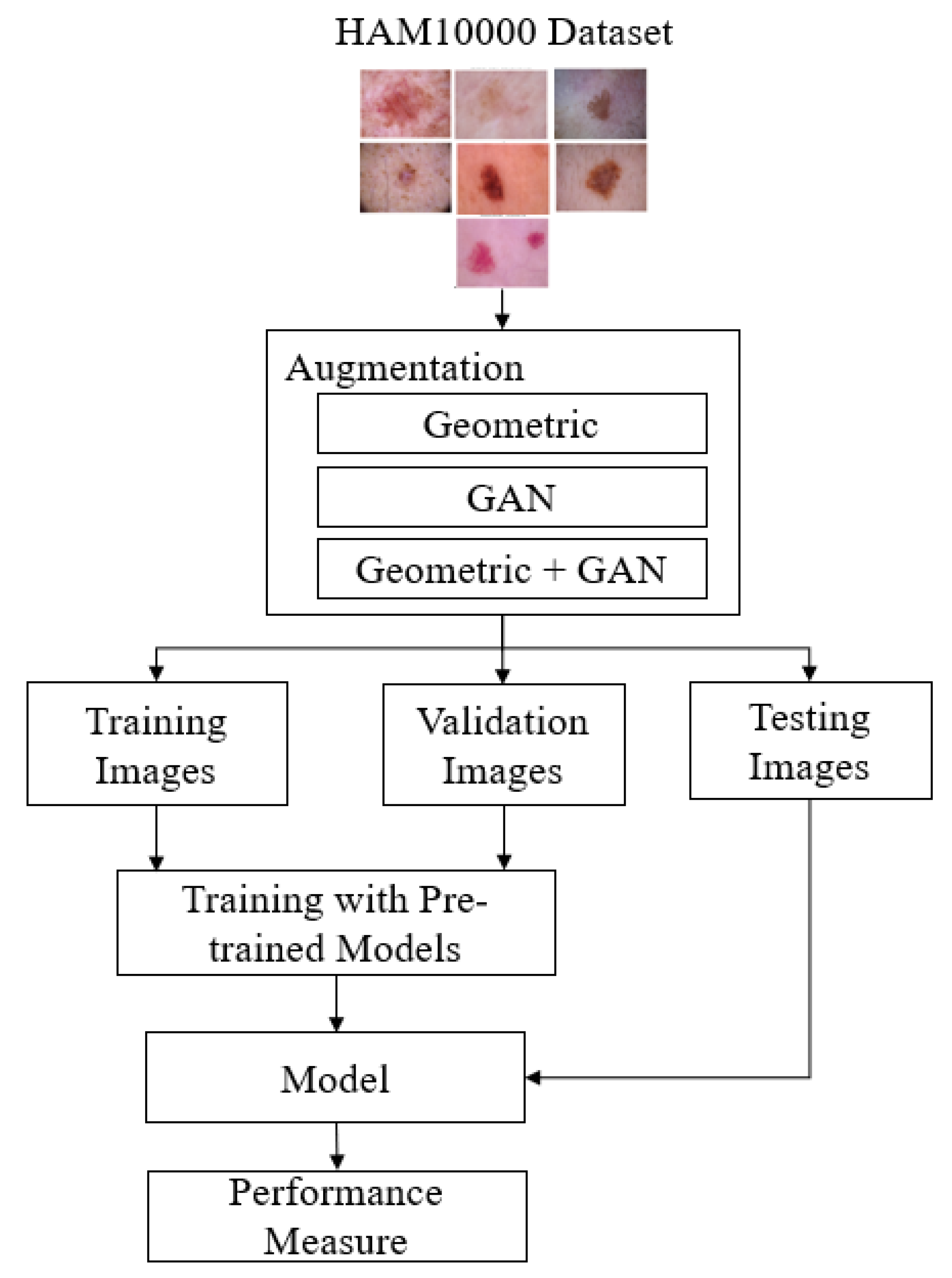



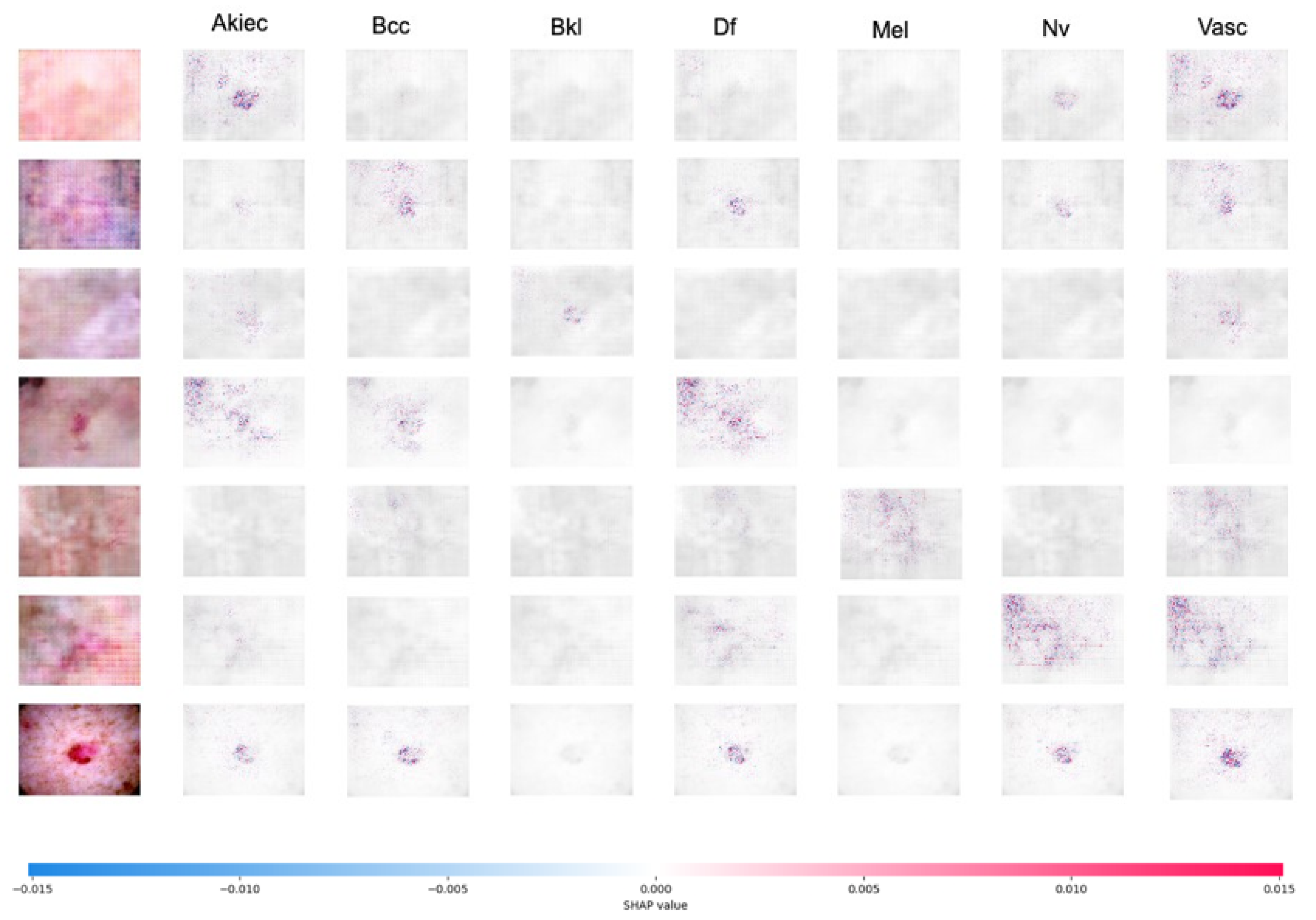
| Parameter | Value |
|---|---|
| rotation_range | 20 |
| width_shift_range | 0.2 |
| height_shift_range | 0.2 |
| shear_range | 0.2 |
| zoom_range | 0.2 |
| horizontal_flip | True |
| brightness_range | (0.8, 1.2) |
| Layer | Activation | |
|---|---|---|
| Discriminator | Conv2D | LeakyReLU |
| Conv2D | LeakyReLU | |
| Conv2D | LeakyReLU | |
| Conv2D | LeakyReLU | |
| Flatten | ||
| Dropout | ||
| Dense | Sigmoid | |
| Generator | Dense | LeakyReLU |
| Conv2DTranspose | LeakyReLU | |
| Conv2DTranspose | LeakyReLU | |
| Conv2DTranspose | LeakyReLU | |
| Conv2D | Tanh |
| Parameter | Value |
|---|---|
| Optimizer | Adam |
| Learning rate | 0.0001 |
| Optimizer parameters | beta_1 = 0.9, beta_2 = 0.999 |
| Epochs | 100 (with early stopping) |
| Layer | Output Shape | Activation |
|---|---|---|
| Dense | (None, 64) | Relu |
| Dense | (None, 32) | Relu |
| Dense | (None, 7) | Softmax |
| Original | Geometric Aug. | GAN | Geometric Aug.+GAN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Category | Train | Test | Val | Train | Test | Val | Train | Test | Val | Train | Test | Val |
| vasc | 110 | 26 | 6 | 4801 | 1350 | 554 | 4843 | 1359 | 503 | 4805 | 1371 | 529 |
| nv | 4822 | 1347 | 536 | 4826 | 1316 | 563 | 4854 | 1302 | 549 | 4856 | 1300 | 549 |
| mel | 792 | 222 | 99 | 4877 | 1303 | 525 | 4858 | 1319 | 528 | 4836 | 1361 | 508 |
| df | 83 | 25 | 7 | 4775 | 1423 | 507 | 4831 | 1325 | 549 | 4831 | 1340 | 534 |
| bkl | 785 | 224 | 90 | 4887 | 1316 | 502 | 4813 | 1329 | 563 | 4831 | 1337 | 537 |
| bcc | 370 | 101 | 43 | 4783 | 1360 | 562 | 4792 | 1387 | 526 | 4798 | 1394 | 513 |
| akiec | 248 | 58 | 21 | 4844 | 1319 | 542 | 4802 | 1366 | 537 | 4836 | 1284 | 585 |
| Num. images | 7210 | 2003 | 802 | 33,793 | 9387 | 3755 | 33,793 | 9387 | 3755 | 33,793 | 9387 | 3755 |
| Total images | 10,015 | 46,935 | 46,935 | 46,935 | ||||||||
| Augmentation Method | Pre-Trained Model | Acc | Prec | Rec | F1 | Sensitivity AtSpecificity | Specificity AtSensitivity | G-Mean | Epoch |
|---|---|---|---|---|---|---|---|---|---|
| Original Data | Xception | 79.93 | 80.70 | 79.53 | 80.11 | 99.16 | 91.71 | 95.36 | 12 |
| Inceptionv3 | 78.88 | 79.51 | 78.48 | 78.99 | 99.31 | 92.31 | 95.75 | 11 | |
| Resnet152v2 | 84.12 | 84.77 | 83.67 | 84.22 | 99.33 | 93.56 | 96.40 | 18 | |
| EfficientnetB7 | 78.03 | 79.63 | 77.28 | 78.44 | 99.49 | 94.91 | 97.17 | 11 | |
| InceptionresnetV2 | 79.63 | 80.20 | 78.68 | 79.44 | 99.28 | 91.76 | 95.45 | 19 | |
| VGG19 | 81.73 | 81.83 | 81.63 | 81.73 | 99.12 | 91.26 | 95.11 | 28 | |
| Geometric | Xception | 97.05 | 97.06 | 97.01 | 97.03 | 99.87 | 99.12 | 99.49 | 19 |
| Inceptionv3 | 97.38 | 97.48 | 97.35 | 97.41 | 99.90 | 99.20 | 99.55 | 31 | |
| Resnet152v2 | 96.90 | 96.95 | 96.86 | 96.90 | 99.85 | 98.93 | 99.39 | 28 | |
| EfficientnetB7 | 97.95 | 98.00 | 97.90 | 97.95 | 99.91 | 99.41 | 99.66 | 19 | |
| InceptionresnetV2 | 97.40 | 97.46 | 97.36 | 97.41 | 99.89 | 99.20 | 99.55 | 28 | |
| VGG19 | 97.22 | 97.24 | 97.20 | 97.22 | 99.83 | 98.84 | 99.33 | 32 | |
| GAN | Xception | 96.08 | 96.35 | 95.96 | 96.16 | 99.86 | 98.70 | 99.28 | 10 |
| Inceptionv3 | 96.50 | 96.62 | 96.45 | 96.53 | 99.86 | 98.64 | 99.25 | 16 | |
| Resnet152v2 | 96.30 | 96.47 | 96.23 | 96.35 | 99.79 | 98.37 | 99.08 | 20 | |
| EfficientnetB7 | 96.48 | 96.59 | 96.44 | 96.51 | 99.79 | 98.25 | 99.02 | 20 | |
| InceptionresnetV2 | 96.22 | 96.32 | 96.20 | 96.26 | 99.82 | 98.44 | 99.13 | 18 | |
| VGG19 | 96.22 | 96.26 | 96.20 | 96.23 | 99.70 | 100.00 | 99.85 | 38 | |
| Geometric + GAN | Xception | 96.21 | 96.51 | 96.04 | 96.27 | 99.94 | 99.22 | 99.58 | 9 |
| Inceptionv3 | 96.45 | 96.56 | 96.39 | 96.48 | 99.86 | 98.56 | 99.21 | 20 | |
| Resnet152v2 | 96.59 | 96.75 | 96.45 | 96.60 | 99.86 | 98.87 | 99.36 | 14 | |
| EfficientnetB7 | 96.50 | 96.61 | 96.43 | 96.52 | 99.89 | 98.92 | 99.40 | 14 | |
| InceptionresnetV2 | 96.71 | 96.82 | 96.67 | 96.74 | 99.85 | 98.62 | 99.23 | 21 | |
| VGG19 | 95.39 | 97.36 | 93.89 | 95.59 | 100.00 | 99.93 | 99.96 | 17 |
| Augmentation Method | Pre-Trained Model | Acc | Prec | Rec | F1 | Sensitivity AtSpecificity | Specificity AtSensitivity | G-Mean | Epoch |
|---|---|---|---|---|---|---|---|---|---|
| Geometric | EfficientnetB7 | 98.07 | 98.10 | 98.06 | 98.08 | 99.92 | 99.46 | 99.69 | 20 |
| GAN | Inceptionv3 | 96.48 | 96.63 | 96.44 | 96.53 | 99.83 | 98.54 | 99.18 | 17 |
| Geometric + GAN | InceptionresnetV2 | 96.90 | 97.07 | 96.87 | 96.97 | 99.86 | 98.90 | 99.38 | 22 |
| Ref. | Method | Acc | Prec | Rec | F1 | Stdev |
|---|---|---|---|---|---|---|
| Alam et al. [17] | AlexNet, InceptionV3, and RegNetY-320 | 91 | - | - | 88.1 | - |
| Kalpana et al. [2] | ESVMKRF-HEAO | 97.4 | 96.3 | 95.9 | 97.4 | 0.7767 |
| Shan et al. [26] | AttDenseNet-121 | 98 | 91.8 | 85.4 | 85.6 | 6.3003 |
| Gomathi et al. [4] | DODL net | 98.76 | 96.02 | 95.37 | 94.32 | 1.7992 |
| Alwakid et al. [27] | InceptionResnet-V2 | 91.26 | 91 | 91 | 91 | 0.1501 |
| Sae-Lim et al. [18] | Modified MobileNet | 83.23 | - | 85 | 82 | - |
| Ameri [28] | AlexNet | 84 | - | - | - | - |
| Chaturvedi et al. [6] | ResNeXt101 | 93.2 | 88 | 88 | - | 3.0022 |
| Shahin Ali et al. [29] | DCNN | 91.43 | 96.57 | 93.66 | 95.09 | 2.5775 |
| Sevli et al. [30] | Custom CNN architecture | 91.51 | - | - | - | - |
| Fraiwan et al. [31] | DenseNet201 | 82.9 | 78.5 | 73.6 | 74.4 | 4.6522 |
| Balambigai et al. [32] | Grid search ensemble | 77.17 | - | - | - | - |
| Shaheen et al. [33] | PSOCNN | 97.82 | - | - | 98 | - |
| This study | Geometric+EfficientnetB7+Custom FC | 98.07 | 98.10 | 98.06 | 98.08 | 0.0002 |
| GAN+InceptionV3 | 96.50 | 96.62 | 96.45 | 96.53 | 0.0009 | |
| Geometric+GAN+InceptionresnetV2+Custom FC | 96.90 | 97.07 | 96.87 | 96.97 | 0.0011 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Supriyanto, C.; Salam, A.; Zeniarja, J.; Wijaya, A. Two-Stage Input-Space Image Augmentation and Interpretable Technique for Accurate and Explainable Skin Cancer Diagnosis. Computation 2023, 11, 246. https://doi.org/10.3390/computation11120246
Supriyanto C, Salam A, Zeniarja J, Wijaya A. Two-Stage Input-Space Image Augmentation and Interpretable Technique for Accurate and Explainable Skin Cancer Diagnosis. Computation. 2023; 11(12):246. https://doi.org/10.3390/computation11120246
Chicago/Turabian StyleSupriyanto, Catur, Abu Salam, Junta Zeniarja, and Adi Wijaya. 2023. "Two-Stage Input-Space Image Augmentation and Interpretable Technique for Accurate and Explainable Skin Cancer Diagnosis" Computation 11, no. 12: 246. https://doi.org/10.3390/computation11120246
APA StyleSupriyanto, C., Salam, A., Zeniarja, J., & Wijaya, A. (2023). Two-Stage Input-Space Image Augmentation and Interpretable Technique for Accurate and Explainable Skin Cancer Diagnosis. Computation, 11(12), 246. https://doi.org/10.3390/computation11120246





