Drug Name Recognition: Approaches and Resources
Abstract
:1. Introduction
2. Challenges of Drug Name Recognition
- The ways of naming drugs vary greatly. For example, the drug “quetiapine” (generic name) has the brand name “Seroquel XR”, while its systematic International Union of Pure and Applied Chemistry (IUPAC) name is “2-[2-(4-dibenzo [b,f][1,4] thiazepin-11-ylpiperazin-1-yl) ethoxy] ethanol”. Furthermore, some drug names and their synonyms are the same as normal English words or phrases. For example, brand names of “oxymetazoline nasal” and “caffeine” are “Duration” and “Stay Awake”, respectively.
- The frequent occurrences of abbreviations and acronyms make it difficult to identify the concepts to which the terms refer to. For example, the abbreviation “PN” can refer to the drug “penicillin” or other concepts such as “pneumonia”, “polyarteritis nodosa” and “polyneuritis”.
- New drugs are constantly and rapidly reported in scientific publications. Moreover, drug names may be misspelled in electronic medical records such as progress notes and discharge summaries. This makes DNR systems that rely only on dictionaries of known drug names not effective.
- Drug names may contain a number of symbols mixed with common words. For example, the IUPAC name of an atypical antipsychotic is “7-{4-[4-(2,3-dichlorophenyl) piperazin-1-yl]butoxy}-3,4-dihydroquinolin-2(1H)-one”. It is difficult to determine the boundaries of such drug names in texts.
- Some drug names may correspond to non-continuous strings of text. For example, “loop diuretics” and “potassium-sparing diuretics” in the sentence “In some patients, the administration of a non-steroidal anti-inflammatory agent can reduce the diuretic, natriuretic, and antihypertensive effects of loop, potassium-sparing and thiazide diuretics”. Such examples pose great difficulties to DNR.
3. Benchmark Datasets
| Dataset | Data Source | URL |
|---|---|---|
| ADE [10] | Medical case reports | https://sites.google.com/site/adecorpus/ |
| PK [11] | Biomedical literature abstracts | http://rweb.compbio.iupui.edu/corpus/ |
| PK-DDI [12] | Drug package inserts | http://purl.org/NET/nlprepository/PI-PK-DDI-Corpus |
| EU-ADR [13] | Biomedical literature abstracts | http://euadr.erasmusmc.nl/sda/euadr_corpus.tgz |
| i2b2 Medication Extraction [4] | Discharge summaries | https://www.i2b2.org/NLP/DataSets/ |
| DrugNer [3] | Biomedical literature abstracts | http://labda.inf.uc3m.es/DrugDDI/DrugNer.html |
| DDIExtraction 2011 [14] | Texts selected from DrugBank | http://labda.inf.uc3m.es/ddicorpus |
| DDIExtraction 2013 [15] | Biomedical literature abstracts and texts selected from DrugBank | http://labda.inf.uc3m.es/ddicorpus |
| CHEMDNER [5] | Biomedical literature abstracts | http://www.biocreative.org/resources/biocreative-iv/chemdner-corpus/ |
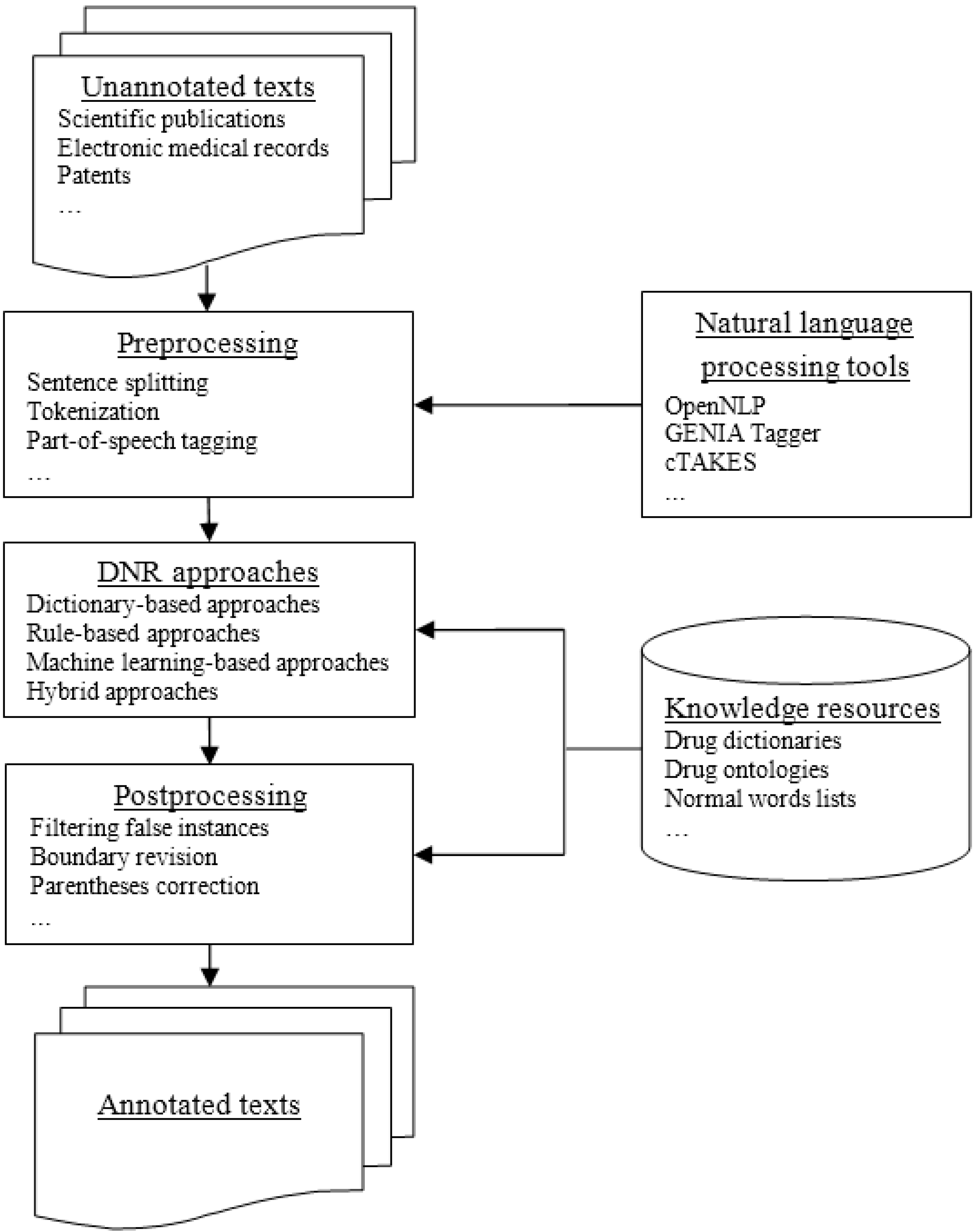
4. General Architecture of Drug Name Recognition Systems
| NLP Toolkit | Target Domain | URL |
|---|---|---|
| OpenNLP | General | http://opennlp.apache.org |
| LingPipe | General | http://alias-i.com/lingpipe |
| NLTK | General | http://www.nltk.org |
| ANNIE | General | https://gate.ac.uk/sale/tao/splitch6.html#chap:annie |
| NaCTeM | General | http://www.nactem.ac.uk/software.php |
| Stanford NLP Toolkit [23] | General | http://www-nlp.stanford.edu/software/ |
| U-Compare [21,22] | General | http://u-compare.org |
| JULIE Lab [24] | General | http://www.julielab.de/Resources |
| GENIA Tagger [25] | Biomedical | http://www.nactem.ac.uk/GENIA/tagger/ |
| GDep [26] | Biomedical | http://people.ict.usc.edu/~sagae/parser/gdep/ |
| Neji [27] | Biomedical | http://bioinformatics.ua.pt/neji/ |
| BioLemmatizer [28] | Biomedical | http://biolemmatizer.sourceforge.net/ |
| cTAKES [29] | Clinical | http://ctakes.apache.org/ |
5. Approaches for Drug Name Recognition
5.1. Dictionary-Based Approaches
| Knowledge Resource | URL |
|---|---|
| DrugBank | http://www.drugbank.ca/ |
| KEGG DRUG | http://www.kegg.jp/kegg/drug/ |
| PharmGKB | http://www.pharmgkb.org/ |
| CTD | http://ctdbase.org/ |
| RxNorm | http://www.nlm.nih.gov/research/umls/rxnorm/ |
| RxTerms | http://wwwcf.nlm.nih.gov/umlslicense/rxtermApp/rxTerm.cfm |
| Drugs@FDA | http://www.fda.gov/Drugs/InformationOnDrugs/ucm135821.htm |
| TTD | http://bidd.nus.edu.sg/group/ttd/ttd.asp |
| ChEBI | http://www.ebi.ac.uk/chebi |
| MeSH | http://www.nlm.nih.gov/mesh/ |
| PubChem | https://pubchem.ncbi.nlm.nih.gov/ |
| UMLS Metathesaurus | http://www.nlm.nih.gov/research/umls/ |
| Jochem | http://www.biosemantics.org/index.php/resources/jochem |
5.2. Rule-Based Approaches
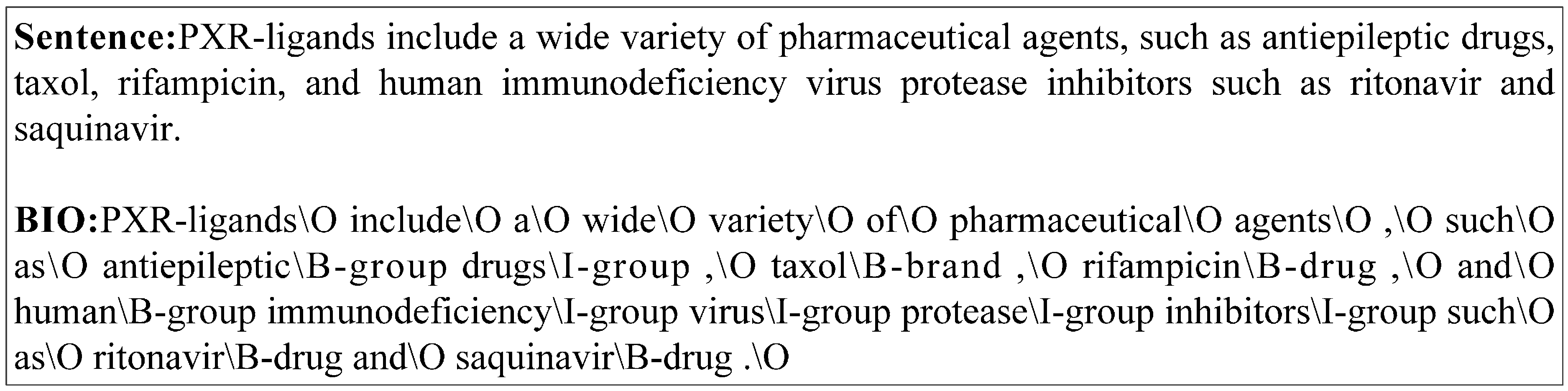
5.3. Machine Learning-Based Approaches
| Toolkit | Machine Learning Models | URL |
|---|---|---|
| MALLET | Naïve Bayes (NB), Decision Trees (DT), ME, HMM, CRF | http://mallet.cs.umass.edu/ |
| WEKA | NB, DT, SVM | http://www.cs.waikato.ac.nz/ml/weka/ |
| CRFsuite | CRF | http://www.chokkan.org/software/crfsuite/ |
| CRF++ | CRF | http://taku910.github.io/crfpp/ |
| LIBSVM | SVM | http://www.csie.ntu.edu.tw/~cjlin/libsvm/ |
| SVMlight | SVM | http://www.cs.cornell.edu/People/tj/svm_light/ |
| Feature | Description | Reference |
|---|---|---|
| Character feature | N-grams of characters in a word. | [17,31,68,70,71,74] |
| Word feature | N-grams of words in a context window. | [17,31,64,68,70,71,74,75] |
| Lemma | N-grams of lemmas of words. | [17,31,68,71,74] |
| Stem | N-grams of stems of words. | [31,74] |
| POS | N-grams of POS tags. | [17,31,64,68,71,74,75] |
| Text chunking | N-grams of text chunking tags. | [17,71,75] |
| Dependency parsing | Dependency parsing results of words in a sentence. | [71] |
| Affix | Suffixes and prefixes of a word. | [17,31,64,68,71,74,75] |
| Orthographic feature | Starting with a uppercase letter, containing only alphanumeric characters, containing a hyphen, digits and capitalized characters counting, etc. | [17,31,64,68,71,74,75] |
| Word shape | Uppercase letters, lowercase letters, digits, and other characters in a word are converted to “A”, “a”, “0” and “O”, respectively. For example, “Phenytoin” is mapped to “Aaaaaaaaa”. | [17,31,68,71,74,75] |
| Dictionary feature | Whether an n-gram matches with part of a drug name in drug dictionaries. | [17,31,64,68,71,74,75] |
| Outputs of NER tools | Features derived from the output of existing chemical NER tools. | [31,68,74] |
| Word representation | Word representation features based on Brown clustering, word2vec, etc. | [70,75] |
| Conjunction feature | Conjunctions of different types of features, e.g., conjunction of lemma and POS features. | [17,71,75] |
5.4. Hybrid Approaches
| Type of Texts | Reference | Category of Approach | Dataset | F-Score Detection | F-Score Classification |
|---|---|---|---|---|---|
| Discharge summaries | [64] | ML | i2b2 Medication Extraction | 89.80% | * |
| [67] | ML | i2b2 Medication Extraction | 88.35% | * | |
| [84] | Hybrid (ML + Rule) | i2b2 Medication Extraction | 87.06% | * | |
| [87] | Dict | i2b2 Medication Extraction | 85.89% | * | |
| [60] | Rule | i2b2 Medication Extraction | 80.00% | * | |
| [59] | Rule | 26 discharge summaries | 87.92% | * | |
| Clinic office visit notes | [54] | Dict | 52 clinic office visit notes | 73.80% | * |
| Biomedical literature abstracts | [31] | ML | CHEMDNER | 87.39% | * |
| [70] | ML | CHEMDNER | 87.11% | * | |
| [58] | Rule | CHEMDNER | 86.86% | * | |
| [88] | Dict | CHEMDNER | 77.91% | * | |
| [82] | Hybrid (Dic + Rule) | CHEMDNER | 77.84% | * | |
| [44] | Dict | 100 abstracts | 50.00% | * | |
| DrugBank documents | [83] | Hybrid (ML + Dic) | DDIExtraction 2011 | 92.54% | * |
| Mix of DrugBank documents and biomedical literature abstracts | [75] | ML | DDIExtraction 2013 | 83.85% | 79.36% |
| [68] | ML | DDIExtraction 2013 | 83.30% | 71.50% | |
| [52] | Dict | DDIExtraction 2013 | 66.70% | * | |
| [89] | Dict | DDIExtraction 2013 | 60.90% | 52.90% |
6. Concluding Remarks and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Segura-Bedmar, I.; Martínez, P.; Pablo-Sánchez, C. Using a shallow linguistic kernel for drug-drug interaction extraction. J. Biomed. Inform. 2011, 44, 789–804. [Google Scholar] [CrossRef] [PubMed]
- Warrer, P.; Hansen, W.; Juhl-Jensen, L.; Aagaard, L. Using text-mining techniques in electronic patient records to identify ADRs from medicine use. Br. J. Clin. Pharmacol. 2012, 73, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Segura-Bedmar, I.; Martínez, P.; Segura-Bedmar, M. Drug name recognition and classification in biomedical texts: A case study outlining approaches underpinning automated systems. Drug Discov. Today 2008, 13, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Uzuner, Ö.; Solti, I.; Cadag, E. Extracting medication information from clinical text. J. Am. Med. Inform. Assoc. 2010, 17, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Krallinger, M.; Leitner, F.; Rabal, O.; Vazquez, M.; Oyarzabal, J.; Valencia, A. CHEMDNER: The drugs and chemical names extraction challenge. J. Cheminform. 2015, 7 (S1), S1. [Google Scholar] [CrossRef] [PubMed]
- Segura-Bedmar, I.; Martínez, P.; Herrero-Zazo, M. SemEval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (DDIExtraction 2013). In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 341–350.
- Vazquez, M.; Krallinger, M.; Leitner, F.; Valencia, A. Text mining for drugs and chemical compounds: Methods, tools and applications. Mol. Inf. 2011, 30, 506–519. [Google Scholar] [CrossRef]
- Gurulingappa, H.; Mudi, A.; Toldo, L. Challenges in mining the literature for chemical information. RSC Adv. 2013, 3, 16194–16211. [Google Scholar] [CrossRef]
- Eltyeb, S.; Salim, N. Chemical named entities recognition: A review on approaches and applications. J Cheminform. 2014, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Gurulingappa, H.; Rajput, A.; Roberts, A.; Fluck, J.; Hofmann-Apitius, M.; Toldo, L. Development of a benchmark corpus to support the automatic extraction of drug-related adverse effects from medical case reports. J. Biomed. Inform. 2012, 45, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Karnik, S.; Subhadarshini, A.; Wang, Z.; Philips, S.; Han, X.; Chiang, C.; Liu, L.; Boustani, M.; Rocha, L.M.; et al. An integrated pharmacokinetics ontology and corpus for text mining. BMC Bioinform. 2013, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Boyce, R.; Gardner, G.; Harkema, H. Using natural language processing to extract drug-drug interaction information from package inserts. In Proceedings of the 2012 Workshop on Biomedical Natural Language Processing, Montreal, QC, Canada, 3–8 June 2012; pp. 206–213.
- Mulligen, E.; Fourrier-Reglat, A.; Gurwitz, D.; Molokhia, M.; Nieto, A.; Trifiro, G.; Kors, J.A.; Furlong, L.I. The EU-ADR corpus: Annotated drugs, diseases, targets, and their relationships. J. Biomed. Inform. 2012, 45, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Segura-Bedmar, I.; Martínez, P.; Sánchez-Cisneros, D. The 1st DDIExtraction-2011 challenge task: Extraction of drug-drug interactions from biomedical texts. In Proceedings of the 1st Challenge Task on Drug-Drug Interaction Extraction, Huelva, Spain, 5 September 2011; pp. 1–9.
- Herrero-Zazo, M.; Segura-Bedmar, I.; Martínez, P.; Declerck, T. The DDI corpus: An annotated corpus with pharmacological substances and drug-drug interactions. J. Biomed. Inform. 2013, 46, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Lai, P.; Chang, Y.; Tsai, R.T. Enhancing of chemical compound and drug name recognition using representative tag scheme and fine-grained tokenization. J. Cheminform. 2015, 7 (S1), S14. [Google Scholar] [CrossRef] [PubMed]
- Batista-Navarro, R.; Rak, R.; Ananiadou, S. Optimising chemical named entity recognition with pre-processing analytics, knowledge-rich features and heuristics. J. Cheminform. 2015, 7 (S1), S6. [Google Scholar] [CrossRef] [PubMed]
- Treebank tokenization. Available online: http://www.cis.upenn.edu/~treebank/tokenization.html (accessed on 24 November 2015).
- Ferrucci, D.; Lally, A. UIMA: An architectural approach to unstructured information processing in the corporate research environment. Nat. Lang. Eng. 2004, 10, 327–348. [Google Scholar] [CrossRef]
- Apache UIMA. Available online: http://uima.apache.org/ (accessed on 24 November 2015).
- Kano, Y.; Baumgartner, W.A., Jr.; McCrohon, L.; Ananiadou, S.; Cohen, K.B.; Hunter, L.; Tsujii, J. U-Compare: Share and compare text mining tools with UIMA. Bioinformatics 2009, 25, 1997–1998. [Google Scholar] [CrossRef] [PubMed]
- Kano, Y.; Miwa, M.; Cohen, K.B.; Hunter, L.E. U-Compare: A modular NLP workflow construction and evaluation system. IBM J. Res. Dev. 2011, 55, 1–11. [Google Scholar] [CrossRef]
- Manning, C.D.; Surdeanu, M.; Bauer, J.; Finkel, J.; Bethard, S.J.; McClosky, D. The Stanford CoreNLP natural language processing toolkit. In Proceedings of 52nd Annual Meeting of the Association for Computational Linguistics: System Demonstrations, Baltimore, MD, USA, 22–27 June 2014; pp. 55–60.
- Hahn, U.; Buyko, E.; Landefeld, R.; Mühlhausen, M.; Poprat, M.; Tomanek, K.; Wermter, J. An overview of JCORE, the JULIE Lab UIMA component repository. In Proceedings of the LREC’08 Workshop “Towards Enhanced Interoperability for Large HLT Systems: UIMA for NLP”, Marrakech, Morocco, 26–27 May 2008; pp. 1–7.
- Tsuruoka, Y.; Tsujii, J. Bidirectional inference with the easiest-first strategy for tagging sequence data. In Proceedings of the Conference on Human Language Technology and Empirical Methods in Natural Language Processing (HLT/EMNLP 2005), Vancouver, BC, Canada, 6–8 October 2005; pp. 467–474.
- Miyao, Y.; Saetre, R.; Sagae, K.; Matsuzaki, T.; Tsujii, J. Task-oriented evaluation of syntactic parsers and their representations. In Proceedings of the 45th Meeting of the Association for Computational Linguistics, Columbus, OH, USA, 19–20 June 2008; pp. 46–54.
- Campos, D.; Matos, S.; Oliveira, J. A modular framework for biomedical concept recognition. BMC Bioinform. 2013, 14, 281. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Christiansen, T.; Baumgartner, W.A., Jr.; Verspoor, K. BioLemmatizer: A lemmatization tool for morphological processing of biomedical text. J. Biomed. Semant. 2012, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Savova, G.; Masanz, J.; Ogren, P.; Zheng, J.; Sohn, S.; Kipper-Schuler, K.C.; Chute, C.G. Mayo clinical text analysis and knowledge extraction system (cTAKES): Architecture, component evaluation and applications. J. Am. Med. Inform. Assoc. 2010, 17, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Grego, T.; Pinto, F.; Couto, F.M. LASIGE: Using conditional random fields and ChEBI ontology. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 660–666.
- Leaman, R.; Wei, C.; Lu, Z. tmChem: A high performance approach for chemical named entity recognition and normalization. J. Cheminform. 2015, 7 (S1), S3. [Google Scholar] [CrossRef] [PubMed]
- Grego, T.; Couto, F.M. Enhancement of chemical entity identification in text using semantic similarity validation. PLoS ONE 2013, 8, e62984. [Google Scholar] [PubMed]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2013, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Boussard, T.; Whirl-Carrillo, M.; Hebert, J.; Gong, L.; Owen, R.; Gong, M.; Gor, W.; Liu, F.; Truong, C.; Whaley, R.; et al. The pharmacogenetics and pharmacogenomics knowledge base: Accentuating the knowledge. Nucleic Acids Res. 2008, 36, D913–D918. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Wiegers, T.C.; Mattingly, C.J. The Comparative Toxicogenomics Database’s 10th year anniversary: Update 2015. Nucleic Acids Res. 2015, 43, D914–D920. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Ma, W.; Moore, R.; Ganesan, V.; Nelson, S. RxNorm: Prescription for electronic drug information exchange. IT Prof. 2005, 7, 17–23. [Google Scholar] [CrossRef]
- Fung, K.; McDonald, C.; Bray, B. RxTerms—A drug interface terminology derived from RxNorm. In Proceedings of the AMIA 2008 Annual Symposium, Washington, DC, USA, 8–12 November 2008; pp. 227–231.
- Qin, C.; Zhang, C.; Zhu, F.; Xu, F.; Chen, S.Y.; Zhang, P.; Li, Y.H.; Yang, S.Y.; Wei, Y.Q.; Tao, L.; et al. Therapeutic target database update 2014: A resource for targeted therapeutics. Nucleic Acids Res. 2014, 42, D1118–D1123. [Google Scholar] [CrossRef] [PubMed]
- Hastings, J.; de Matos, P.; Dekker, A.; Ennis, M.; Harsha, B.; Kale, N.; Muthukrishnan, V.; Owen, G.; Turner, S.; Williams, M.; et al. The ChEBI reference database and ontology for biologically relevant chemistry: Enhancements for 2013. Nucleic Acids Res. 2013, 41, D456–D463. [Google Scholar] [CrossRef] [PubMed]
- Lipscomb, C. Medical Subject Headings (MeSH). Bull. Med. Libr. Assoc. 2000, 88, 265–266. [Google Scholar] [PubMed]
- Li, Q.; Cheng, T.; Wang, Y.; Bryant, S.H. PubChem as a public resource for drug discovery. Drug Discov. Today 2010, 15, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Bodenreider, O. The Unified Medical Language System (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Hettne, K.; Stierum, R.; Schuemie, M.; Hendriksen, P.J.; Schijvenaars, B.J.; Mulligen, E.M.; Kleinjans, J.; Kors, J.A. A dictionary to identify small molecules and drugs in free text. Bioinformatics 2009, 25, 2983–2991. [Google Scholar] [CrossRef] [PubMed]
- Kolářik, C.; Hofmann-Apitius, M.; Zimmermann, M.; Fluck, J. Identification of new drug classification terms in textual resources. Bioinformatics 2007, 23, i264–i272. [Google Scholar] [CrossRef] [PubMed]
- Chhieng, D.; Day, T.; Gordon, G.; Hicks, J. Use of natural language programming to extract medication from unstructured electronic medical records. In Proceedings of the AMIA 2007 Annual Symposium, Chicago, IL, USA, 10–14 November 2007; p. 908.
- Wanger, R.; Fischer, M. The string-to-string correction problem. J. ACM 1974, 21, 168–173. [Google Scholar]
- Hall, P.; Dowling, G. Approximate string matching. Comput. Surv. 1980, 12, 381–402. [Google Scholar] [CrossRef]
- Philips, L. Hanging on the Metaphone. Comput. Lang. 1990, 7, 12. [Google Scholar]
- Levin, M.; Krol, M.; Doshi, A.; Reich, D. Extraction and mapping of drug names from free text to a standardized nomenclature. In Proceedings of the AMIA 2007 Annual Symposium, Chicago, IL, USA, 10–14 November 2007; pp. 438–442.
- Rindflesch, T.; Tanabe, L.; Weinstein, J.; Hunter, L. EDGAR: Extraction of drugs, genes and relations from the biomedical literature. In Proceedings of the Pacific Symposium on Biocomputing 2000 (PSB 2000), Honolulu, HI, USA, 5–9 January 2000; pp. 517–528.
- Sanchez-Cisneros, D.; Martínez, P.; Segura-Bedmar, I. Combining dictionaries and ontologies for drug name recognition in biomedical texts. In Proceedings of the 7th International Workshop on Data and Text Mining in Biomedical Informatics, Miami, FL, USA, 4–7 December 2013; pp. 27–30.
- Aronson, A. Effective mapping of biomedical text to the UMLS Metathesaurus: The metamap program. In Proceedings of the AMIA 2001 Annual Symposium, Washington, DC, USA, 3–7 November 2001; pp. 17–21.
- Sirohi, E.; Peissig, P. Study of effect of drug lexicons on medication extraction from electronic medical records. In Proceedings of the Pacific Symposium on Biocomputing 2005, Big Island of Hawaii, HI, USA, 4–8 January 2005; pp. 308–318.
- Xu, H.; Stenner, S.; Doan, S.; Johnson, K.B.; Waitman, L.R.; Denny, J.C. MedEx: A medication information extraction system for clinical narratives. J. Am. Med. Inform. Assoc. 2010, 17, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Ata, C.; Can, T. DBCHEM: A database query based solution for the chemical compound and drug name recognition task. In Proceedings of the 4th BioCreative Challenge Evaluation Workshop, Bethesda, MD, USA, 7–9 October 2013; pp. 42–46.
- SCOWL (And Friends). Available online: http://wordlist.aspell.net/ (accessed on 24 November 2015).
- Lowe, D.; Sayle, R. LeadMine: A grammar and dictionary driven approach to entity recognition. J. Cheminform. 2015, 7 (S1), S5. [Google Scholar] [CrossRef] [PubMed]
- Gold, S.; Elhadad, N.; Zhu, X.; Cinimo, J.J.; Hripcsak, G. Extracting structured medication event information from discharge summaries. In Proceedings of the AMIA 2008 Annual Symposium, Washington, DC, USA, 8–12 November 2008; pp. 237–241.
- Hamon, T.; Grabar, N. Linguistic approach for identification of medication names and related information in clinical narratives. J. Am. Med. Inform. Assoc. 2010, 17, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Morgan, A.; Das, A.; Garber, A. Investigation of unsupervised pattern learning techniques for bootstrap construction of a medical treatment lexicon. In Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing, Stroudsburg, PA, USA, 4–5 June 2009; pp. 63–70.
- Coden, A.; Gruhl, D.; Lewis, N.; Tanenblatt, M.; Terdiman, J. SPOT the drug! An unsupervised pattern matching method to extract drug names from very large clinical corpora. In Proceedings of the IEEE 2nd International Conference on Healthcare Informatics, Imaging and Systems Biology, San Diego, CA, USA, 27–28 September 2012; pp. 33–39.
- Zhao, H.; Huang, C.; Li, M.; Lu, B. A unified character-based tagging framework for Chinese word segmentation. ACM Trans. Asian Lang. Inf. Process. 2010, 9, 1–32. [Google Scholar] [CrossRef]
- Halgrim, S.; Xia, F.; Solti, I.; Cadag, E.; Uzuner, O. A cascade of classifiers for extracting medication information from discharge summaries. J. Biomed. Semant. 2011, 2 (S3), S2. [Google Scholar] [CrossRef] [PubMed]
- Björne, J.; Kaewphan, S.; Salakoski, T. UTurku: Drug named entity detection and drug-drug interaction extraction using SVM classification and domain knowledge. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 651–659.
- Malyszko, J.; Filipowska, A. Lexicon-free and context-free drug names identification methods using hidden markov models and pointwise mutual information. In Proceedings of the 6th International Workshop on Data and Text Mining in Biomedical Informatics, Maui, HI, USA, 29 October–2 November 2012; pp. 9–12.
- Patrick, J.; Li, M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J. Am. Med. Inform. Assoc. 2010, 17, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Rocktäschel, T.; Huber, T.; Weidlich, M.; Leser, U. WBI-NER: The impact of domain-specific features on the performance of identifying and classifying mentions of drugs. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 356–363.
- Abacha, A.B.; Chowdhury, M.F.M.; Karanasiou, A.; Mrabet, Y.; Lavelli, A.; Zweigenbaum, P. Text mining for pharmacovigilance: Using machine learning for drug name recognition and drug-drug interaction extraction and classification. J. Biomed. Inform. 2015, 58, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ji, D.; Yao, X.; Wei, X.; Liang, X. CHEMDNER system with mixed conditional random fields and multi-scale word clustering. J. Cheminform. 2015, 7 (S1), S4. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.; Matos, S.; Oliveira, J. A document processing pipeline for annotating chemical entities in scientific documents. J. Cheminform. 2015, 7 (S1), S7. [Google Scholar] [CrossRef] [PubMed]
- Lamurias, A.; Grego, T.; Couto, F.M. Chemical compound and drug name recognition using CRFs and semantic similarity based on ChEBI. In Proceedings of the 4th BioCreative Challenge Evaluation Workshop, Bethesda, MD, USA, 7–9 October 2013; pp. 75–81.
- Sikdar, U.K.; Ekbal, A.; Saha, S. Domain-independent model for chemical compound and drug name recognition. In Proceedings of the 4th BioCreative Challenge Evaluation Workshop, Bethesda, MD, USA, 7–9 October 2013; pp. 158–161.
- Huber, T.; Rocktäschel, T.; Weidlich, M.; Thomas, P.; Leser, U. Extended feature set for chemical named entity recognition and indexing. In Proceedings of the 4th BioCreative Challenge Evaluation Workshop, Bethesda, MD, USA, 7–9 October 2013; pp. 88–91.
- Liu, S.; Tang, B.; Chen, Q.; Wang, X.; Fan, X. Feature engineering for drug name recognition in biomedical texts: Feature conjunction and feature selection. Comput. Math. Method Med. 2015. [Google Scholar] [CrossRef] [PubMed]
- Rocktäschel, T.; Weidlich, M.; Leser, U. ChemSpot: A hybrid system for chemical named entity recognition. Bioinformatics 2012, 28, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.F.; de Souza, P.V.; Mercer, R.L.; Pietra, V.; Lai, J. Class-based N-gram models of natural language. Comput. Linguist. 1992, 18, 467–479. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of the 27th Annual Conference on Neural Information Processing Systems, Lake Tahoe, CA, USA, 5–10 December 2013; pp. 3111–3119.
- Forman, G. An extensive empirical study of feature selection metrics for text classification. J. Mach. Learn. Res. 2003, 3, 1289–1305. [Google Scholar]
- Yang, Y.; Pedersen, J. A comparative study on feature selection in text categorization. In Proceedings of the 14th International Conference on Machine Learning, Nashville, TN, USA, 8–12 July 1997; pp. 412–420.
- Zheng, Z.; Wu, X.; Srihari, R. Feature selection for text categorization on imbalanced data. ACM SIGKDD Explor. Newslett. 2004, 6, 80–89. [Google Scholar] [CrossRef]
- Akhondi, S.; Hettne, K.; Horst, E.; van Mulligen, E.M.; Kors, J.A. Recognition of chemical entities: Combining dictionary-based and grammar-based approaches. J Cheminform. 2015, 7 (S1), S10. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yang, Z.; Lin, H.; Li, Y. Drug name recognition in biomedical texts: A machine-learning-based method. Drug Discov. Today 2014, 19, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Tikk, D.; Solt, I. Improving textual medication extraction using combined conditional random fields and rule-based systems. J. Am. Med. Inform. Assoc. 2010, 17, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Korkontzelos, I.; Piliouras, D.; Dowsey, A.W.; Ananiadou, S. Boosting drug named entity recognition using an aggregate classifier. Artif. Intell. Med. 2015, 65, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Usié, A.; Cruz, J.; Comas, J.; Solsona, F.; Alves, R. A tool for the identification of chemical entities (CheNER-BioC). In Proceedings of the 4th BioCreative Challenge Evaluation Workshop, Bethesda, MD, USA, 7–9 October 2013; pp. 66–69.
- Yang, H. Automatic extraction of medication information from medical discharge summaries. J. Am. Med. Inform. Assoc. 2010, 17, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Irmer, M.; Bobach, C.; Böhme, T.; Laube, U.; Püschel, A.; Weber, L. Chemical named entity recognition with OCMiner. In Proceedings of the 4th BioCreative Challenge Evaluation Workshop, Bethesda, MD, USA, 7–9 October 2013; pp. 92–96.
- Sanchez-Cisneros, D.; Gali, F.A. UEM-UC3M: An ontology-based named entity recognition system for biomedical texts. In Proceedings of the 7th International Workshop on Semantic Evaluation, Atlanta, GA, USA, 14–15 June 2013; pp. 622–627.
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural. Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Hinton, G.; Deng, L.; Dahl, E.; Fadiga, L.; Metta, G. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal. Process. Mag. 2012, 29, 82–97. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G. ImageNet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems 25 (NIPS 2012), Lake Tahoe, NV, USA, 3–6 December 2012.
- Liu, X.; Zhang, S.; Wei, F.; Zhou, M. Recognizing named entity in tweets. In Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics, Portland, OR, USA, 19–24 June 2011; pp. 359–367.
- Majumder, M.; Barman, U.; Prasad, R.; Saurabh, K.; Saha, S. A novel technique for name identification from homeopathy diagnosis discussion forum. Proc. Technol. 2012, 6, 379–386. [Google Scholar] [CrossRef]
- Yan, S.; Spangler, W.; Chen, Y. Chemical name extraction based on automatic training data generation and rich feature set. IEEE-ACM Trans. Comput. Biol. Bioinform. 2013, 10, 1218–1233. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Wu, Y.; Jiang, M.; Denny, J.; Xu, H. Recognizing and encoding disorder concepts in clinical text using machine learning and vector space model. In Proceedings of the Online Working Notes of the CLEF 2013 Evaluation Labs and Workshop, Valencia, Spain, 23–26 September 2013.
- Cogley, J.; Stokes, N.; Carthy, J. Medical disorder recognition with structural support vector machines. In Proceedings of the Online Working Notes of the CLEF 2013 Evaluation Labs and Workshop, Valencia, Spain, 23–26 September 2013.
- Leal, A.; Martins, B.; Couto, F.M. ULisboa: Recognition and normalization of medical concepts. In Proceedings of the 9th International Workshop on Semantic Evaluation, Denver, CO, USA, 4–5 June 2015; pp. 406–411.
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Tang, B.; Chen, Q.; Wang, X. Drug Name Recognition: Approaches and Resources. Information 2015, 6, 790-810. https://doi.org/10.3390/info6040790
Liu S, Tang B, Chen Q, Wang X. Drug Name Recognition: Approaches and Resources. Information. 2015; 6(4):790-810. https://doi.org/10.3390/info6040790
Chicago/Turabian StyleLiu, Shengyu, Buzhou Tang, Qingcai Chen, and Xiaolong Wang. 2015. "Drug Name Recognition: Approaches and Resources" Information 6, no. 4: 790-810. https://doi.org/10.3390/info6040790
APA StyleLiu, S., Tang, B., Chen, Q., & Wang, X. (2015). Drug Name Recognition: Approaches and Resources. Information, 6(4), 790-810. https://doi.org/10.3390/info6040790





