Feature Selection and Recognition Methods for Discovering Physiological and Bioinformatics RESTful Services
Abstract
:1. Introduction
2. Related Work
3. Datasets and Tools
3.1. Dependent Libraries
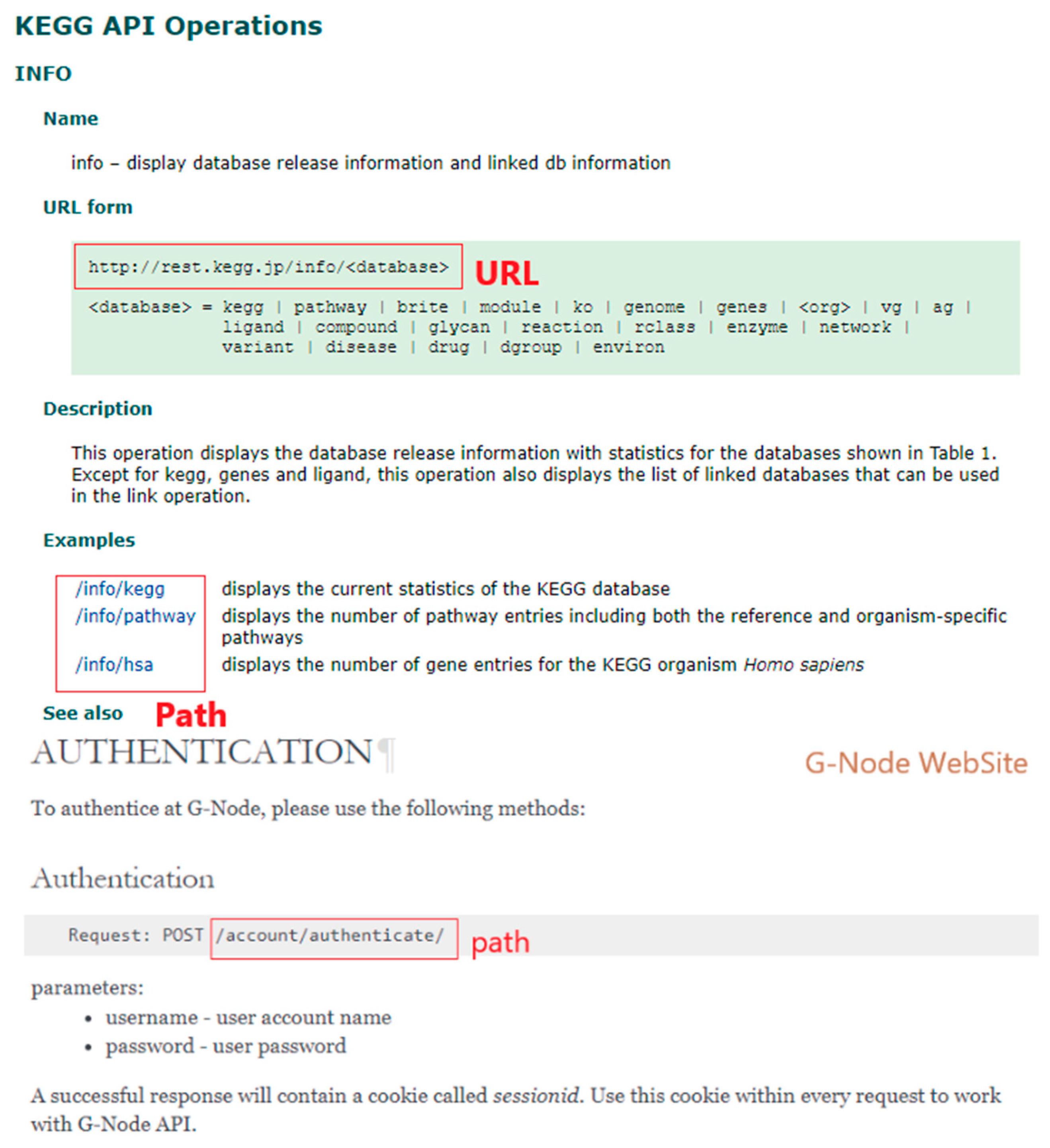
3.2. Data Acquisition
3.3. Datasets
4. Implementation Details
4.1. Web Crawler
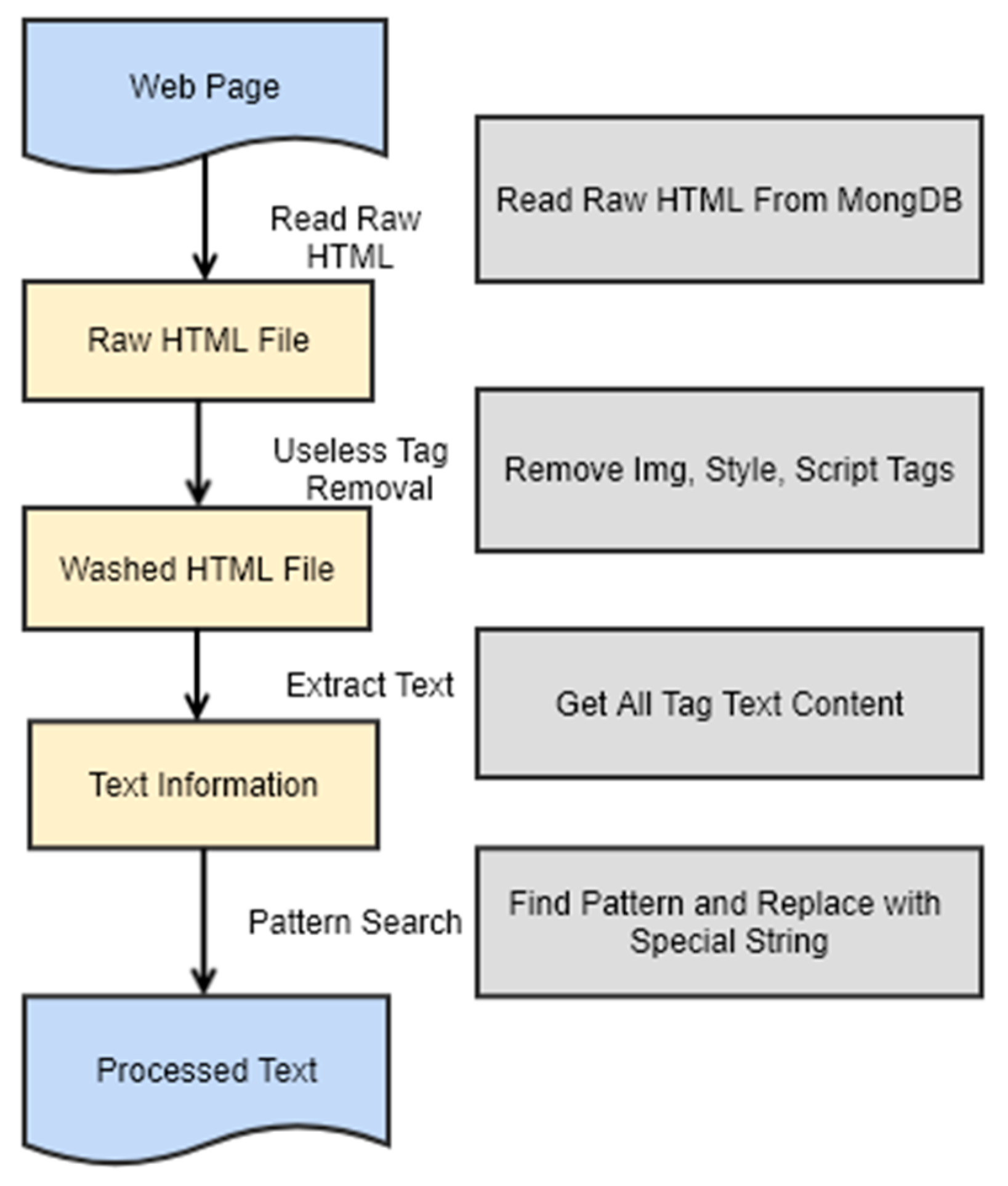
4.2. Preprocessing of Web Pages
4.3. Feature Extraction and Selection
| Algorithm 1 The detail process of dictionary initialization |
| Input: the text T, stop words , the Threshold of DF , Output: the set of words |
| for each text : lowercase () tokenize () end create for each words array + = set () end for each word in , : document_frequency () if or : remove from endif end return |
4.4. Classification Method
5. Results and Discussion
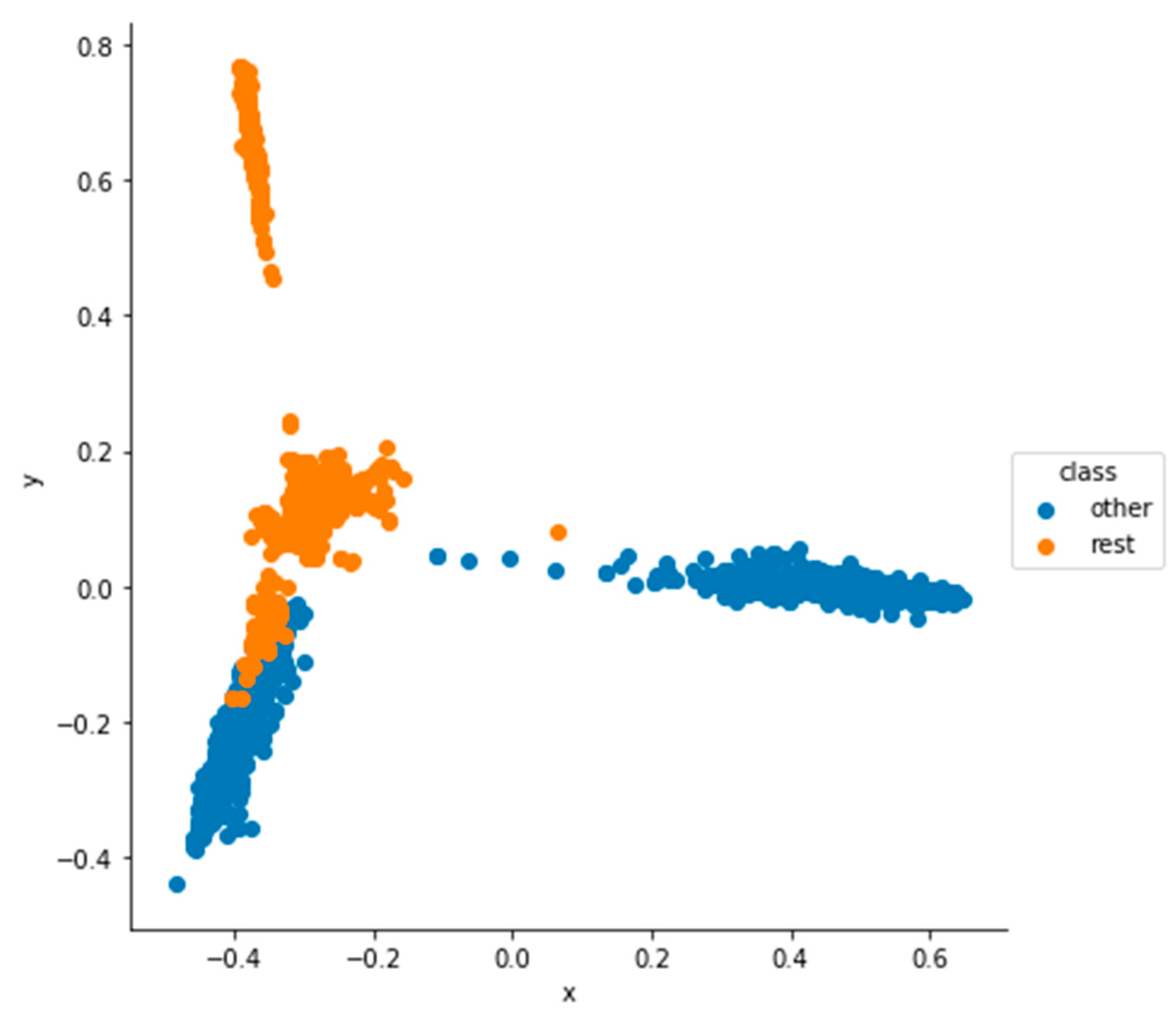
5.1. Dimensionality Reduction and Visualization Results
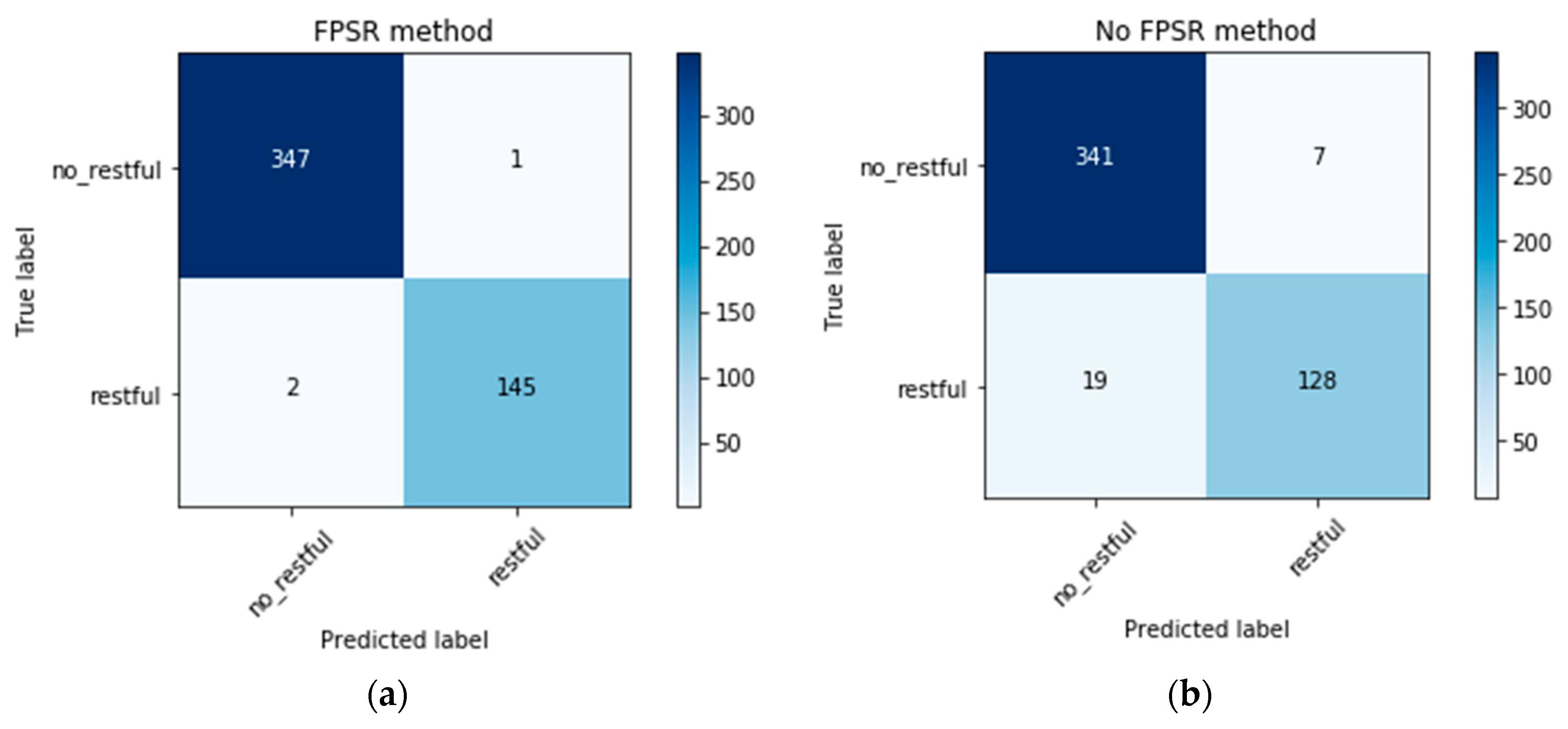
5.2. Classification Results
6. Conclusions and Future Work
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lamine, S.B.A.B.; Zghal, H.B.; Mrissa, M.; Guegan, C.G. An ontology-based approach for personalized restful web service discovery. Procedia Comput. Sci. 2017, 112, 2127–2136. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, J.; He, K.; Li, Z.; Huang, Y. Mining and clustering service goals for restful service discovery. Knowl. Inf. Syst. 2018, 1–32. [Google Scholar] [CrossRef]
- Fielding, R.T.; Taylor, R.N. Architectural Styles and the Design of Network-Based Software Architectures. Ph.D. Thesis, University of California, Irvine, CA, USA, 2000. [Google Scholar]
- Haupt, F.; Fischer, M.; Karastoyanova, D.; Leymann, F.; Vukojevichaupt, K. Service composition for REST. In Proceedings of the IEEE 18th International Enterprise Distributed Object Computing Conference (EDOC 2014), Ulm, Germany, 3–5 September 2014; pp. 110–119. [Google Scholar]
- Crasso, M.; Rodriguez, J.M.; Zunino, A.; Campo, M. Revising WSDL documents: Why and how. IEEE Internet Comput. 2010, 14, 48–56. [Google Scholar] [CrossRef]
- Sheth, A.P.; Gomadam, K.; Lathem, J. Sa-rest: Semantically interoperable and easier-to-use services and mashups. IEEE Internet Comput. 2007, 11, 91–94. [Google Scholar] [CrossRef]
- Pedrinaci, C.; Domingue, J. Toward the next wave of services: Linked services for the web of data. J. UCS 2010, 16, 1694–1719. [Google Scholar]
- Roman, D.; Kopecký, J.; Vitvar, T.; Domingue, J.; Fensel, D. Wsmo-lite and hrests: Lightweight semantic annotations for web services and restful apis. Web Semant. Sci. Serv. Agents World Wide Web 2015, 31, 39–58. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Dong, L.; Lin, R.; Yan, D.; Li, J. Towards effectively identifying restful web services. In Proceedings of the 2014 IEEE 21st International Conference on Web Services (ICWS 2014), Anchorage, AK, USA, 27 June–2 July 2014; pp. 518–525. [Google Scholar]
- Qi, X.; Davison, B.D. Web page classification: Features and algorithms. ACM Comput. Surv. (CSUR) 2009, 41, 12. [Google Scholar] [CrossRef]
- Asatkar, B.P.; Wagh, K.; Chatur, P. Classification of web pages using naive bayesian approach. In Proceedings of the International Conference on Recent Trends in Engineering Sciences 2017 (ICRTES 2017), Namakkal, India, 25 March 2017; p. 21. [Google Scholar]
- Belmouhcine, A.; Benkhalifa, M. Implicit links-based techniques to enrich k-nearest neighbors and naive bayes algorithms for web page classification. In Proceedings of the 9th International Conference on Computer Recognition Systems (CORES 2015), Wroclaw, Poland, 25–27 May 2015; pp. 755–766. [Google Scholar]
- Mohamed, T.A.A.A.D.; Abu-Kresha, T.; Bakry, A.N.R. An efficient method for web page classification based on text. Int. J. Eng. Comput. Sci. 2017, 6, 21854–21860. [Google Scholar]
- Kan, M.Y.; Thi, H.O.N. Fast webpage classification using url features. In Proceedings of the 14th ACM Conference on Information and Knowledge Management (CIKM 2005), Bremen, Germany, 31 October–5 November, 2005; pp. 325–326. [Google Scholar]
- Hu, R.; Xiao, L.; Zheng, W. Drug related webpages classification using images and text information based on multi-kernel learning. In Proceedings of the SPIE 9th International Symposium on Multispectral Image Processing and Pattern Recognition (MIPPR 2015), Enshi, China, 31 October–1 November 2015. [Google Scholar]
- Rajalakshmi, R.; Xaviar, S. Experimental study of feature weighting techniques for url based webpage classification. Procedia Comput. Sci. 2017, 115, 218–225. [Google Scholar] [CrossRef]
- Altay, B.; Dokeroglu, T.; Cosar, A. Context-sensitive and keyword density-based supervised machine learning techniques for malicious webpage detection. Soft Comput. 2018, 1–15. [Google Scholar] [CrossRef]
- Siddiqui, A.; Adnan, M.; Siddiqui, R.A.; Mubeen, T. A comparative study of web pages classification methods applied to health consumer web pages. In Proceedings of the 2015 Second International Conference on Computing Technology and Information Management (ICCTIM 2015), Johor, Malaysia, 21–23 April 2015; pp. 43–48. [Google Scholar]
- Onan, A. Classifier and feature set ensembles for web page classification. J. Inf. Sci. 2016, 42, 150–165. [Google Scholar] [CrossRef]
- Kiziloluk, S.; Ozer, A.B. Web pages classification with parliamentary optimization algorithm. Int. J. Softw. Eng. Knowl. Eng. 2017, 27, 499–513. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, H.; Li, W.; Uludag, M.; Squizzato, S.; Park, Y.M.; Buso, N.; Cowley, A.P.; Lopez, R. Analysis tool web services from the embl-ebi. Nucleic Acids Res. 2013, 41, W597–W600. [Google Scholar] [CrossRef] [PubMed]
- Herz, A.V.; Meier, R.; Nawrot, M.P.; Schiegel, W.; Zito, T. G-node: An integrated tool-sharing platform to support cellular and systems neurophysiology in the age of global neuroinformatics. Neural Netw. 2008, 21, 1070–1075. [Google Scholar] [CrossRef] [PubMed]
- Uğuz, H. A two-stage feature selection method for text categorization by using information gain, principal component analysis and genetic algorithm. Knowl. Based Syst. 2011, 24, 1024–1032. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Wang, W.; Wang, B.; Yang, B.; Liu, Y. A method for topic classification of web pages using LDA-SVM model. In Proceedings of the 2017 Chinese Intelligent Automation Conference (CIAC’17), Tianjin, China, 27–29 October 2017; pp. 589–596. [Google Scholar]
- Basnet, R.; Mukkamala, S.; Sung, A.H. Detection of phishing attacks: A machine learning approach. In Soft Computing Applications in Industry; Springer: Berlin/Heidelberg, Germany, 2008; pp. 373–383. [Google Scholar]



| Feature Pattern | Regular Expression | Custom String |
|---|---|---|
| http://locahost:8080/path1/path2 | (https?)://[-A-Za-z0-9+&@#/%?=~_|!:,.;]+[-A-Za-z0-9+&@#/%=~_|] | httpaddr |
| username@email.com | ([a-zA-Z0-9_.+-]+@[a-zA-RZ0-9-]+.[a-zA-Z]+) | emailaddr |
| /path1/path2 | s(/[w-]+)+/? | pathaddr |
| Precision | Recall | f1-Socre | |
|---|---|---|---|
| no_restful | 0.994 | 0.997 | 0.996 |
| restful | 0.993 | 0.986 | 0.990 |
| avg/total | 0.994 | 0.994 | 0.994 |
| Precision | Recall | f1-Socre | |
|---|---|---|---|
| no_restful | 0.947 | 0.980 | 0.963 |
| restful | 0.948 | 0.871 | 0.908 |
| avg/total | 0.947 | 0.947 | 0.947 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, C.; Gao, X.; Wang, Y.; Li, J.; Fan, L.; Qin, X.; Zhou, Q.; Wang, Z.; Huang, L. Feature Selection and Recognition Methods for Discovering Physiological and Bioinformatics RESTful Services. Information 2018, 9, 227. https://doi.org/10.3390/info9090227
Song C, Gao X, Wang Y, Li J, Fan L, Qin X, Zhou Q, Wang Z, Huang L. Feature Selection and Recognition Methods for Discovering Physiological and Bioinformatics RESTful Services. Information. 2018; 9(9):227. https://doi.org/10.3390/info9090227
Chicago/Turabian StyleSong, Chao, Xinyu Gao, Yongqian Wang, Jinhai Li, Lifeng Fan, Xiaohuang Qin, Qiao Zhou, Zhongyi Wang, and Lan Huang. 2018. "Feature Selection and Recognition Methods for Discovering Physiological and Bioinformatics RESTful Services" Information 9, no. 9: 227. https://doi.org/10.3390/info9090227
APA StyleSong, C., Gao, X., Wang, Y., Li, J., Fan, L., Qin, X., Zhou, Q., Wang, Z., & Huang, L. (2018). Feature Selection and Recognition Methods for Discovering Physiological and Bioinformatics RESTful Services. Information, 9(9), 227. https://doi.org/10.3390/info9090227





