Complexity-Driven Adversarial Validation for Corrupted Medical Imaging Data
Abstract
1. Introduction
- Extension to diverse medical datasets: The proposed methodology for evaluating distribution disturbances was extended to twelve 2D medical datasets in MedMNIST encompassing both binary and multiclass classification tasks, as well as grayscale and RGB image modalities: PathMNIST, ChestMNIST, DermaMNIST, OCTMNIST, PneumoniaMNIST, RetinaMNIST, BreastMNIST, BloodMNIST, TissueMNIST, OrganAMNIST, OrganCMNIST, and OrganSMNIST.
- Clinically inspired distribution shift simulation: We apply distortions by simulating distribution shifts induced by clinically relevant, real-world factors, including motion blur, additive noise, brightness and contrast variation, and sharpness variation. These perturbations represent common variations in medical imaging that result from differences in devices and acquisition protocols. Three severity levels were applied to each distortion type across all benchmark datasets, generating 12 corrupted variants per dataset. By combining all four distortion types, a total of 996 perturbed datasets were generated, allowing a comprehensive analysis of the effect of different distortions on distribution shift by dataset type (grayscale or color), number of classes (binary or multiclass), and distortion level.
- Intra-Class Separability Analysis: The CSG metric is used to quantify distortion-induced alterations in intra-class separability, highlighting variations across perturbation categories.
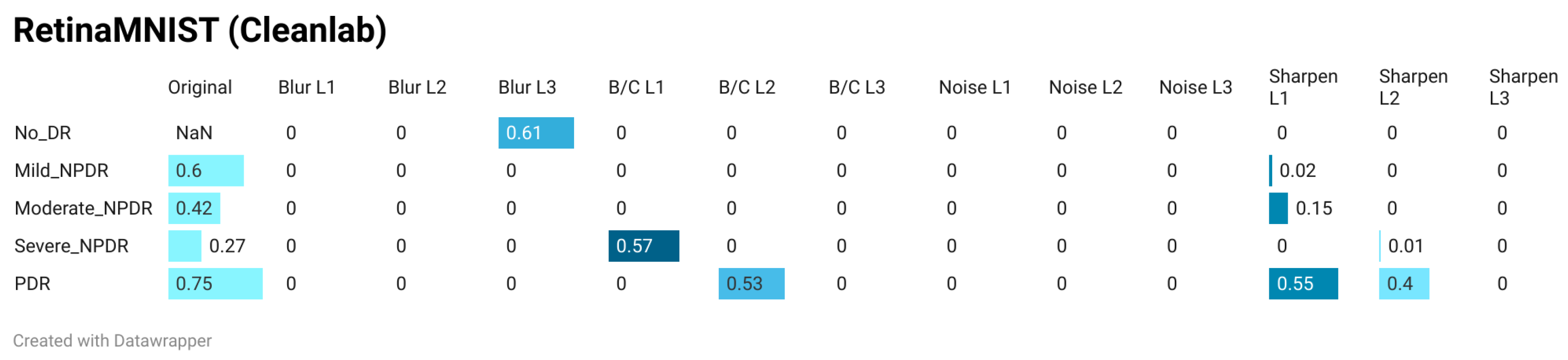
- Comparison with a widely adopted i.i.d. assumption metric: A comprehensive comparison between the results of the proposed methodology and those obtained using Cleanlab’s Non-IID score was conducted on the RetinaMNIST dataset using a pre-trained ResNet-50 architecture. A first comparison included a class-based evaluation of the Non-IID score and the CSG score across the four distortion types and three distortion levels. Another comparison quantified the correlation coefficient between the CSG value vector for the five dataset classes and the corresponding vector obtained using the Non-IID score for each distortion.
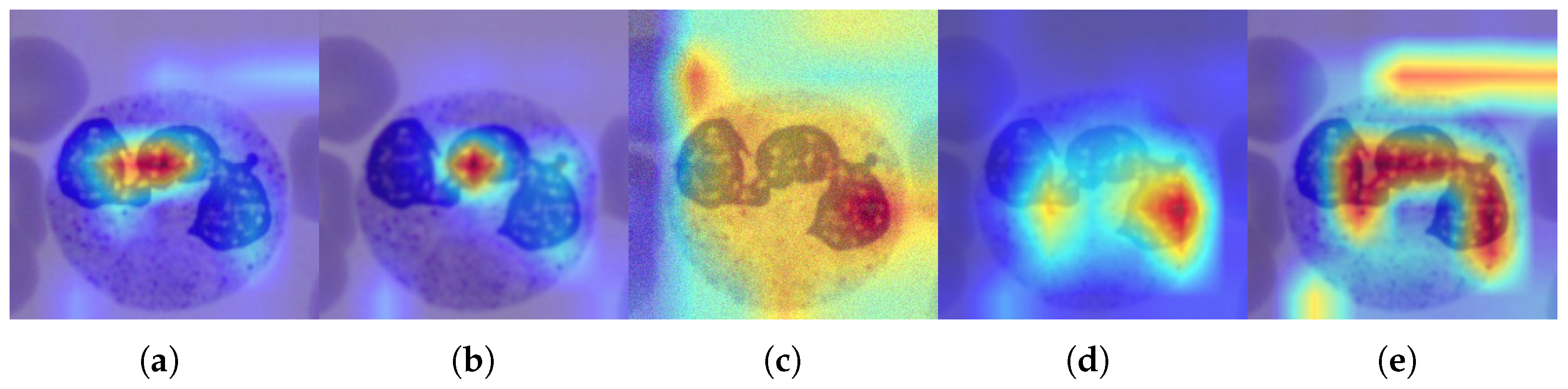
- Integration of interpretability analysis: Training and inference of a ResNet-50 architecture were conducted on the BloodMNIST dataset and its distorted versions, enabling the computation of class activation maps to support and interpret the experimental results.
2. Materials and Methods
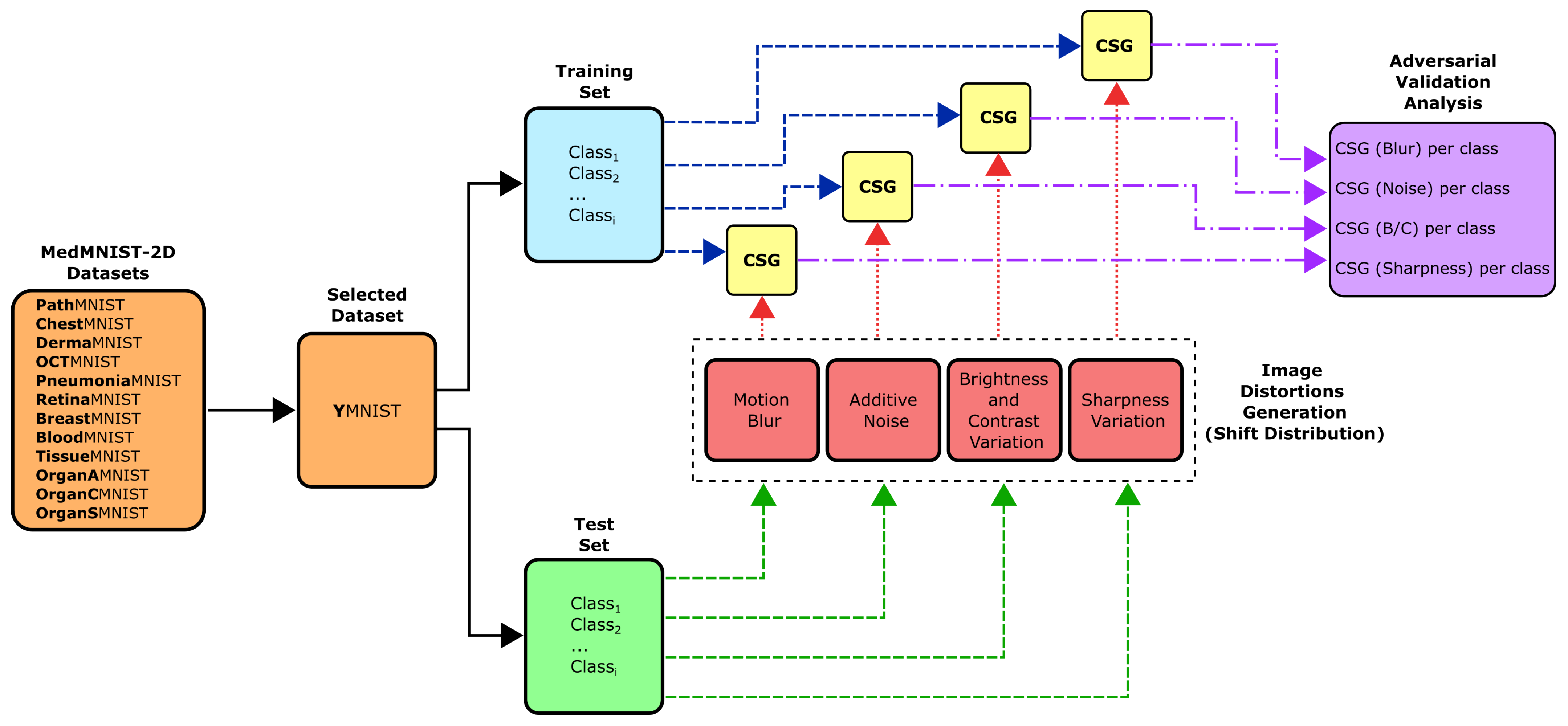
2.1. Methodology Overview
2.2. Dataset Complexity Measure
- 1.
- Degree matrix D: A diagonal matrix in which each element represents the total connectivity of class i, and which is equal to the sum of the weights, , for each j.
- 2.
- Laplacian matrix : Encode the topological structure of class relationships.
2.3. MedMNIST-2D
2.4. Distribution Shift
- 1.
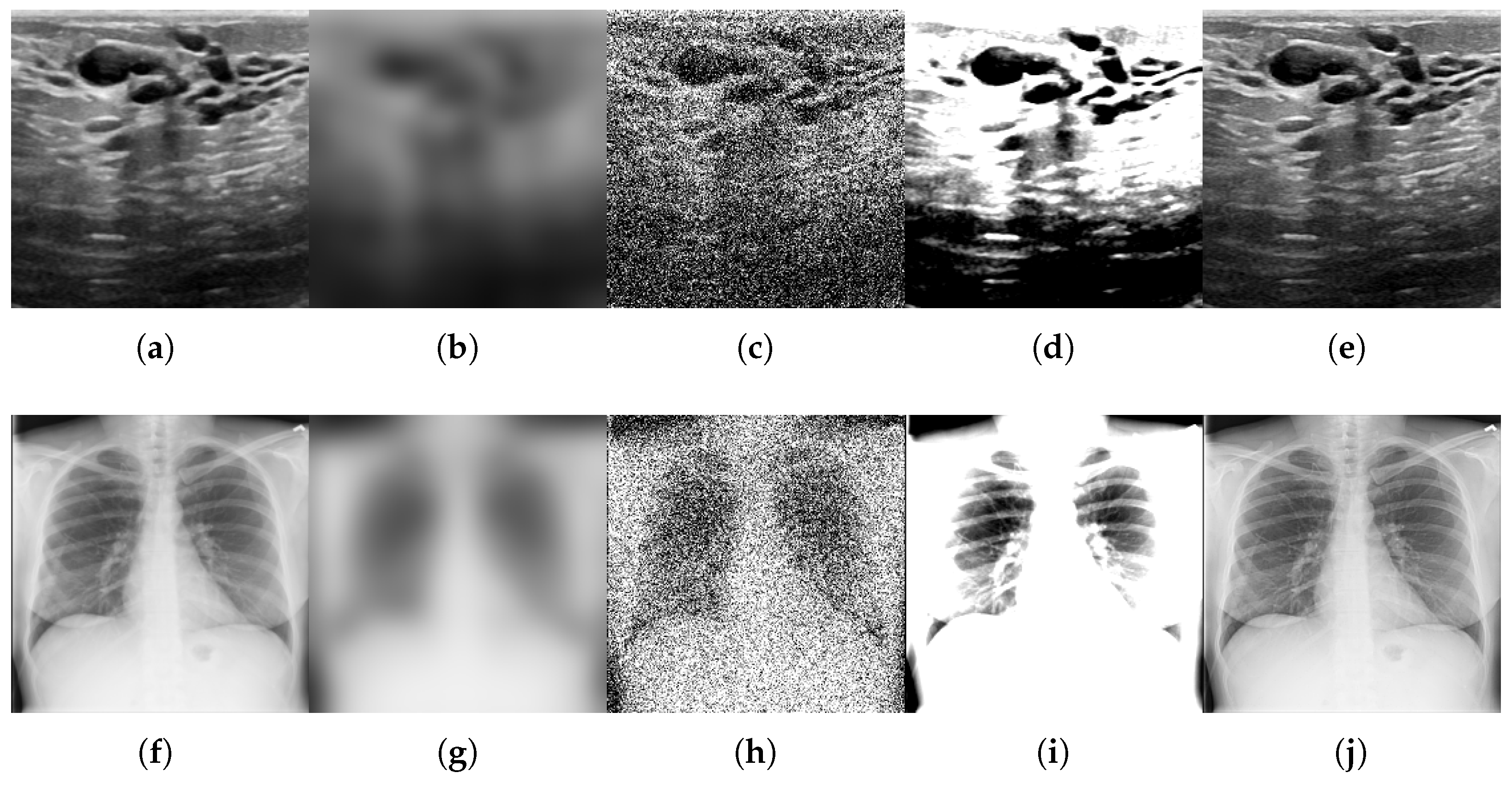
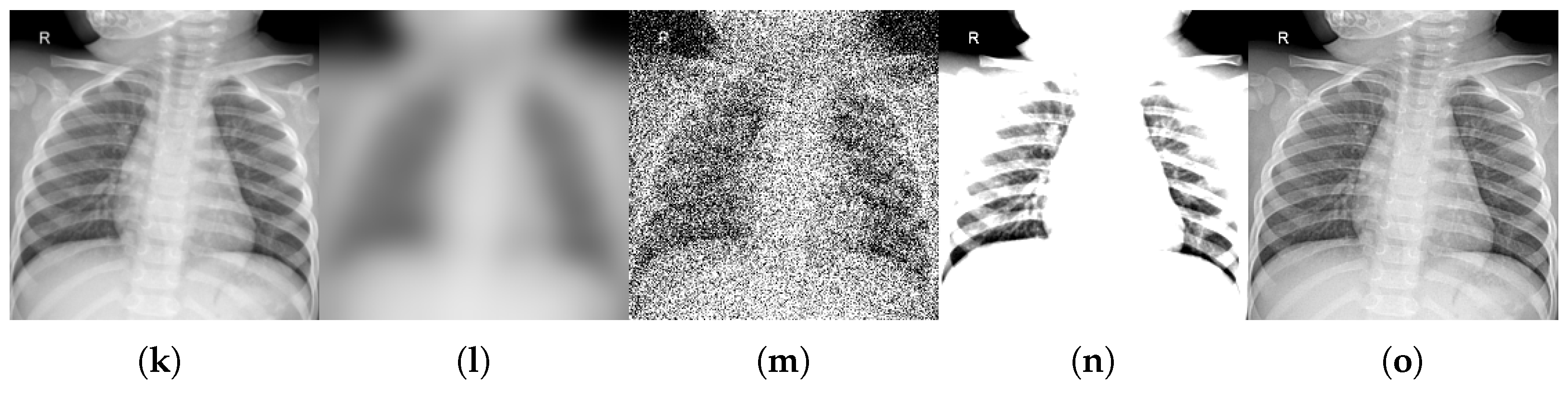
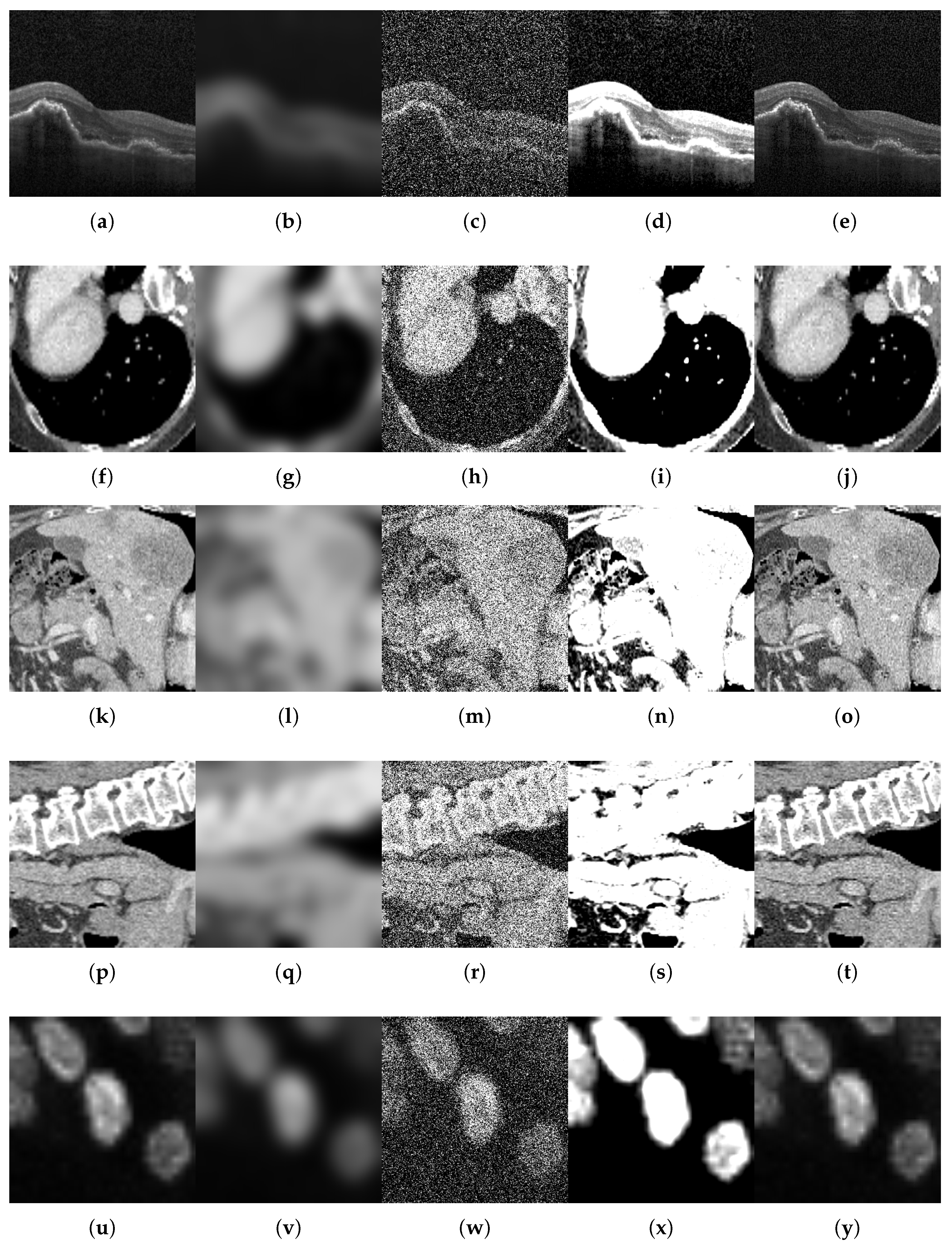
- Motion blur, which simulates patient movement during acquisition, is implemented via Gaussian kernel convolution with a radius of R pixels in the range of . This simulates increasing levels of patient movement while taking into account that motion artifacts degrade edge sharpness.
- 2.
- Additive noise, representing sensor or electronic degradation, is introduced by zero-mean Gaussian noise with standard deviations in to emulate the effects of sensor degradation, introducing stochastic signal corruption.
- 3.
- Brightness and contrast variation, which emulates exposure miscalibration, is achieved by adjusting both parameters simultaneously using the following pairs: . These pairs represent scanner exposure miscalibrations, where variations in illumination alter the perceived tissue contrast.
- 4.
- Sharpness variation used to adjust the sharpness of the image in order to modify image details and simulate artifacts that enhance the resolution of high-frequency details. Three values of enhancement factor were used: f in , where a value less than 1.0 gives a blurred image, a factor of 1.0 gives the original image, and a value greater than 1.0 gives a sharper image.
2.5. Adversarial Validation Analysis
3. Experimental Results
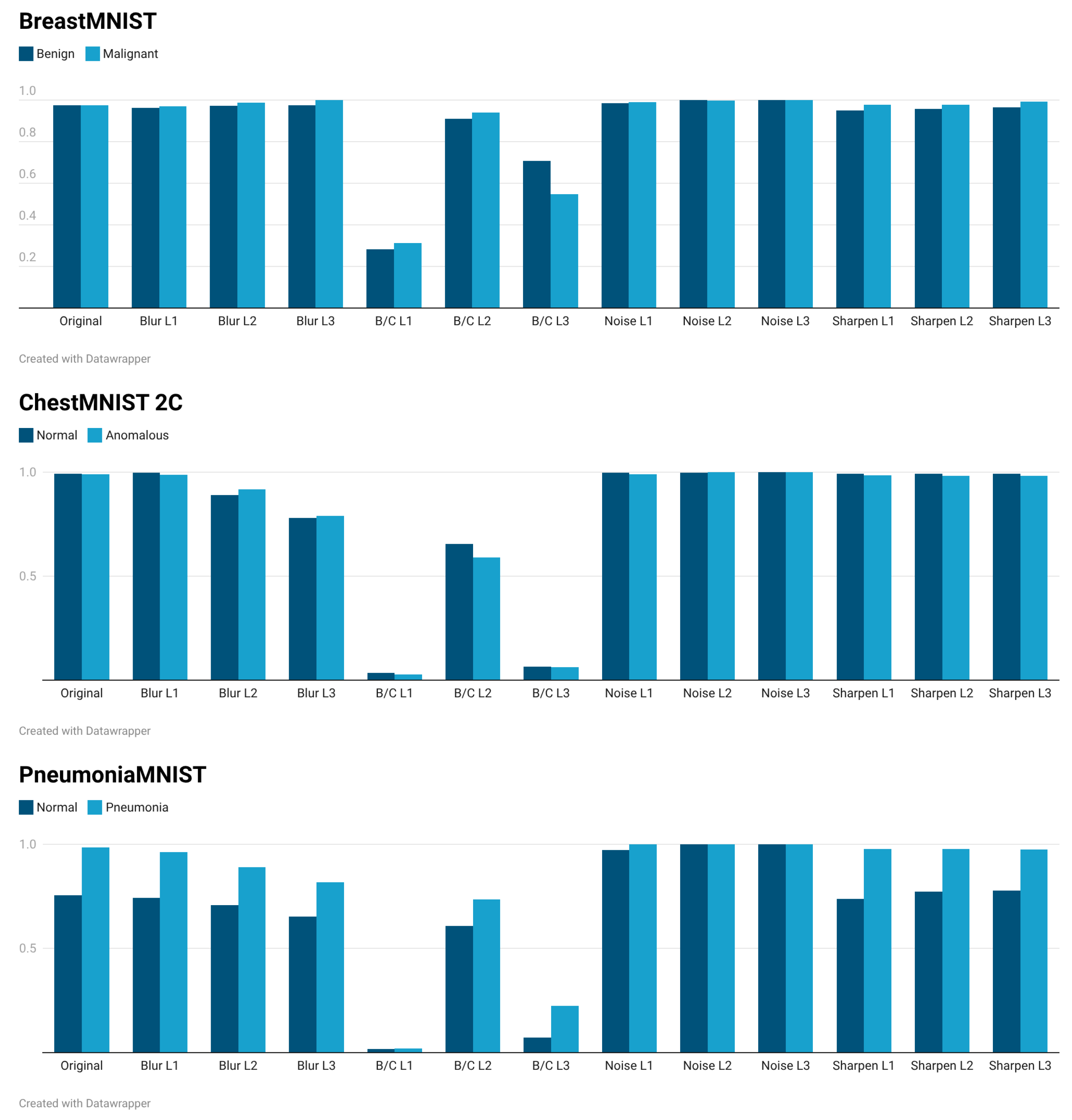
3.1. Adversarial Validation Results in Grayscale Images
3.2. Adversarial Validation Results in Color Images
3.3. Overall Analysis
3.4. Comparison of Results
3.5. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | Computed Tomography |
| CNN | Convolutional Neural Network |
| CSG | Cumulative Spectral Gradient |
| DS | Distribution Shift |
| i.i.d. | independent and identical distribution |
| ID | In-Distribution |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| NIH | National Institutes of Health |
| OOD | Out-Of-Distribution |
| PET | Positron Emission Tomography |
| US | Ultrasound |
References
- Clement David-Olawade, A.; Olawade, D.B.; Vanderbloemen, L.; Rotifa, O.B.; Fidelis, S.C.; Egbon, E.; Akpan, A.O.; Adeleke, S.; Ghose, A.; Boussios, S. AI-Driven Advances in Low-Dose Imaging and Enhancement—A Review. Diagnostics 2025, 15, 689. [Google Scholar]
- Abhisheka, B.; Biswas, S.K.; Purkayastha, B.; Das, D.; Escargueil, A. Recent trend in medical imaging modalities and their applications in disease diagnosis: A review. Multimed. Tools Appl. 2024, 83, 43035–43070. [Google Scholar]
- Melazzini, L.; Bortolotto, C.; Brizzi, L.; Achilli, M.; Basla, N.; D’Onorio De Meo, A.; Gerbasi, A.; Bottinelli, O.M.; Bellazzi, R.; Preda, L. AI for image quality and patient safety in CT and MRI. Eur. Radiol. Exp. 2025, 9, 28. [Google Scholar] [CrossRef]
- Bian, Y.; Li, J.; Ye, C.; Jia, X.; Yang, Q. Artificial intelligence in medical imaging: From task-specific models to large-scale foundation models. Chin. Med. J. 2025, 138, 651–663. [Google Scholar]
- Jiménez-Sánchez, A.; Avlona, N.; Boer, S.; Campello, V.; Feragen, A.; Ferrante, E.; Ganz, M.; Gichoya, J.; Gonzalez, C.; Groefsema, S.; et al. In the picture: Medical imaging datasets, artifacts, and their living review. In Proceedings of the 2025 ACM Conference on Fairness, Accountability, and Transparency (FAccT ’25), Athens, Greece, 23–26 June 2025; Association for Computing Machinery: New York, NY, USA, 2025; pp. 511–531. [Google Scholar]
- Daneshjou, R.; Vodrahalli, K.; Novoa, R.; Jenkins, M.; Liang, W.; Rotemberg, V.; Ko, J.; Swetter, S.; Bailey, E.; Gevaert, O.; et al. Disparities in dermatology AI performance on a diverse, curated clinical image set. Sci. Adv. 2022, 8, eabq6147. [Google Scholar] [CrossRef]
- Yao, H.; Choi, C.; Cao, B.; Lee, Y.; Koh, P.; Finn, C. Wild-time: A benchmark of in-the-wild distribution shift over time. In Proceedings of the 36th International Conference on Neural Information Processing Systems, New Orleans, LA, USA, 28 November–9 December 2022. [Google Scholar]
- Zhang, H.; Singh, H.; Ghassemi, M.; Joshi, S. “Why did the model fail?”: Attributing model performance changes to distribution shifts. In Proceedings of the 40th International Conference on Machine Learning, Honolulu, HI, USA, 23–29 July 2023. [Google Scholar]
- Kim, T.; Park, S.; Lim, S.; Jung, Y.; Muandet, K.; Song, K. Sufficient Invariant Learning for Distribution Shift. In Proceedings of the 2025 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 11–15 June 2025; pp. 4958–4967. [Google Scholar]
- Taori, R.; Dave, A.; Shankar, V.; Carlini, N.; Recht, B.; Schmidt, L. Measuring robustness to natural distribution shifts in image classification. In Proceedings of the 34th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 6–12 December 2020. [Google Scholar]
- Izzo, Z.; Ying, L.; Zou, J. How to learn when data reacts to your model: Performative gradient descent. In Proceedings of the 38th International Conference on Machine Learning, Online, 18–24 July 2021; pp. 4641–4650. [Google Scholar]
- Renza, D.; Moya-Albor, E.; Chavarro, A. Adversarial Validation in Image Classification Datasets by Means of Cumulative Spectral Gradient. Algorithms 2024, 17, 531. [Google Scholar] [CrossRef]
- Koh, P.; Sagawa, S.; Marklund, H.; Xie, S.; Zhang, M.; Balsubramani, A.; Hu, W.; Yasunaga, M.; Phillips, R.; Gao, I.; et al. WILDS: A benchmark of in-the-wild distribution shifts. In Proceedings of the 38th International Conference on Machine Learning, Online, 18–24 July 2021; pp. 5637–5664. [Google Scholar]
- Chen, A.; Lee, Y.; Setlur, A.; Levine, S.; Finn, C. Confidence-based model selection: When to take shortcuts for subpopulation shifts. arXiv 2023, arXiv:2306.11120. [Google Scholar] [CrossRef]
- Northcutt, C.; Jiang, L.; Chuang, I. Confident Learning: Estimating Uncertainty in Dataset Labels. J. Artif. Intell. Res. JAIR 2021, 70, 1373–1411. [Google Scholar]
- Yao, H.; Wang, Y.; Li, S.; Zhang, L.; Liang, W.; Zou, J.; Finn, C. Improving out-of-distribution robustness via selective augmentation. In Proceedings of the 39th International Conference on Machine Learning, Baltimore, MD, USA, 17–23 July 2022; pp. 25407–25437. [Google Scholar]
- Liang, W.; Yang, X.; Zou, J. Metashift: A dataset of datasets for evaluating contextual distribution shifts. arXiv 2022, arXiv:2202.06523. [Google Scholar] [CrossRef]
- Öztürk, Ş.; Duran, O.; Çukur, T. DenoMamba: A fused state-space model for low-dose CT denoising. IEEE J. Biomed. Health Inform. 2025. [Google Scholar] [CrossRef]
- Kabas, B.; Arslan, F.; Nezhad, V.; Ozturk, S.; Saritas, E.; Çukur, T. Physics-Driven Autoregressive State Space Models for Medical Image Reconstruction. arXiv 2025, arXiv:2412.09331. [Google Scholar]
- Zhang, S.; Zhang, X.; Shen, L.; Wan, S.; Ren, W. Wavelet-based physically guided normalization network for real-time traffic dehazing. Pattern Recognit. 2026, 172, 112451. [Google Scholar]
- Liu, Y.; Wang, X.; Hu, E.; Wang, A.; Shiri, B.; Lin, W. VNDHR: Variational Single Nighttime Image Dehazing for Enhancing Visibility in Intelligent Transportation Systems via Hybrid Regularization. IEEE Trans. Intell. Transp. Syst. 2025, 26, 10189–10203. [Google Scholar] [CrossRef]
- Aguilera-González, S.; Renza, D.; Moya-Albor, E. Evaluation of Dataset Distribution in Biomedical Image Classification Against Image Acquisition Distortions. In Proceedings of the 20th International Symposium on Medical Information Processing and Analysis (SIPAIM), Antigua, Guatemala, 13–15 November 2024; pp. 1–6. [Google Scholar]
- Branchaud-Charron, F.; Achkar, A.; Jodoin, P. Spectral Metric for Dataset Complexity Assessment. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 3210–3219. [Google Scholar]
- González-Santoyo, C.; Renza, D.; Moya-Albor, E. Identifying and Mitigating Label Noise in Deep Learning for Image Classification. Technologies 2025, 13, 132. [Google Scholar] [CrossRef]
- Yang, J.; Shi, R.; Wei, D.; Liu, Z.; Zhao, L.; Ke, B.; Pfister, H.; Ni, B. MedMNIST v2-A large-scale lightweight benchmark for 2D and 3D biomedical image classification. Sci. Data 2023, 10, 41. [Google Scholar] [PubMed]
- Yang, J.; Shi, R.; Ni, B. MedMNIST Classification Decathlon: A Lightweight AutoML Benchmark for Medical Image Analysis. In Proceedings of the IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 191–195. [Google Scholar]
- Cummings, J.; Snorrason, E.; Mueller, J. Detecting Dataset Drift and Non-IID Sampling via k-Nearest Neighbors. In Proceedings of the ICML 2023 Data-centric Machine Learning Research Workshop, Honolulu, HI, USA, 23–29 July 2023. [Google Scholar]
- Wightman, R.; Touvron, H.; Jégou, H. Resnet strikes back: An improved training procedure in timm. arXiv 2021, arXiv:2110.00476. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Haga, A.; Takahashi, W.; Aoki, S.; Nawa, K.; Yamashita, H.; Abe, O.; Nakagawa, K. Standardization of imaging features for radiomics analysis. J. Med. Investig. 2019, 66, 35–37. [Google Scholar]
- Shinohara, R.; Sweeney, E.; Goldsmith, J.; Shiee, N.; Mateen, F.; Calabresi, P.; Jarso, S.; Pham, D.; Reich, D.; Crainiceanu, C.; et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014, 6, 9–19. [Google Scholar]
- Kanakaraj, P.; Yao, T.; Cai, L.; Lee, H.; Newlin, N.; Kim, M.; Gao, C.; Pechman, K.; Archer, D.; Hohman, T.; et al. Deepn4: Learning N4ITK bias field correction for T1-weighted images. Neuroinformatics 2024, 22, 193–205. [Google Scholar] [PubMed]
- Xu, C.; Sun, Y.; Zhang, Y.; Liu, T.; Wang, X.; Hu, D.; Huang, S.; Li, J.; Zhang, F.; Li, G. Stain Normalization of Histopathological Images Based on Deep Learning: A Review. Diagnostics 2025, 15, 1032. [Google Scholar] [CrossRef] [PubMed]
- Bhati, D.; Neha, F.; Amiruzzaman, M. A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging. J. Imaging 2024, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Chavarro, A.; Renza, D.; Moya-Albor, E. ConvNext as a Basis for Interpretability in Coffee Leaf Rust Classification. Mathematics 2024, 12, 2668. [Google Scholar] [CrossRef]




| Dataset | Modality | Classes | Size (Train/Val/Test) | Original Resolution | Description | Example Application |
|---|---|---|---|---|---|---|
| Breast | Histology (Grayscale) | 2 | 546/78/156 | 50 × 50 to 500 × 500 | Breast biopsy images for benign/malignant classification | Breast cancer diagnosis |
| Chest | X-ray (Grayscale) | 2 | 78,468/11,219/22,433 | 1024 × 1024 | Chest X-rays for detecting multiple lung conditions | Pneumonia, atelectasis detection, etc. |
| Pneumonia | X-ray (Grayscale) | 2 | 4708/524/624 | 1024 × 1024 | Chest X-rays for binary classification (pneumonia vs. normal) | Pneumonia diagnosis |
| Dataset | Modality | Classes | Size (Train/Val/Test) | Original Resolution | Description | Example Application |
|---|---|---|---|---|---|---|
| OCT | OCT (Grayscale) | 4 | 97,477/10,832/1000 | 384 × 384 | Optical coherence tomography images for retinal diseases. | Macular degeneration detection |
| OrganA | CT (Grayscale) | 11 | 34,581/6491/17,778 | Variable | Axial-view CT slices for multi-organ segmentation | Abdominal organ localization |
| OrganC | CT (Grayscale) | 11 | 13,000/2392/8266 | Variable | Coronal-view CT slices for multi-organ segmentation | Cross-sectional organ analysis |
| OrganS | CT (Grayscale) | 11 | 13,940/2452/8829 | Variable | Sagittal-view CT slices for multi-organ segmentation | Lateral organ structure assessment |
| Tissue | Histology (Grayscale) | 8 | 165,466/23,640/47,280 | 20 × 20 to 60 × 60 | Human tissue images for cell type classification | Biopsy analysis |
| Dataset | Modality | Classes | Size (Train/Val/Test) | Original Resolution | Description | Example Application |
|---|---|---|---|---|---|---|
| Blood | Microscopy (RGB) | 8 | 11,959/1712/3421 | 300 × 200 to 500 × 500 | Peripheral blood cells for cell type classification | Blood disorder identification |
| Derma | Dermatoscopy (RGB) | 7 | 7007/1003/2005 | 600 × 450 | Skin lesion images for dermatological disease classification. | Melanoma diagnosis |
| Path | Histology (RGB) | 9 | 89,996/10,004/7180 | Variable | Colorectal tissue images for pathological pattern classification | Colorectal cancer diagnosis |
| Retina | Fundus (RGB) | 5 | 1080/120/400 | Variable | Retina images for diabetic retinopathy grading | Diabetic retinopathy detection |
| Variation | Correlation |
|---|---|
| Original | −0.7247 |
| Blur L1 | 0.1398 |
| Blur L2 | 0.4916 |
| Blur L3 | 0.6898 |
| B/C L1 | −0.1909 |
| B/C L2 | 0.7138 |
| B/C L3 | 0.9475 |
| Noise L1 | −0.9911 |
| Noise L2 | −0.9895 |
| Noise L3 | −1.0000 |
| Sharpen L1 | −0.9560 |
| Sharpen L2 | −0.9928 |
| Sharpen L3 | −0.9874 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Renza, D.; Brieva, J.; Moya-Albor, E. Complexity-Driven Adversarial Validation for Corrupted Medical Imaging Data. Information 2026, 17, 125. https://doi.org/10.3390/info17020125
Renza D, Brieva J, Moya-Albor E. Complexity-Driven Adversarial Validation for Corrupted Medical Imaging Data. Information. 2026; 17(2):125. https://doi.org/10.3390/info17020125
Chicago/Turabian StyleRenza, Diego, Jorge Brieva, and Ernesto Moya-Albor. 2026. "Complexity-Driven Adversarial Validation for Corrupted Medical Imaging Data" Information 17, no. 2: 125. https://doi.org/10.3390/info17020125
APA StyleRenza, D., Brieva, J., & Moya-Albor, E. (2026). Complexity-Driven Adversarial Validation for Corrupted Medical Imaging Data. Information, 17(2), 125. https://doi.org/10.3390/info17020125








