Effects of Semantic Features on Machine Learning-Based Drug Name Recognition Systems: Word Embeddings vs. Manually Constructed Dictionaries
Abstract
:1. Introduction
2. Related Work
2.1. Drug Name Recognition
2.2. Word Embeddings Learning Algorithms
3. Methods
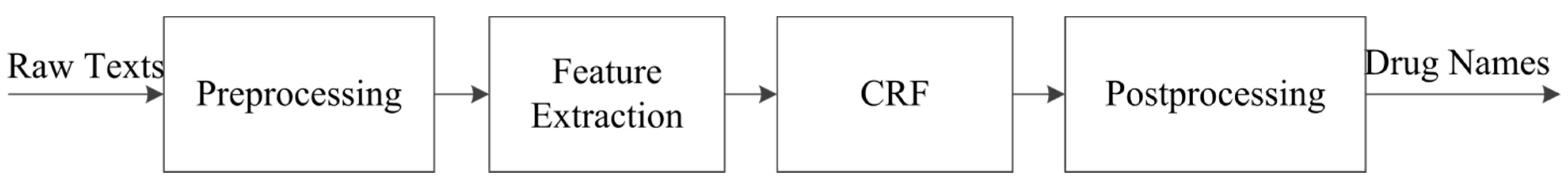
3.1. Drug Name Recognition
3.2. Conditional Random Fields
3.3. Skip-Gram Model
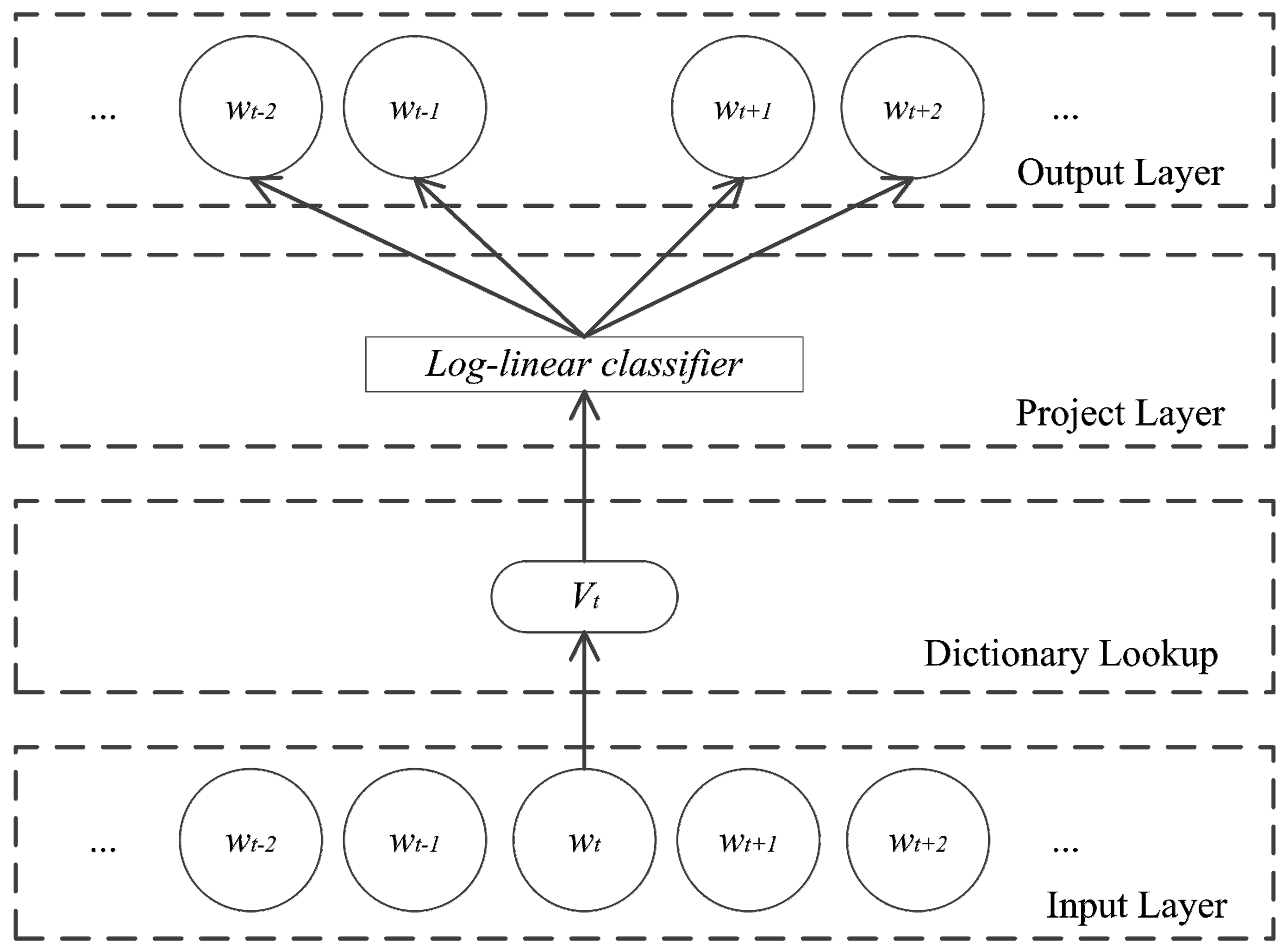
3.4. Features
3.4.1. General Features
- 1)
- Word feature: The word itself.
- 2)
- 3)
- Chunk feature: The chunk information of a word generated by the GENIA toolkit, where 11 chunking types defined in the CoNLL-2000 shared task [52] are used.
- 4)
- Affix feature: Prefixes and suffixes of the length of 3, 4, and 5.
- 5)
- Orthographical feature: Each word is classified into one of the four classes {“Is-capitalized”, “All-capitalized”, “Alphanumeric”, “All-digits”} based on regular expressions. The class label of a word is used as its feature. In addition, we check whether a word contains a hyphen, and denote whether a word contains a hyphen or not by {“Y”, “N”}.
- 6)
- Word shape feature: We used two types of word shapes: “generalized word class” and “brief word class” similar to [9]. The generalized word class of a token is generated by mapping any uppercase letter, lowercase letter, digit and other character in this token to “X”, “x”, “0” and “O”, respectively, while the brief word class of a token is generated by mapping consecutive uppercase letters, lowercase letters, digits and other characters to “X”, “x”, “0” and “O”, respectively. For example, word shape features of “Aprepitant” are “Xxxxxxxxxx” and “Xx”, respectively.
3.4.2. Semantic Features Based on Drug Dictionaries
- 1)
- 2)
- Drugs@FDA: Drugs@FDA [54] is a database provided by U.S. FDA. It contains information about FDA-approved drug names, generic prescription, etc. We extract the Drugname and Activeingred fields in Drugs@FDA and totally 8391 drug names are extracted to build a drug dictionary.
- 3)
- Jochem: Jochem [20] is a joint chemical dictionary for the identification of small molecules and drugs in text. We extract 1,527,751 concepts from Jochem.
3.4.3. Semantic Features Based on Word Embeddings
4. Experiments
4.1. Dataset
- Drug: Names of chemical agents used in the treatment, cure, prevention or diagnosis of diseases that have been approved for human use.
- Brand: Brand names of drugs specified by pharmaceutical companies.
- Group: Terms in texts designating chemical or pharmacological relationships among a group of drugs.
- No-human: Names of chemical agents that affect living organism, but have not been approved to be used in humans for a medical purpose.
| DrugBank | MEDLINE | |||||
|---|---|---|---|---|---|---|
| Training | Test | Total | Training | Test | Total | |
| Documents | 572 | 54 | 626 | 142 | 58 | 200 |
| Sentences | 5675 | 145 | 5820 | 1301 | 520 | 1821 |
| Drug | 8197 | 180 | 8377 | 1228 | 171 | 1399 |
| Group | 3206 | 65 | 3271 | 193 | 90 | 283 |
| Brand | 1423 | 53 | 1476 | 14 | 6 | 20 |
| No-human | 103 | 5 | 108 | 401 | 115 | 516 |
4.2. Evaluation Metrics
- Strict matching: A tagged drug name is correct only if its boundary and class exactly match with a gold drug name.
- Exact boundary matching: A tagged drug name is correct if its boundary matches with a gold drug name regardless of its class.
- Type matching: A tagged drug name is correct if there is some overlap between it and a gold drug name of the same class.
- Partial boundary matching: A tagged drug name is correct if there is some overlap between it and a gold drug name regardless of its class.
4.3. Experimental Results
| Feature | P | R | F1 |
|---|---|---|---|
| Fg | 78.41 | 67.78 | 72.71 |
| Fg + FWE | 82.70 | 69.68 | 75.63 |
| Fg + FFDA | 83.19 | 69.24 | 75.58 |
| Fg + FDrugBank | 85.51 | 68.80 | 76.25 |
| Fg + FJochem | 77.71 | 72.16 | 74.83 |
| Fg + FFDA + FWE | 85.59 | 70.12 | 77.09 |
| Fg + FDrugBank + FWE | 86.24 | 71.08 | 78.05 |
| Fg + FJochem + FWE | 79.64 | 71.28 | 75.23 |
| Fg + FFDA + FDrugBank + FJochem | 83.84 | 71.87 | 77.39 |
| Fg + FFDA + FDrugBank + FJochem + FWE | 84.75 | 72.89 | 78.37 |
4.4. Performance Comparison between Our System and Participating Systems of DDIExtraction 2013
| System | Strict | ||
|---|---|---|---|
| P | R | F1 | |
| Our system | 84.75 | 72.89 | 78.37 |
| WBI [57] | 73.40 | 69.80 | 71.50 |
| NLM_LHC | 73.20 | 67.90 | 70.40 |
| LASIGE [58] | 69.60 | 62.10 | 65.60 |
| UTurku [6] | 73.70 | 57.90 | 64.80 |
| UC3M [59] | 51.70 | 54.20 | 52.90 |
| UMCC_DLSI_DDI [60] | 19.50 | 46.50 | 27.50 |
| WBI | Our System | ||||||
|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | ΔF1 | |
| Strict | 73.40 | 69.80 | 71.50 | 84.75 | 72.89 | 78.37 | +6.87 |
| Exact | 85.50 | 81.30 | 83.30 | 90.68 | 77.99 | 83.86 | +0.56 |
| Type | 76.70 | 73.00 | 74.80 | 87.46 | 75.22 | 80.88 | +6.08 |
| Partial | 87.70 | 83.50 | 85.60 | 92.37 | 79.45 | 85.42 | −0.18 |
| Drug (strict) | 73.60 | 85.20 | 79.00 | 92.02 | 85.47 | 88.62 | +9.62 |
| Brand (strict) | 81.00 | 86.40 | 83.60 | 100.00 | 94.92 | 97.39 | +13.79 |
| Group (strict) | 79.20 | 76.10 | 77.60 | 89.44 | 81.94 | 85.53 | +7.93 |
| No-human (strict) | 31.40 | 9.10 | 14.10 | 89.47 | 14.05 | 24.29 | +10.19 |
| DrugBank (strict) | 88.10 | 87.50 | 87.80 | 90.60 | 88.82 | 89.70 | +1.90 |
| MEDLINE (strict) | 60.70 | 55.80 | 58.10 | 78.77 | 60.21 | 68.25 | +10.15 |
5. Discussion
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Segura-Bedmar, I.; Martínez, P.; Segura-Bedmar, M. Drug name recognition and classification in biomedical texts: A case study outlining approaches underpinning automated systems. Drug Discov. Today 2008, 13, 816–823. [Google Scholar]
- Segura-Bedmar, I.; Martínez, P.; Herrero-Zazo, M. SemEval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (DDIExtraction 2013). In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 341–350.
- Sanchez-Cisneros, D.; Martínez, P.; Segura-Bedmar, I. Combining dictionaries and ontologies for drug name recognition in biomedical texts. In Proceedings of the 7th International Workshop on Data and Text Mining in Biomedical Informatics, San Francisco, CA, USA, 1 November 2013; pp. 27–30.
- He, L.; Yang, Z.; Lin, H.; Li, Y. Drug name recognition in biomedical texts: A machine-learning-based method. Drug Discov. Today 2014, 19, 610–617. [Google Scholar] [PubMed]
- Krallinger, M.; Leitner, F.; Rabal, O.; Vazquez, M.; Oyarzabal, J.; Valencia, A. CHEMDNER: The drugs and chemical names extraction challenge. J. Cheminformatics 2015, 7 (Suppl. 1), S1. [Google Scholar] [CrossRef] [PubMed]
- Björe, J.; Kaewphan, S.; Salakoski, T. UTurku: Drug named entity detection and drug-drug interaction extraction using SVM classification and domain knowledge. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 651–659.
- Finkel, J.; Grenager, T.; Manning, C. Incorporating non-local information into information extraction systems by gibbs sampling. In Proceedings of the 43rd Annual Meeting of the Association for Computational Linguistics, Ann Arbor, MI, USA, 25–30 June 2005; pp. 363–370.
- Tkachenko, M.; Simanovsky, A. Named entity recognition: Exploring features. In Proceedings of the KONVENS 2012, Vienna, Austria, 19–21 September 2012; pp. 118–127.
- Settles, B. Biomedical named entity recognition using conditional random fields and rich feature sets. In Proceedings of the COLING 2004 International Joint Workshop on Natural Language Processing in Biomedicine and its Applications, Geneva, Switzerland, 23–27 August 2004; pp. 104–107.
- McDonald, R.; Pereira, F. Identifying gene and protein mentions in text using conditional random fields. BMC Bioinform. 2005, 6 (Suppl. 1), S6. [Google Scholar] [CrossRef] [PubMed]
- Patrick, J.; Li, M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J. Am. Med. Inform. Assoc. 2010, 17, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Chen, Y.; Liu, M.; Rosenbloom, S.T.; Mani, S.; Denny, J.C.; Xu, H. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J. Am. Med. Inform. Assoc. 2011, 18, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Turian, J.; Ratinov, L.; Bengio, Y. Word representations: A simple and general method for semi-supervised learning. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Uppsala, Sweden, 11–16 July 2010; pp. 384–394.
- Collobert, R.; Weston, J.; Bottou, L.; Karlen, M.; Kavukcuoglu, K.; Kuksa, P. Natural language processing (almost) from scratch. J. Mach. Learn. Res. 2011, 12, 2493–2537. [Google Scholar]
- Tang, B.; Cao, H.; Wang, X.; Chen, Q.; Xu, H. Evaluating word representation features in biomedical named entity recognition tasks. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Passos, A.; Kumar, V.; McCallum, A. Lexicon Infused Phrase Embeddings for Named Entity Resolution. In Proceedings of the 18th Conference on Computational Language Learning, Baltimore, MD, USA, 26–27 June 2014; pp. 78–86.
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. In Proceedings of the Workshop at ICLR, Scottsdale, AZ, USA, 2–4 May 2013.
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of the 27th Annual Conference on Neural Information Processing Systems, Lake Tahoe, CA, USA, 5–10 December 2013; pp. 3111–3119.
- Knox, C.; Law, V.; Jewison, T.; Liu, P.; Ly, S.; Frolkis, A.; Pon, A.; Banco, K.; Mak, C.; Neveu, V.; et al. DrugBank 3.0: A comprehensive resource for “omics” research on drugs. Nucleic Acids Res. 2011, 39, D1035–D1041. [Google Scholar] [CrossRef] [PubMed]
- Hettne, K.; Stierum, R.; Schuemie, M.; Hendriksen, P.; Schijvenaars, B.; van Mulligen, E.; Kleinjans, J.; Kors, J. A dictionary to identify small molecules and drugs in free text. Bioinformatics 2009, 25, 2983–2991. [Google Scholar] [CrossRef] [PubMed]
- Aronson, A.; Bodenreider, O.; Chang, H.; Humphrey, S.; Mork, J.; Nelson, S.; Rindflesch, T.; Wilbur, W. The NLM indexing initiative. In Proceedings of the AMIA Annual Symposium, Los Angeles, CA, USA, 4–8 November 2000; pp. 17–21.
- Segura-Bedmar, I.; Martínez, P.; Sánchez-Cisneros, D. The 1st DDIExtraction-2011 challenge task: Extraction of drug-drug interactions from biomedical texts. In Proceedings of the 1st Challenge Task on Drug-Drug Interaction Extraction, Huelva, Spain, 7 September 2011; pp. 1–9.
- Leaman, R.; Wei, C.; Lu, Z. tmChem: A high performance approach for chemical named entity recognition and normalization. J. Cheminformatics 2015, 7 (Suppl. 1), S3. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Feng, Y.; Wang, X.; Wu, Y.; Zhang, Y.; Jiang, M.; Wang, J. A comparison of conditional random fields and structured support vector machines for chemical entity recognition in biomedical literature. J. Cheminformatics 2015, 7 (Suppl. 1), S8. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ji, D.; Yao, X.; Wei, X.; Liang, X. CHEMDNER system with mixed conditional random fields and multi-scale word clustering. J. Cheminformatics 2015, 7 (Suppl. 1), S4. [Google Scholar] [CrossRef] [PubMed]
- Batista-Navarro, R.; Rak, R.; Ananiadou, S. Optimising chemical named entity recognition with pre-processing analytics, knowledge-rich features and heuristics. J. Cheminformatics 2015, 7 (Suppl. 1), S6. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.; Matos, S.; Oliveira, J. A document processing pipeline for annotating chemical entities in scientific documents. J. Cheminformatics 2015, 7 (Suppl. 1), S7. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Stenner, S.; Doan, S.; Johnson, K.B.; Waitman, L.R.; Denny, J.C. MedEx: A medication information extraction system for clinical narratives. J. Am. Med. Inform. Assoc. 2010, 17, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Doan, S.; Collier, N.; Xu, H.; Duy, P.; Phuong, T. Recognition of medication information from discharge summaries using ensembles of classifiers. BMC Med. Inform. Decis. Mak. 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- Halgrim, S.; Xia, F.; Solti, I.; Cadag, E.; Uzuner, Ö. A cascade of classifiers for extracting medication information from discharge summaries. J. Biomed. Semant. 2011, 2 (Suppl. 3), S2. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, A.; Kvist, M.; Dalianis, H.; Duneld, M. Identifying adverse drug event information in clinical notes with distributional semantic representations of context. J. Biomed. Inform. 2015, 57, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Skeppstedt, M.; Kvist, M.; Nilsson, G.; Dalianis, H. Automatic recognition of disorders, findings, pharmaceuticals and body structures from clinical text: An annotation and machine learning study. J. Biomed. Inform. 2014, 49, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; de Souza, P.; Mercer, R.; Pietra, V.; Lai, J. Class-based n-gram models of natural language. Comput. Linguist. 1992, 18, 467–479. [Google Scholar]
- Landauer, T.; Foltz, P.; Laham, D. An introduction to latent semantic analysis. Discourse Process. 1998, 25, 259–284. [Google Scholar] [CrossRef]
- Lund, K.; Burgess, C.; Atchley, R. Semantic and associative priming in high dimensional semantic space. In Proceedings of the 17th Annual Conference of the Cognitive Science Society, Pittsburgh, PA, USA, 22–25 July 1995; pp. 660–665.
- Jonnalagadda, S.; Cohen, T.; Wub, S.; Gonzalez, G. Enhancing clinical concept extraction with distributional semantics. J. Biomed. Inform. 2012, 45, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Bengio, Y.; Ducharme, R.; Vincent, P.; Jauvin, C. A neural probabilistic language model. J. Mach. Learn. Res. 2003, 3, 1137–1155. [Google Scholar]
- Morin, F.; Bengio, Y. Hierarchical probabilistic neural network language model. In Proceedings of the 10th International Workshop on Artificial Intelligence and Statistics, Bridgetown, Barbados, 6–8 January 2005; pp. 246–252.
- Mnih, A.; Hinton, G. A scalable hierarchical distributed language model. In Proceedings of the 22nd Annual Conference on Neural Information Processing Systems, Vancouver, Canada, 8–11 December 2008; pp. 1081–1088.
- Huang, E.; Socher, R.; Manning, C.; Ng, A. Improving word representations via global context and multiple word prototypes. In Proceedings of the 50th Annual Meeting of the Association for Computational Linguistics, Jeju, Korea, 8–14 July 2012; pp. 873–882.
- Mikolov, T.; Karafiát, M.; Burget, L.; Cernocky, J.; Khudanpur, S. Recurrent neural network based language model. In Proceedings of the 11th Annual Conference of the International Speech Communication Association, Makuhari, Japan, 26–30 September 2010; pp. 1045–1048.
- Natural Language Toolkit. Available online: http://www.nltk.org/ (accessed on 11 December 2015).
- Ratinov, L.; Roth, D. Design challenges and misconceptions in named entity recognition. In Proceedings of the 13th Conference on Computational Natural Language Learning, Boulder, CO, USA, 4 June 2009; pp. 147–155.
- Lafferty, J.; McCallum, A.; Pereira, F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Proceedings of the 18th International Conference on Machine Learning, Williamstown, MA, USA, 28 June–1 July 2001; pp. 282–289.
- McCallum, A.; Li, W. Early results for named entity recognition with conditional random fields, feature induction and web-enhanced lexicons. In Proceedings of the 7th Conference on Natural Language Learning, Geneva, Switzerland, 23–27 August 2003; pp. 188–191.
- Sutton, C.; McCallum, A.; Rohanimanesh, K. Dynamic conditional random fields: Factorized probabilistic models for labeling and segmenting sequence data. J. Mach. Learn. Res. 2007, 8, 693–723. [Google Scholar]
- Peng, F.; Feng, F.; McCallum, A. Chinese segmentation and new word detection using conditional random fields. In Proceedings of the 20th International Conference on Computational Linguistics, Geneva, Switzerland, 23–27 August 2004; pp. 562–568.
- CRFsuite. Available online: http://www.chokkan.org/software/crfsuite/ (accessed on 11 December 2015).
- word2vec. Available online: https://code.google.com/p/word2vec/ (accessed on 11 December 2015).
- GENIA Tagger. Available online: http://www.nactem.ac.uk/tsujii/GENIA/tagger/ (accessed on 11 December 2015).
- Marcus, M.; Santorini, B.; Marcinkiewicz, M. Building a large annotated corpus of English: The penn treebank. Comput. Linguist. 1993, 19, 313–330. [Google Scholar]
- Sang, E.; Buchholz, S. Introduction to the CoNLL-2000 shared task: Chunking. In Proceedings of the CoNLL-2000, Lisbon, Portugal, 13–14 September 2000; pp. 127–132.
- DrugBank. Available online: http://www.drugbank.ca/downloads (accessed on 11 December 2015).
- Drugs@FDA Data Files. Available online: http://www.fda.gov/Drugs/InformationOnDrugs/ ucm079750.htm (accessed on 11 December 2015).
- Leasing Journal Citations (MEDLINE®/PubMed® including OLDMEDLINE). Available online: http://www.nlm.nih.gov/databases/journal.html (accessed on 11 December 2015).
- Lai, S.; Liu, K.; Xu, L.; Zhao, J. How to generate a good word embedding? 2015; arXiv:1507.05523. [Google Scholar]
- Rocktäschel, T.; Huber, T.; Weidlich, M.; Leser, U. WBI-NER: The impact of domain-specific features on the performance of identifying and classifying mentions of drugs. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 356–363.
- Grego, T.; Pinto, F.; Couto, F. LASIGE: Using conditional random fields and ChEBI ontology. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 660–666.
- Sanchez-Cisneros, D.; Gali, F. UEM-UC3M: An ontology-based named entity recognition system for biomedical texts. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 622–627.
- Collazo, A.; Ceballo, A.; Puig, D.; Gutiérrez, Y.; Abreu, J.; Pérez, R.; Orquín, A.; Montoyo, A.; Muñoz, R.; Camara, F. UMCC_DLSI: Semantic and lexical features for detection and classification drugs in biomedical texts. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 636–643.
- Sang, E.; Meulder, F. Introduction to the CoNLL-2003 shared task: Language-independent named entity recognition. In Proceedings of the 7th Conference on Natural Language Learning, Edmonton, Canada, 31 May–1 June 2003; pp. 142–147.
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Tang, B.; Chen, Q.; Wang, X. Effects of Semantic Features on Machine Learning-Based Drug Name Recognition Systems: Word Embeddings vs. Manually Constructed Dictionaries. Information 2015, 6, 848-865. https://doi.org/10.3390/info6040848
Liu S, Tang B, Chen Q, Wang X. Effects of Semantic Features on Machine Learning-Based Drug Name Recognition Systems: Word Embeddings vs. Manually Constructed Dictionaries. Information. 2015; 6(4):848-865. https://doi.org/10.3390/info6040848
Chicago/Turabian StyleLiu, Shengyu, Buzhou Tang, Qingcai Chen, and Xiaolong Wang. 2015. "Effects of Semantic Features on Machine Learning-Based Drug Name Recognition Systems: Word Embeddings vs. Manually Constructed Dictionaries" Information 6, no. 4: 848-865. https://doi.org/10.3390/info6040848
APA StyleLiu, S., Tang, B., Chen, Q., & Wang, X. (2015). Effects of Semantic Features on Machine Learning-Based Drug Name Recognition Systems: Word Embeddings vs. Manually Constructed Dictionaries. Information, 6(4), 848-865. https://doi.org/10.3390/info6040848





