Automated Detection of Liver Histopathological Findings Based on Biopsy Image Processing
Abstract
:1. Introduction
1.1. Types of Fatty Liver
1.2. Causes of Liver Steatosis
- (1)
- Drugs. Cortisone, synthetic estrogens, contraceptives, amiodarone (Angoron), tamoxifen, and tetracyclines when consumed for a long time may cause hepatic steatosis [5].
- (2)
- Diabetes. Chances for fat deposition in the liver increases in cases where diabetes remains unregulated [5].
- (3)
- Obesity. Liver steatosis is caused by central obesity characterized by increased fat deposition in the abdomen [5].
- (4)
- Sudden weight loss. Crash diets leading to rapid weight loss can also cause fat deposition in the liver.
- (5)
- Rare causes. A series of diseases such as hepatitis C, Crohn’s disease, ulcerative colitis, Wilson’s disease and avitalipoproteinaimia are also considered rare causes for hepatic steatosis.
1.3. Diagnosis and Identification of Steatosis
1.4. Related Work
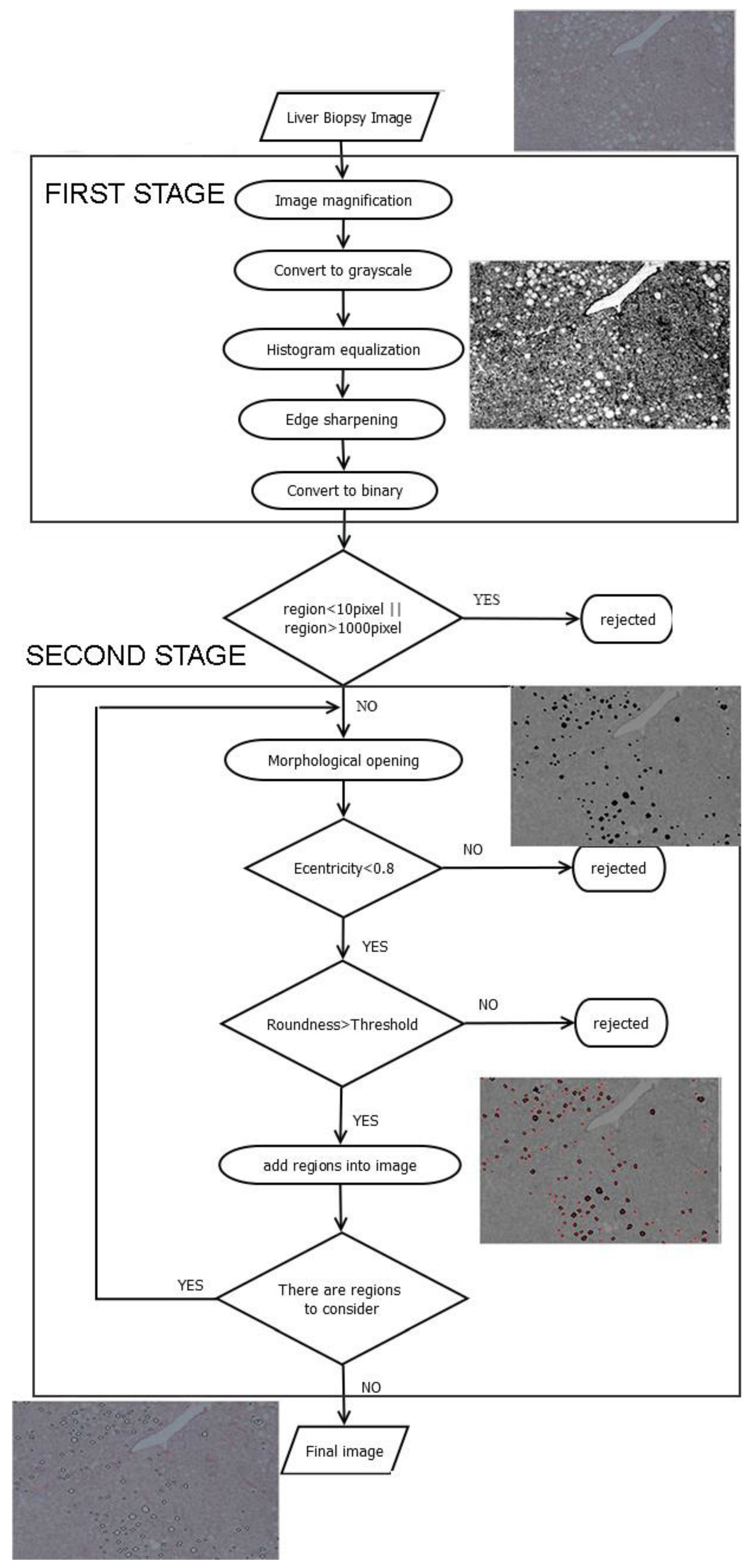
2. Description of Methodology
2.1. Image Preprocessing
- i
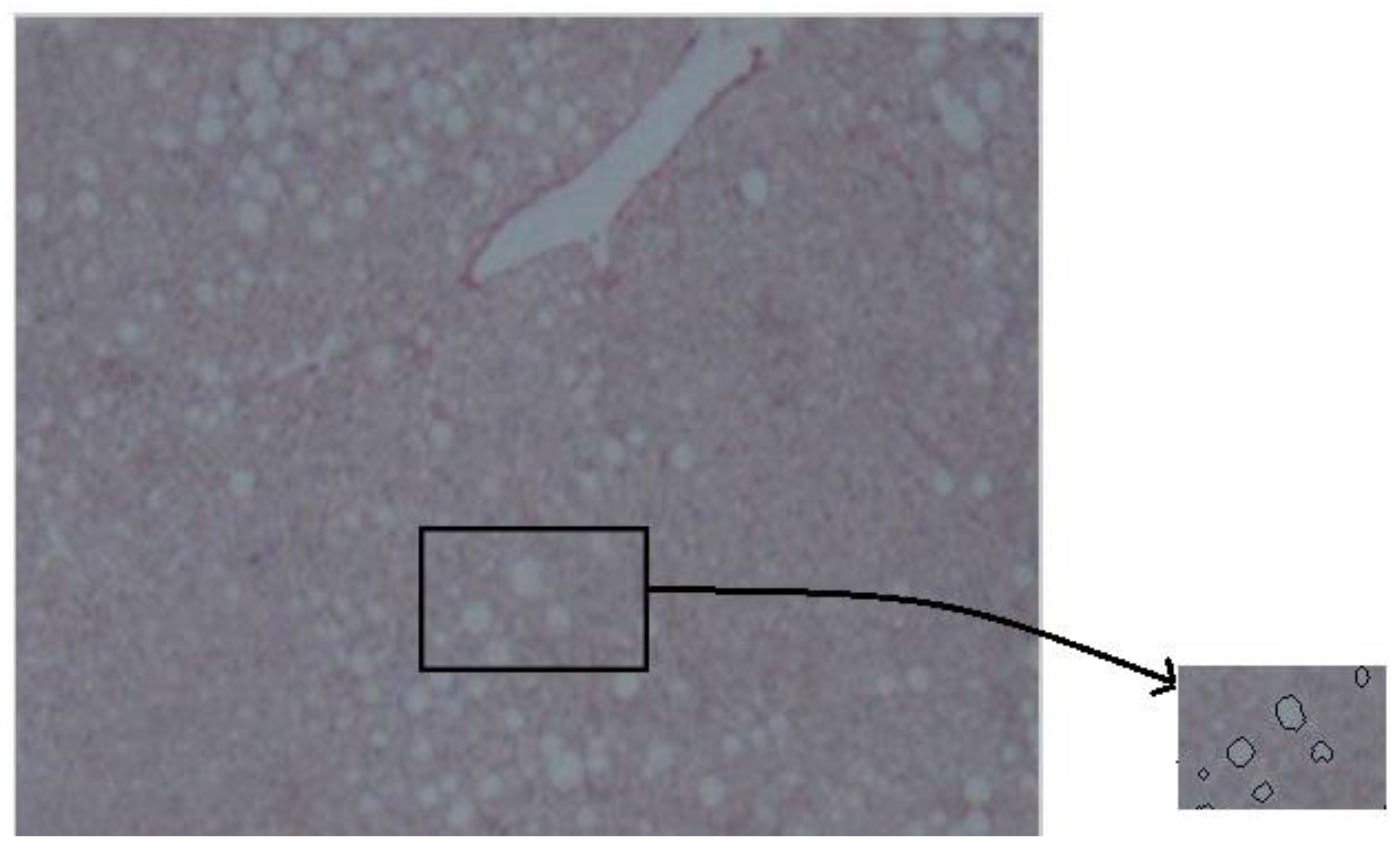
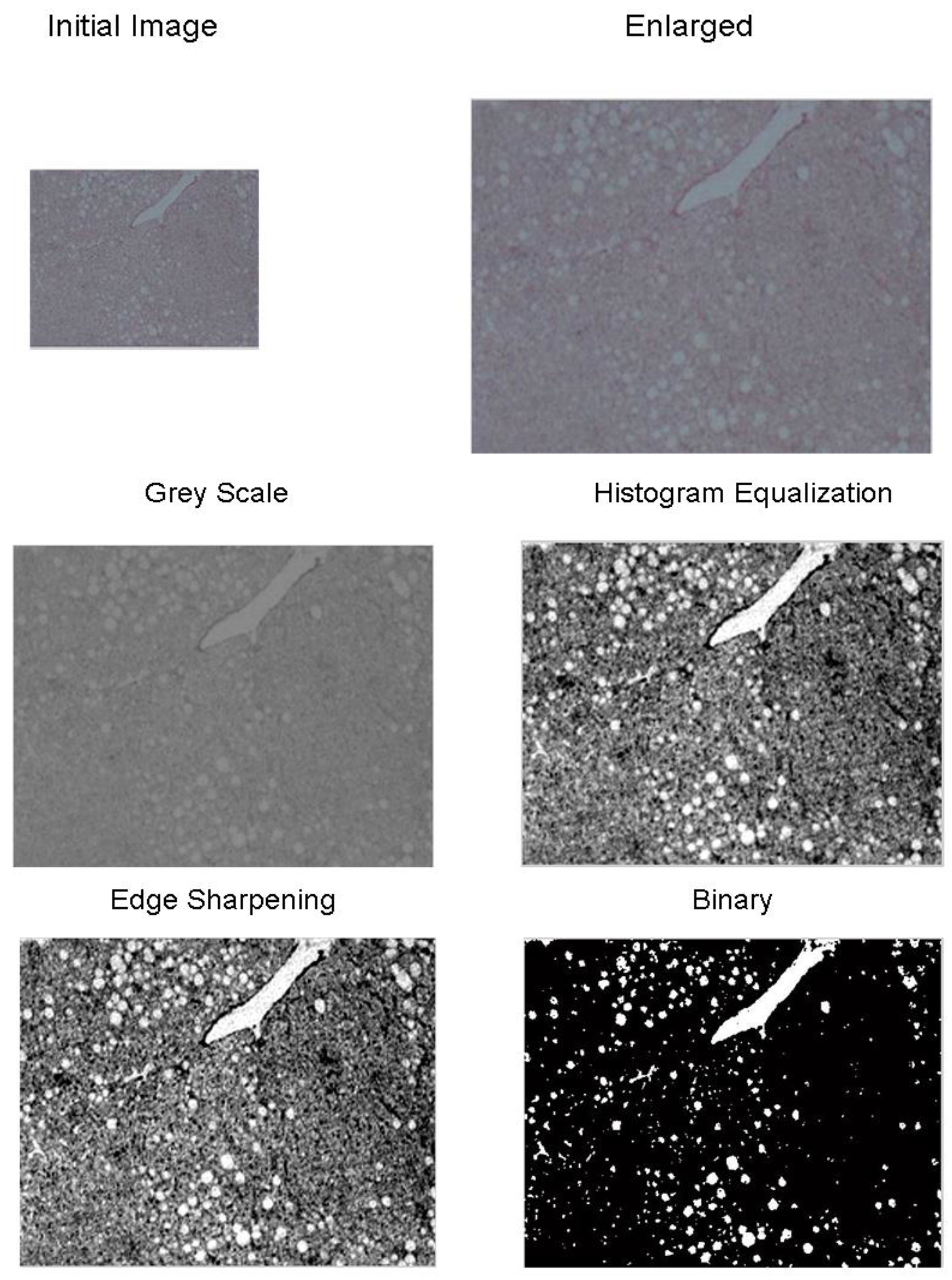
- Image magnification. The methodology is designed to process low-resolution images. Thus, in the first step of the preprocessing stage the image is enlarged by 2×, to make more visible the joined regions. Bicubic interpolation is employed to calculate the additional pixels. A weighted average of pixels in the nearest 4-by-4 neighborhood is the new value of each pixel.
- ii
- Convert to grayscale. The image is converted from red, green, blue (RGB) to grayscale, using a weighted sum of R, G and Β:where I_grayscale is the grayscale image and R, G, and B are the intensity values of each RGB channel, respectively [25].
- iii
- Histogram equalization. Histogram normalization is used to adjust the brightness of the image.
- iv
- Edge sharpening. This step is done by using the unsharp masking method which returns an upgraded version of the grayscale image, where the edges and features have been sharpened.
- v
- Convert to binary. Finally, the image is converted to binary, using histogram thresholding. The threshold was defined based on a trial-and-error approach, and it was set to 200.
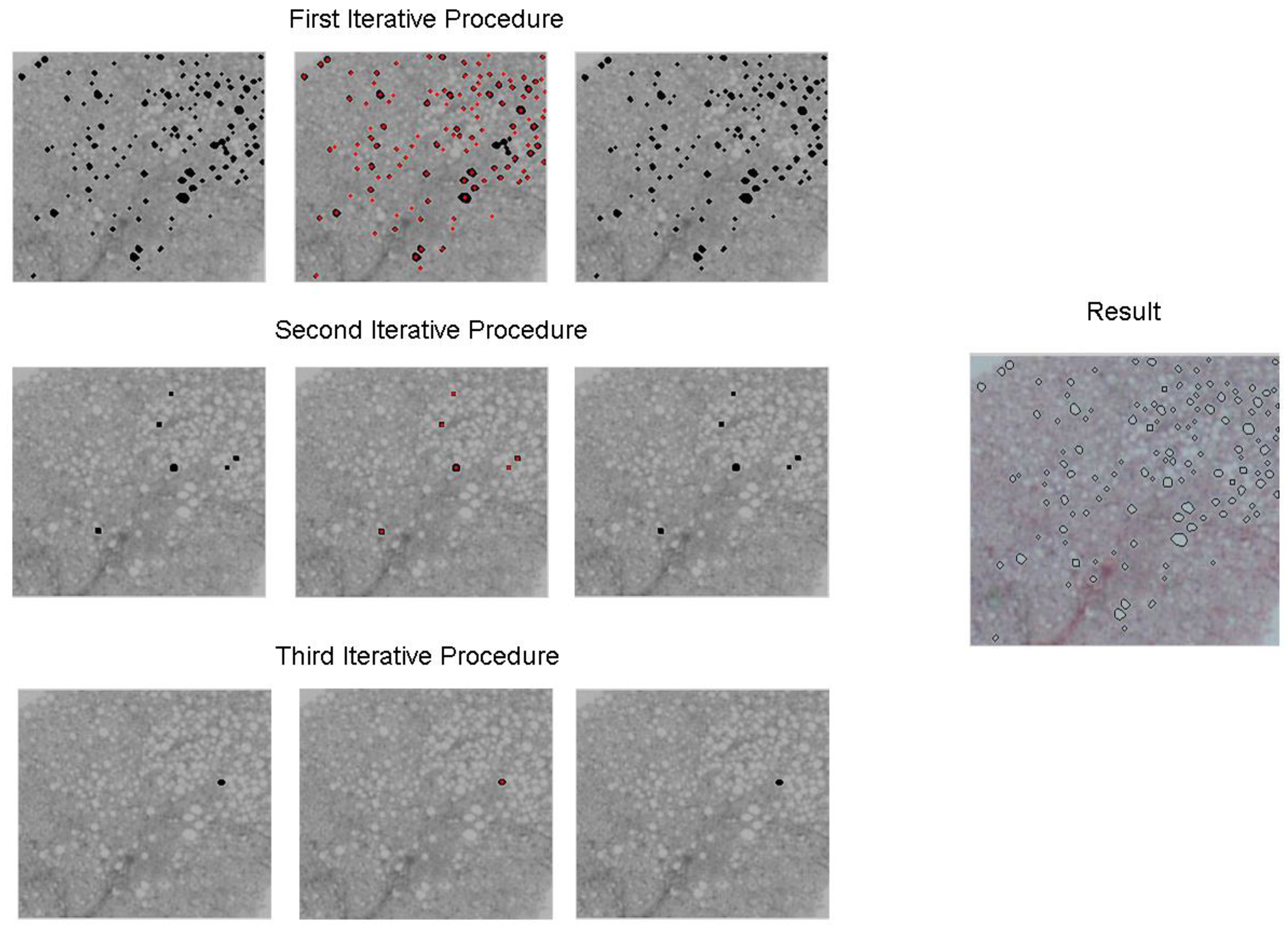
2.2. Second Stage
3. Results
Data Set
4. Discussion and Conclusions
Author Contributions
Conflicts of Interest
References
- “Hepatic Steatosis” in Gale Encyclopedia of Medicine. 2008. Available online: http://medical-dictionary.thefreedictionary.com/Hepatic+steatosis (accessed on 15 March 2017).
- Kong, J.; Lee, M.J.; Bagci, P.; Sharma, P.; Martin, D.; Adsay, N.V.; Saltz, J.H.; Farris, A.B. Computer-based Image Analysis of Liver Steatosis with Large-scale Microscopy Imagery and Correlation with Magnetic Resonance Imaging Lipid Analysis. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine, Atlanta, GA, USA, 12–15 November 2011.
- Kim, S. Fatty Liver. Available online: http://www.healthline.com/health/fatty-liver#Overview1 (accessed on 2 October 2015).
- Di Muzio, B.; Gaillard, F. Diffuse Hepatic Steatosis. Available online: http://radiopaedia.org/articles/diffuse-hepatic-steatosis (accessed on 24 June 2015).
- Cortez-Pinto, H.; Camilo, M.E. Non-alcoholic fatty liver disease/non-alcoholic steatohepatitis (NAFLD/NASH): Diagnosis and clinical course. Best Pract. Res. Clin. Gastroenterol. 2004, 18, 1089–1104. [Google Scholar] [CrossRef] [PubMed]
- Vuppalanchi, R.; Ünalp, A.; van Natta, M.; Cummings, O.W.; Sandrasegaran, K.E. Effects of Liver Biopsy Sample Length and Number of Readings on sampling variability in nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 2009, 7, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Eldad, B.; Ezenekwe, A.; Brunt, E.; Collins, B.; Brent, P.; Kirke, B.; Dibisceglie, A. Comparison of Liver Biopsy and Noninvasive Methods for Diagnosis of Hepatocellular Carcinoma. Clin. Gastroenterol. Hepatol. 2006, 4, 361–368. [Google Scholar]
- Doycheva, I.; Patel, N.; Peterson, M.; Loomba, R. Prognostic implication of liver histology in patients with nonalcoholic fatty liver disease in diabetes. J. Diabetes Its Complicat. 2013, 27, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Kim, P.N.; Kim, K.W.; Lee, S.W.; Yoon, S.E.; Park, S.W.; Ha, H.K.; Lee, M.G.; Hwang, S.; Lee, S.G.; et al. Macrovesicular hepatic steatosis in living liver donors: Use of CT for quantitative and qualitative assessment. Radiology 2006, 239, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Mala, K.; Sadasivam, V. Classification of fatty and cirrhosis liver using wavelet-based statistical texture features and neural network classifier. Int. J. Softw. Inform. 2010, 4, 151–163. [Google Scholar]
- Weijers, G.; Starke, A.; Thijssen, J.M.; Haudum, A.; Wohlsein, P.; Rehage, J.; de Korte, C.L. Transcutaneous vs. intraoperative quantitative ultrasound for staging bovine hepatic steatosis. Ultrasound Med. Biol. 2012, 38, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Choi, J.W.; Kim, K.A.; Seo, T.S.; Lee, J.M.; Park, C.M. Usefulness of standard deviation on the histogram of ultrasound as a quantitative value for hepatic parenchymal echo texture; preliminary study. Ultrasound Med. Biol. 2006, 32, 1817–1826. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Fishbein, M.H.; Rigsby, C.K.; Zhang, G.; Schoeneman, S.E.; Donaldson, J.S. Quantitative MRI for hepatic fat fraction and T2* measurement in pediatric patients with non-alcoholic fatty liver disease. Pediatr. Radiol. 2014, 44, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Goceri, E.; Shah, Z.K.; Layman, R.; Jiang, X.; Gurcan, M.N. Quantification of liver fat: A comprehensive review. Comput. Biol. Med. 2016, 71, 174–189. [Google Scholar] [CrossRef] [PubMed]
- Zaitoun, A.M.; AlMardini, H.; Awad, S.; Ukabam, S.; Makadisi, S.; Record, C.O. Quantitative assessment of fibrosis and steatosis in liver biopsies from patients with chronic hepatitis C. J. Clin. Pathol. 2001, 54, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Marsman, H.; Matsushita, T.; Dierkhising, R.; Kremers, W.; Rosen, C.; Burgart, L.; Nyberg, S.L. Assessment of donor liver steatosis: Pathologist or automated software. Hum. Pathol. 2004, 35, 430–435. [Google Scholar] [CrossRef] [PubMed]
- El-Badry, A.M.; Breitenstein, S.; Jochum, W.; Washington, K.; Paradis, V.; Rubbia-Brandt, L.; Puhan, M.A.; Slankamenac, K.; Graf, R.; Clavien, P.A. Assessment of hepatic steatosis by expert pathologists: The end of a gold standard. Ann. Surg. 2009, 250, 691–697. [Google Scholar] [CrossRef] [PubMed]
- Turlin, B.; Ramm, G.A.; Purdie, D.M.; Lainé, F.; Perrin, M.; Deugnier, Y.; Macdonald, G.A. Assessment of hepatic steatosis: Comparison of quantitative and semi quantitative methods in 108 liver biopsies. J. Liver Int. 2009, 29, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Liquori, G.E.; Giuseppe, C.; Cascella, D.; Mastrodonato, M.; Portincasa, P.; Ferri, D. An innovative methodology for the automated morphometric and quantitative estimation of liver steatosis. Histol. Histopathol. 2009, 24, 49–60. [Google Scholar] [PubMed]
- Roullier, V.; Cavaro-Menard, C.; Guillaume, C.; Aube, C. Fuzzy algorithms to extract vacuoles of steatosis on liver histological color images. In Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 23–26 August 2007.
- Nativ, N.I.; Chen, A.I.; Yarmush, G.; Henry, S.D.; Lefkowitch, J.H.; Klein, K.M.; Maguire, T.J.; Schloss, R.; Guarrera, J.V.; Berthiaume, F.; et al. Automated image analysis method for detecting and quantifying macrovesicular steatosis in hematoxylin and eosin–stained histology images of human livers. Liver Transplant. 2014, 20, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Sciarabba, M.; Vertemati, M.; Moscheni, C.; Cossa, M.; Vizzotto, L. Automated lipid droplets recognition in human steatotic liver: Some preliminary results. In Proceedings of the Medical Image Understanding and Analysis (MIUA) Conference, London, UK, 20–24 September 2009.
- Vanderbeck, S.; Bockhorst, J.; Komorowski, R.; Kleiner, D.E.; Gawrieh, S. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum. Pathol. 2014, 45, 785–792. [Google Scholar] [CrossRef] [PubMed]
- Nazre, B. Detection and spatial analysis of hepatic steatosis in histopathology images using sparse linear models. In Proceedings of the 6th International Conference on Image Processing Theory Tools and Applications (IPTA), Oulu, Finland, 12–15 December 2016.
- MATLAB, version 2016; The MathWorks, Inc.: Natick, MA, USA, 2016.
- MacQueen, J.B. Some Methods for classification and Analysis of Multivariate Observations. In Proceedings of the 5th Berkeley Symposium on Mathematical Statistics and Probability; University of California Press: Berkeley, CA, USA, 1967; Volume 1, pp. 281–297. [Google Scholar]
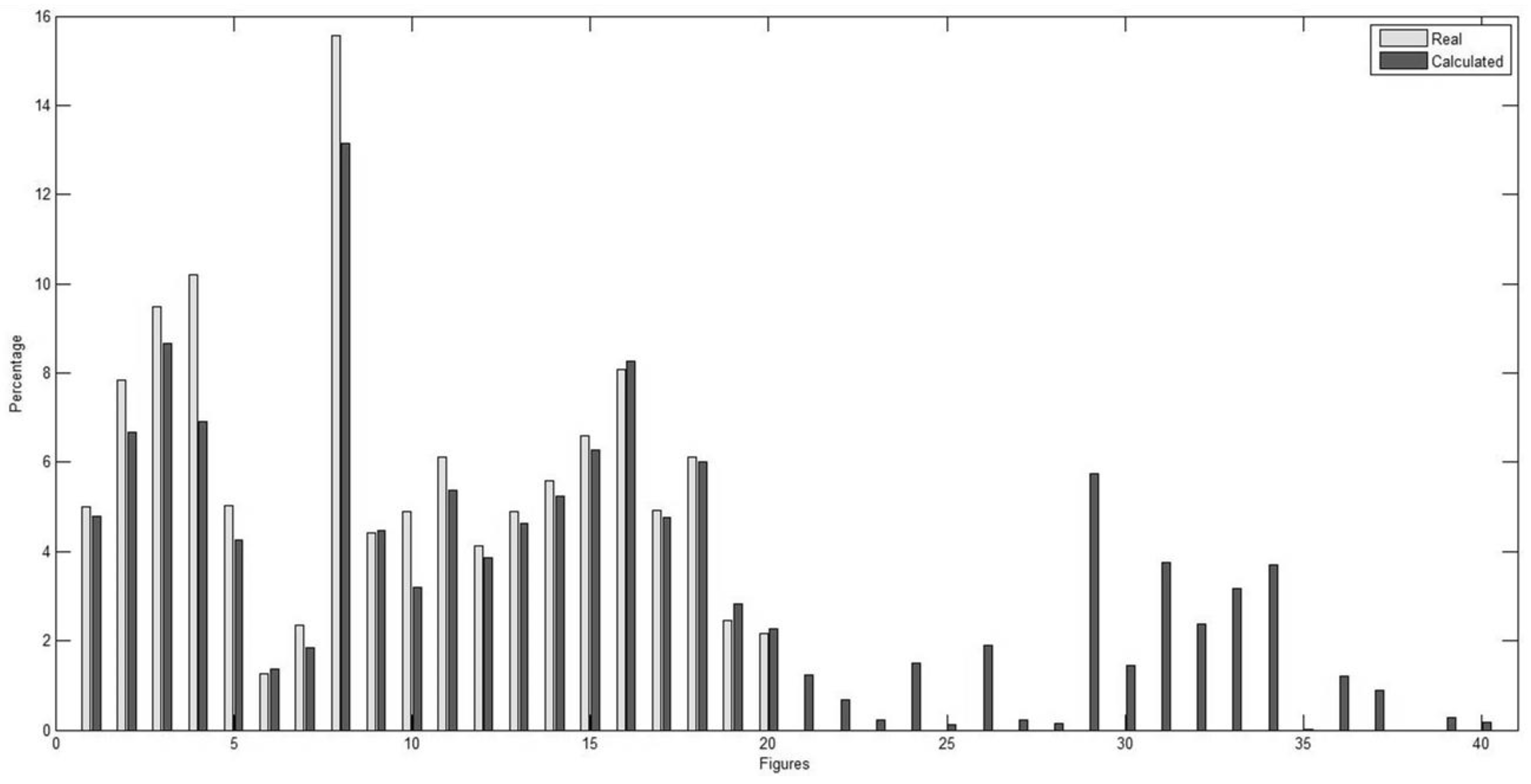
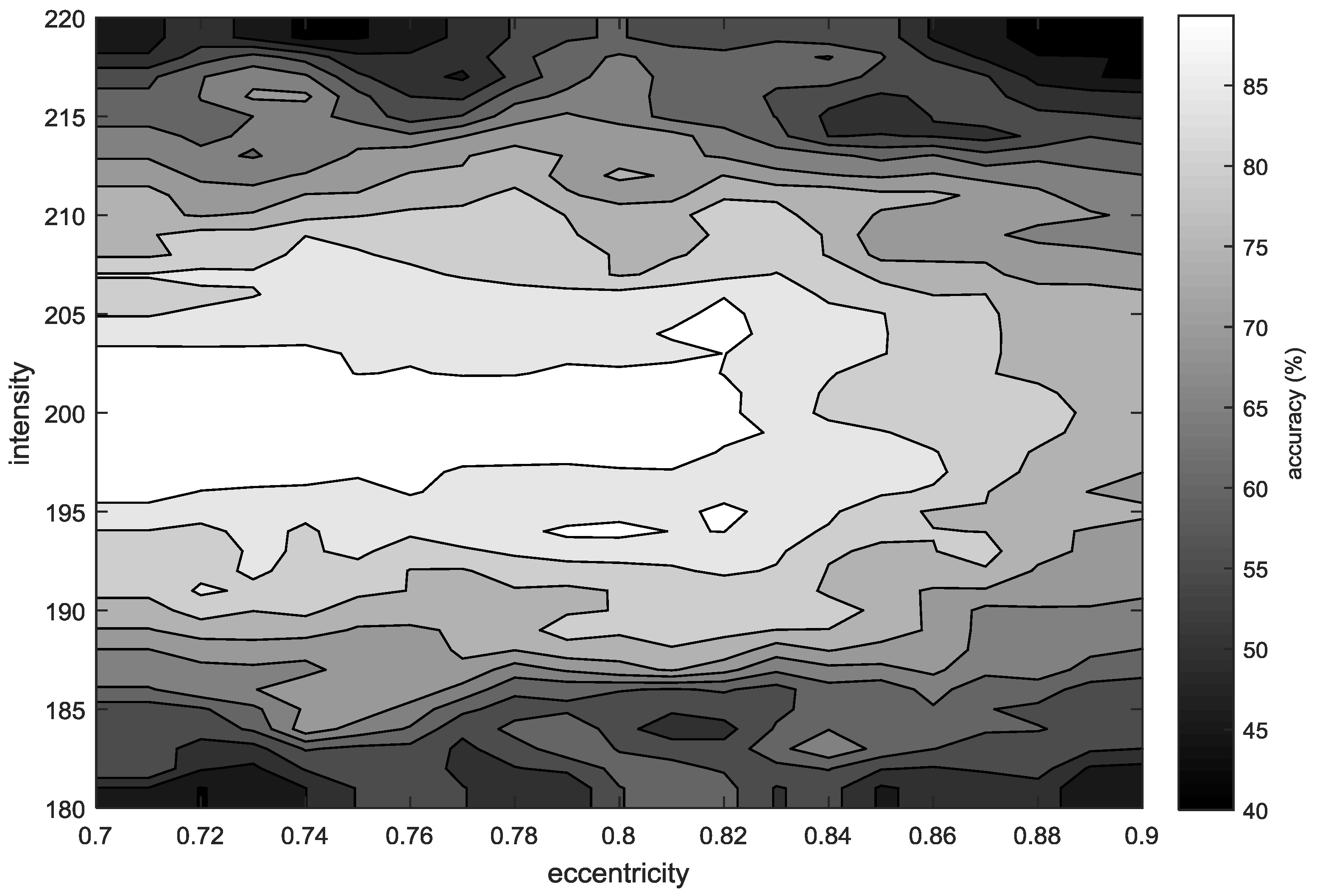
| # | Accuracy (%) | Sensitivity (%) | PPV (%) | # | Accuracy (%) | Sensitivity (%) | PPV (%) |
|---|---|---|---|---|---|---|---|
| 1 | 93.05 | 96.56 | 96.23 | 11 | 88.11 | 88.11 | 100 |
| 2 | 92.97 | 94.44 | 98.35 | 12 | 94 | 94 | 100 |
| 3 | 93.18 | 93.18 | 100 | 13 | 91.89 | 91.89 | 100 |
| 4 | 86.22 | 89.05 | 96.44 | 14 | 92.97 | 92.97 | 100 |
| 5 | 97.78 | 97.78 | 100 | 15 | 97.11 | 97.11 | 100 |
| 6 | 93.44 | 100 | 93.44 | 16 | 94.70 | 98.42 | 96.15 |
| 7 | 90.41 | 90.41 | 100 | 17 | 97.62 | 97.61 | 100 |
| 8 | 79.35 | 86.90 | 90.12 | 18 | 98.75 | 98.75 | 100 |
| 9 | 93.18 | 95.35 | 97.62 | 19 | 86.57 | 100 | 86.56 |
| 10 | 91.84 | 91.84 | 100 | 20 | 95.45 | 100 | 95.45 |
| # | Annotated Steatosis (%) | Calculated Steatosis (%) | Absolute Error % | # | Annotated Steatosis (%) | Calculated Steatosis (%) | Absolute Error % |
|---|---|---|---|---|---|---|---|
| 1 | 4.99 | 4.79 | 0.20 | 21 | 0 | 1.23 | 1.23 |
| 2 | 7.84 | 6.68 | 1.16 | 22 | 0 | 0.67 | 0.67 |
| 3 | 9.48 | 8.65 | 0.83 | 23 | 0 | 0.21 | 0.21 |
| 4 | 10.20 | 6.91 | 3.29 | 24 | 0 | 1.51 | 1.51 |
| 5 | 5.03 | 4.25 | 0.78 | 25 | 0 | 0.11 | 0.11 |
| 6 | 1.25 | 1.36 | 0.11 | 26 | 0 | 1.90 | 1.90 |
| 7 | 2.34 | 1.85 | 0.49 | 27 | 0 | 0.22 | 0.22 |
| 8 | 15.56 | 13.14 | 2.42 | 28 | 0 | 0.13 | 0.13 |
| 9 | 4.41 | 4.48 | 0.07 | 29 | 0 | 5.75 | 5.75 |
| 10 | 4.89 | 3.19 | 1.70 | 30 | 0 | 1.43 | 1.43 |
| 11 | 6.11 | 5.38 | 0.73 | 31 | 0 | 3.76 | 3.76 |
| 12 | 4.12 | 3.85 | 0.27 | 32 | 0 | 2.37 | 2.37 |
| 13 | 4.90 | 4.63 | 0.27 | 33 | 0 | 3.18 | 3.18 |
| 14 | 5.59 | 5.25 | 0.34 | 34 | 0 | 3.69 | 3.69 |
| 15 | 6.59 | 6.28 | 0.31 | 35 | 0 | 0.02 | 0.02 |
| 16 | 8.07 | 8.27 | 0.20 | 36 | 0 | 1.20 | 1.20 |
| 17 | 4.92 | 4.75 | 0.17 | 37 | 0 | 0.89 | 0.89 |
| 18 | 6.12 | 6 | 0.12 | 38 | 0 | 0 | 0 |
| 19 | 2.46 | 2.83 | 0.37 | 39 | 0 | 0.28 | 0.28 |
| 20 | 2.16 | 2.28 | 0.12 | 40 | 0 | 0.17 | 0.17 |
| Author/Year | Sample | Method | Results |
|---|---|---|---|
| Marsman et al., 2004 [16] | 46 High-definition biopsy images | No details for image analysis. Correlation between the measurement of fat using automated software and the assessment of Pathologists. | High correlation value (r = 0.97) |
| Roullier et al., 2007 [20] | 37 Images | Modification of Fuzzy C-Means Algorithm for pixel clustering. | High correlation with pathologist assessment (r2 > 0.85) |
| El Badry 2009 [17] | 46 Images | Thresholding for white area detection and roundness criteria for lipid droplets. | Poor correlation with four pathologists (Spearman rank correlation coefficient: 0.82, 0.22, 0.28, 0.38) |
| Liquori et al., 2009 [19] | Biopsy images from rats | Morphology image preprocessing. Detect fat regions based on color and circular shape. | No method evaluation. Follow-up results for fat development during diet in rats |
| Turlin et al., 2009 [18] | 97 Biopsy images | Image analysis using Image Pro Plus. Filtering and thresholding. | Strong correlation with pathologist’s grading (r = 0.89) |
| Kong et al., 2011 [2] | 21,900 Steatosis regions | Image preprocessing. Separation of bonded areas, image rotation and deletion of small points. | High Pearson Correlation value with MRI (ρ = 0.92) |
| Nativ et al., 2014 [21] | 54 Histological Images | K-means clustering and feature extraction using Decision trees. | Sensitivity 97% Specificity 60% |
| Vanderbeck et al., 2014 [23] | 59 Biopsy images | Image preprocessing. Clustering pixel using the k-means algorithm. Supervised machine learning classifiers. | The overall accuracy of the classification algorithm is greater than 89% |
| Sciarabba et al., 2015 [22] | 15 Images | Clustering using K-means and thresholding in shape features. | Detected steatosis 91% False positive ratio 5% |
| Nazre, 2016 [24] | 38 High resolution images | Morphological filtering and sparse linear models. | Pearson’s correlation with pathologists (ρ = 0.90) |
| Proposed methodology | 40 Low-resolution biopsy images | Image preprocessing. Examination of regions according to their eccentricity and roundness. | Region detection (accuracy > 90%) Steatosis assessment (Abs. Error: 1.07% ± 1.29%) Concordance Correlation Coefficient (CCC = 0.87) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsiplakidou, M.; Tsipouras, M.G.; Giannakeas, N.; Tzallas, A.T.; Manousou, P. Automated Detection of Liver Histopathological Findings Based on Biopsy Image Processing. Information 2017, 8, 36. https://doi.org/10.3390/info8010036
Tsiplakidou M, Tsipouras MG, Giannakeas N, Tzallas AT, Manousou P. Automated Detection of Liver Histopathological Findings Based on Biopsy Image Processing. Information. 2017; 8(1):36. https://doi.org/10.3390/info8010036
Chicago/Turabian StyleTsiplakidou, Maria, Markos G. Tsipouras, Nikolaos Giannakeas, Alexandros T. Tzallas, and Pinelopi Manousou. 2017. "Automated Detection of Liver Histopathological Findings Based on Biopsy Image Processing" Information 8, no. 1: 36. https://doi.org/10.3390/info8010036
APA StyleTsiplakidou, M., Tsipouras, M. G., Giannakeas, N., Tzallas, A. T., & Manousou, P. (2017). Automated Detection of Liver Histopathological Findings Based on Biopsy Image Processing. Information, 8(1), 36. https://doi.org/10.3390/info8010036







