Prior Knowledge Shapes Success When Large Language Models Are Fine-Tuned for Biomedical Term Normalization
Abstract
1. Introduction
- Latent probabilistic knowledge. Hidden or partially accessible knowledge revealed through probabilistic querying of the model, even when deterministic (greedy) decoding fails [36]. For example, if an LLM is queried 100 times and returns the correct ontology ID only 5% of the time, this indicates latent probabilistic knowledge, distinct from not known at all.
- Partial subtoken knowledge. Incomplete but non-random knowledge of the subtoken sequences comprising ontology identifiers, reflected in deterministic outputs that are close to, but not exactly, correct. For example, an LLM that predicts HP:0001259 for Ataxia instead of the correct HP:0001251 demonstrates partial subtoken knowledge, even though greedy decoding produces an incorrect response.
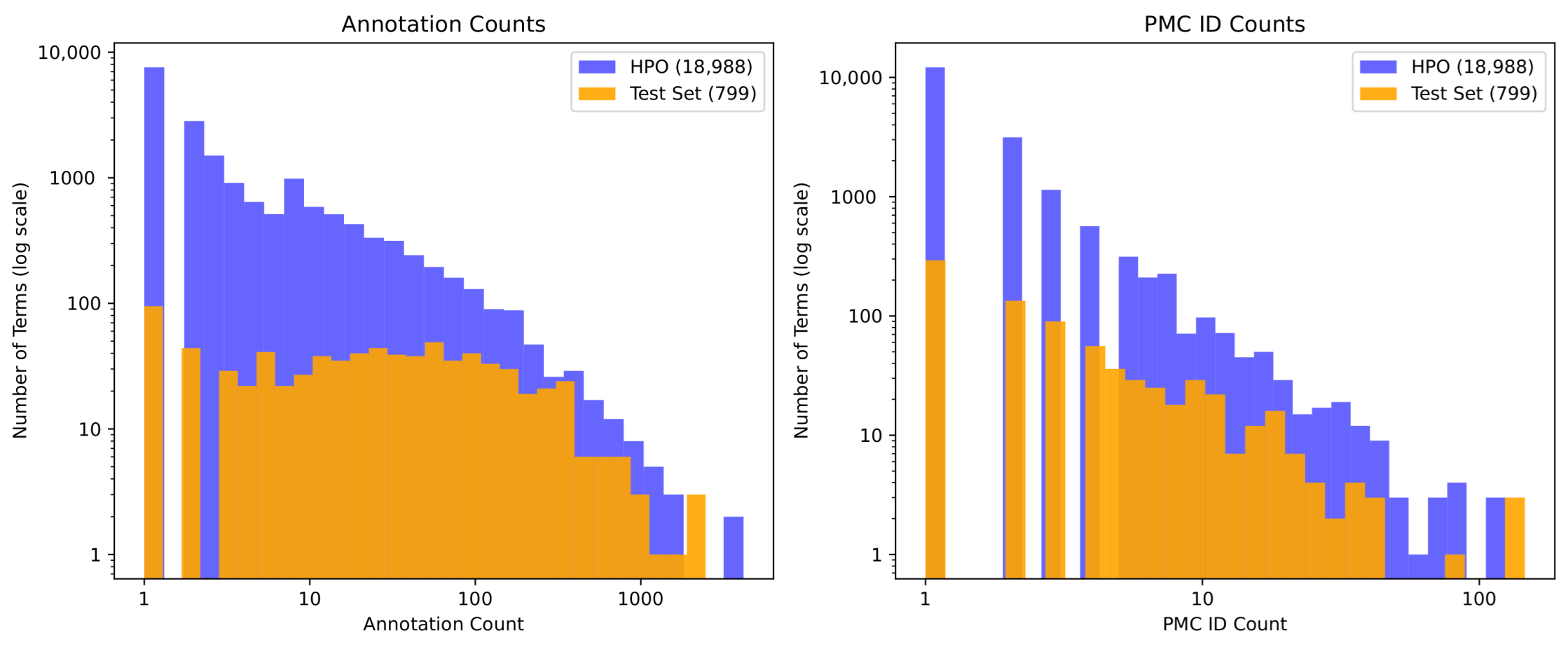
- Term familiarity. The likely exposure of the model to specific term–identifier pairs during pre-training, estimated using external proxies such as annotation frequency in OMIM and Orphanet [37,38], and identifier frequency in the PubMed Central (PMC) corpus [39]. For example, the LLM is more likely to be familiar with the term Hypotonia (decreased tone), which has 1783 disease annotations in HPO, than with Mydriasis (small pupils), which has only 25 annotations.
2. Materials and Methods
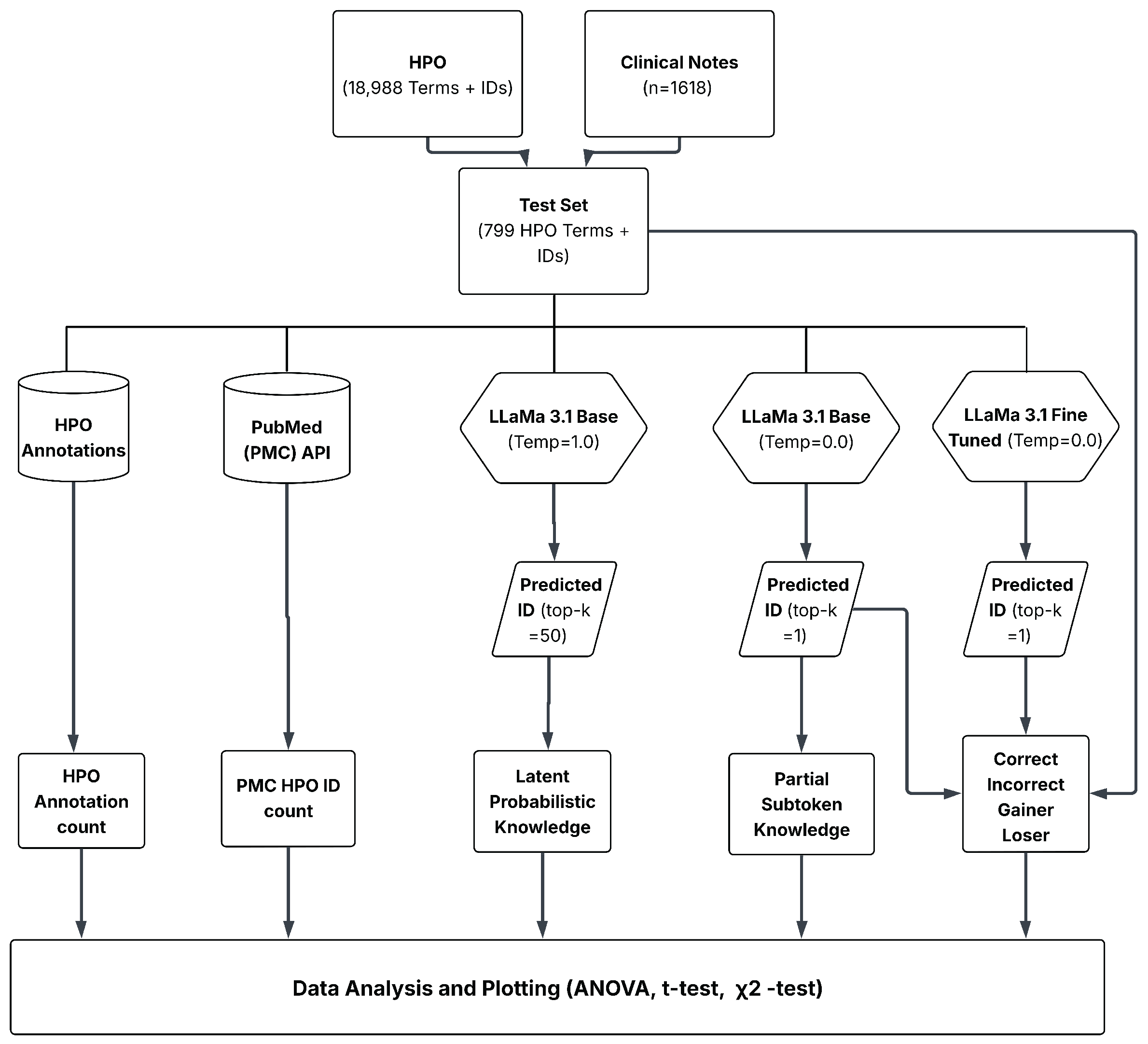
2.1. Workflow
2.2. HPO Dataset
2.3. Test Terms
2.4. Large Language Models and Fine-Tuning
2.5. Top-1 Accuracy (Deterministic Inference)
2.6. Outcome Classification for Fine-Tuning
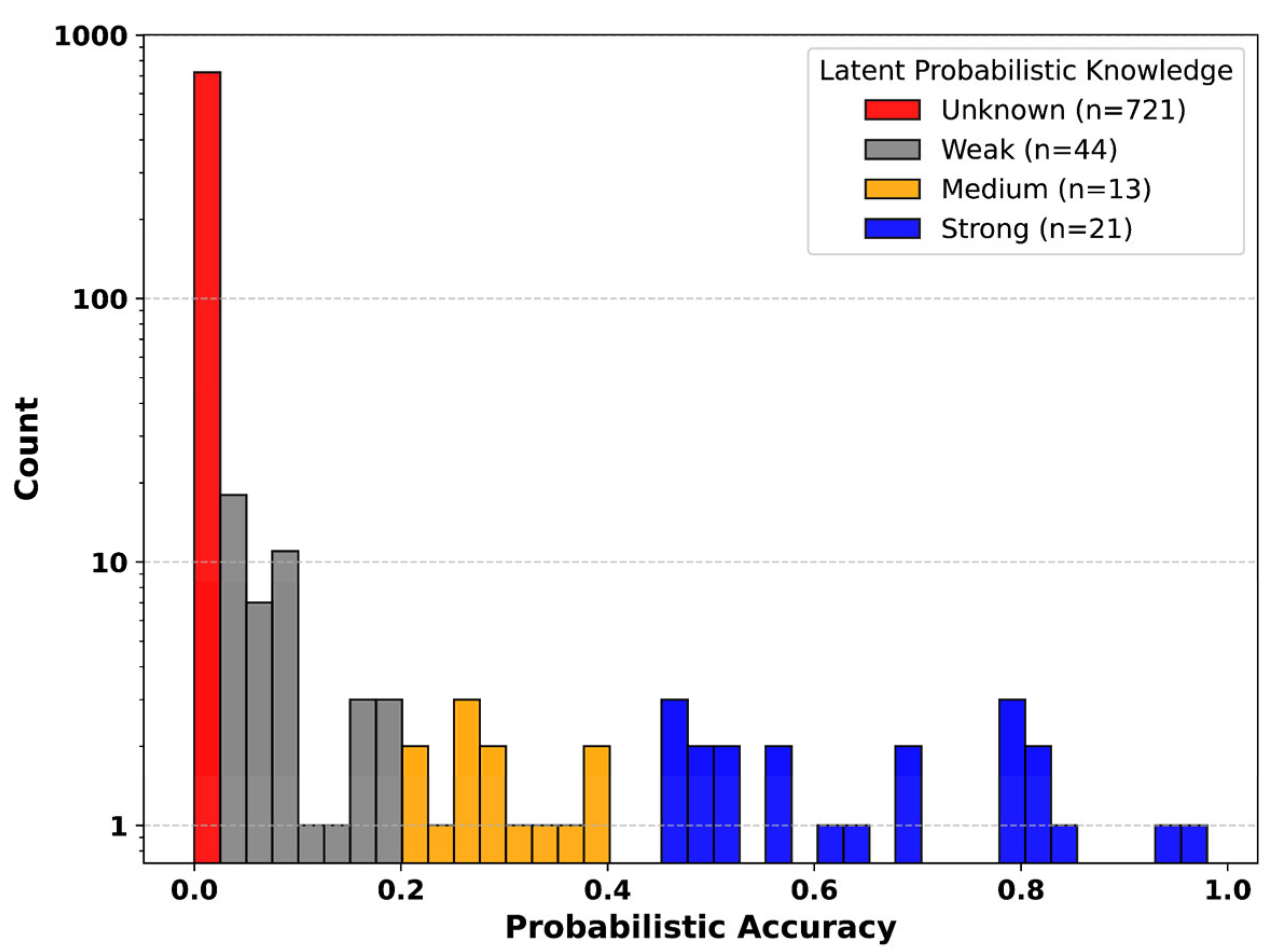
2.7. Latent Probabilistic Knowledge
2.8. Partial Subtoken Knowledge
2.9. Term Familiarity
2.10. Statistical Analysis
3. Results
3.1. Baseline and Fine-Tuned Performance of LLaMA 3.1 8B on HPO Term Normalization
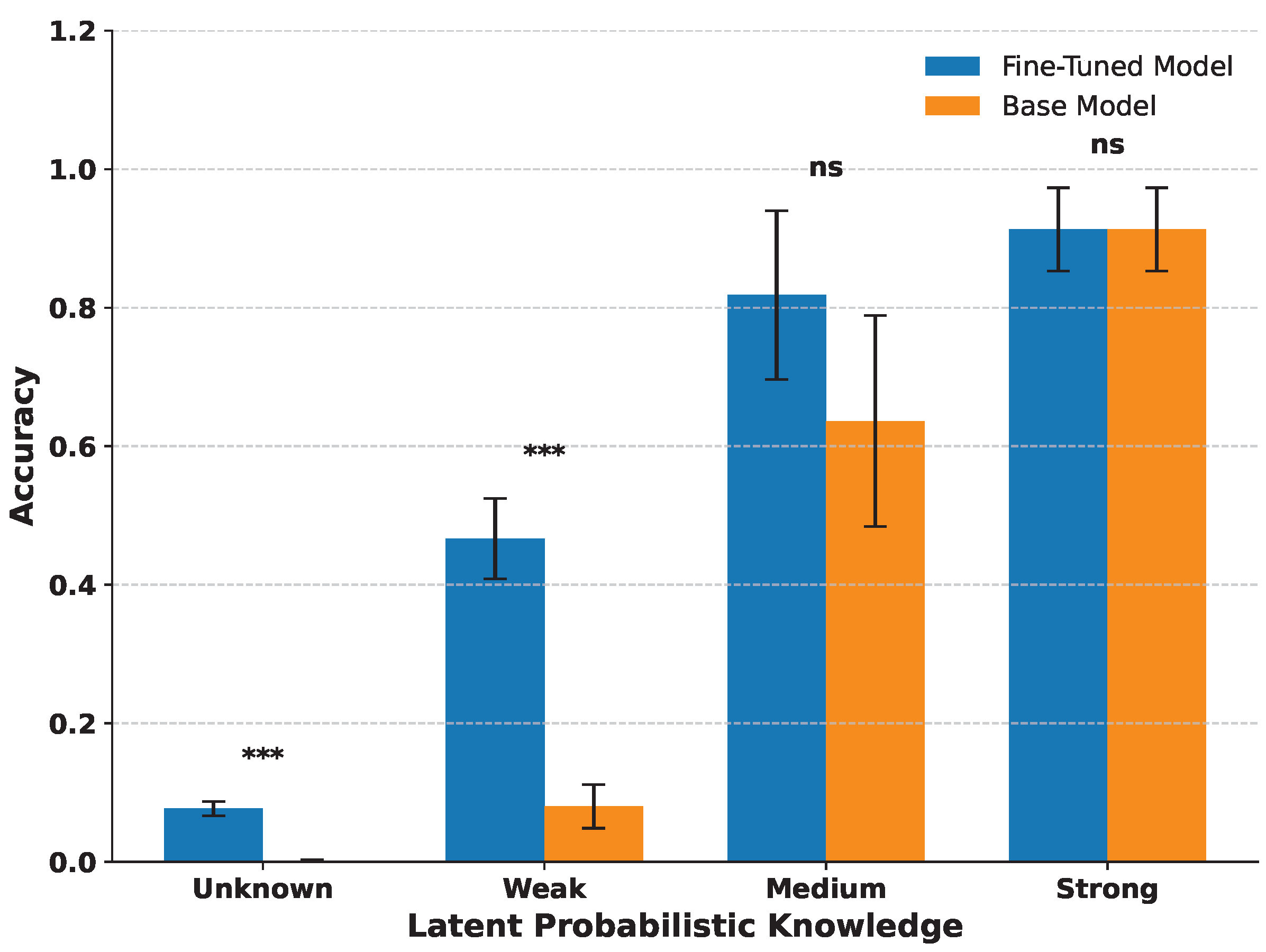
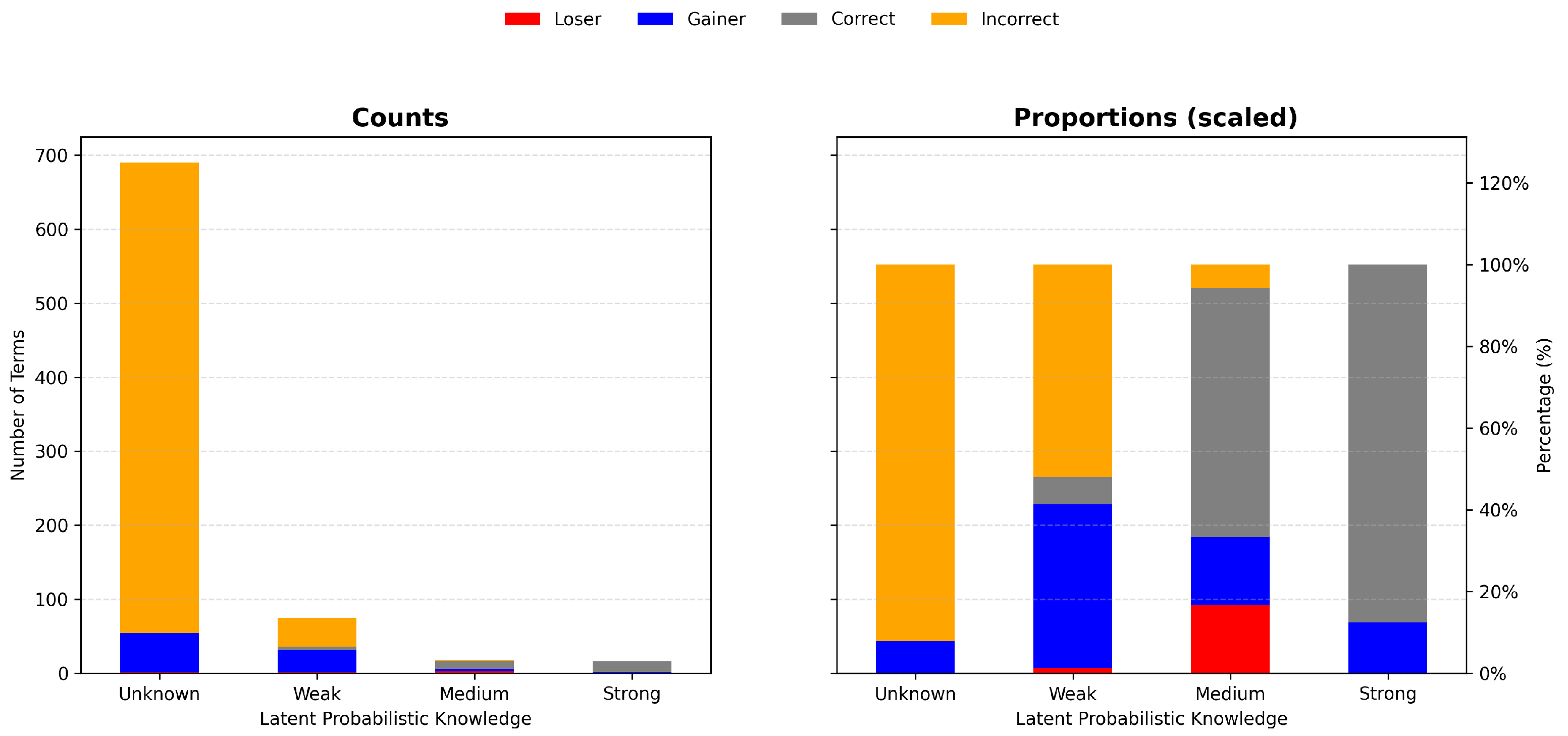
3.2. Latent Probabilistic Knowledge Predicts Fine-Tuning Success
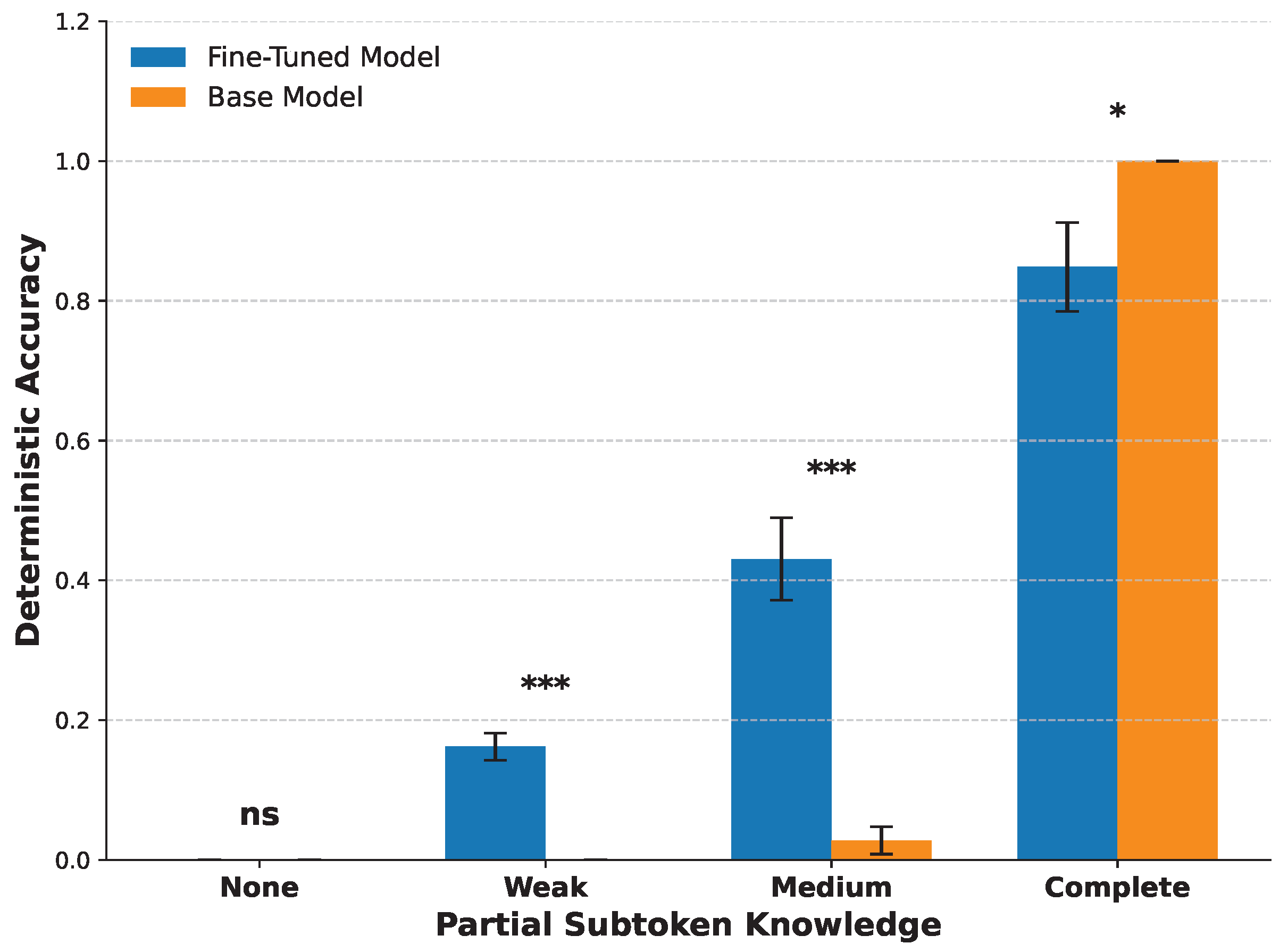
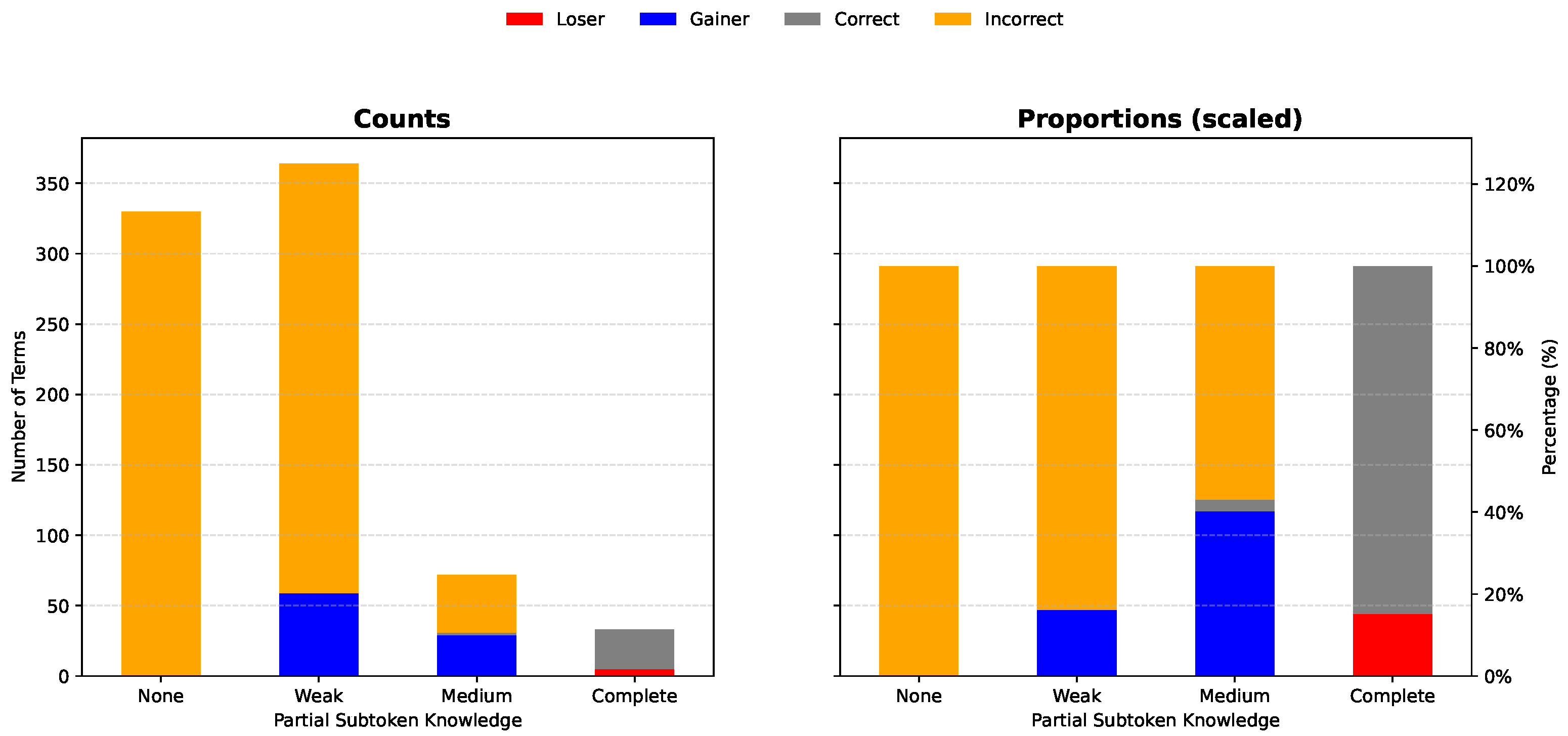
3.3. Partial Subtoken Knowledge Predicts Fine-Tuning Success
3.4. Term Familiarity
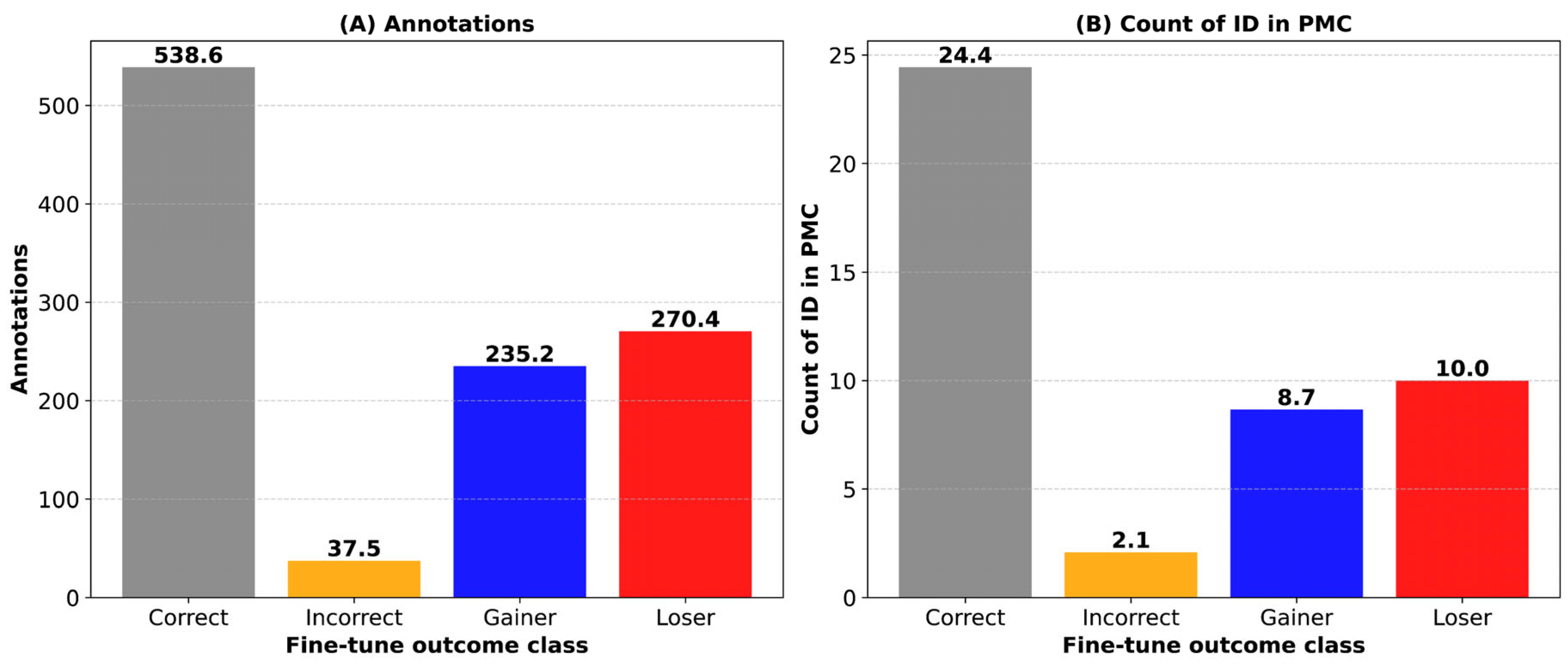
3.5. Positive and Negative Knowledge Flows During Fine-Tuning
4. Discussion
4.1. Limitations
4.2. Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Groza, T.; Köhler, S.; Doelken, S.; Collier, N.; Oellrich, A.; Smedley, D.; Couto, F.M.; Baynam, G.; Zankl, A.; Robinson, P.N. Automatic concept recognition using the human phenotype ontology reference and test suite corpora. Database 2015, 2015, bav005. [Google Scholar] [CrossRef]
- Fu, S.; Chen, D.; He, H.; Liu, S.; Moon, S.; Peterson, K.J.; Shen, F.; Wang, L.; Wang, Y.; Wen, A.; et al. Clinical concept extraction: A methodology review. J. Biomed. Inform. 2020, 109, 103526. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.F.; Henry, S.; Wang, Y.; Shen, F.; Uzuner, O.; Rumshisky, A. The 2019 n2c2/UMass Lowell shared task on clinical concept normalization. J. Am. Med. Inform. Assoc. 2020, 27, 1529-e1. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Rastegar-Mojarad, M.; Moon, S.; Shen, F.; Afzal, N.; Liu, S.; Zeng, Y.; Mehrabi, S.; Sohn, S.; et al. Clinical information extraction applications: A literature review. J. Biomed. Inform. 2018, 77, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.G.; Howsmon, D.; Zhang, B.; Hahn, J.; McGuinness, D.; Hendler, J.; Ji, H. Entity linking for biomedical literature. BMC Med. Inform. Decis. Mak. 2015, 15, 1–9. [Google Scholar] [CrossRef]
- Krauthammer, M.; Nenadic, G. Term identification in the biomedical literature. J. Biomed. Inform. 2004, 37, 512–526. [Google Scholar] [CrossRef]
- Robinson, P.N. Deep phenotyping for precision medicine. Hum. Mutat. 2012, 33, 777–780. [Google Scholar] [CrossRef]
- Bispo, L.G.M.; Amaral, F.G.; da Silva, J.M.N.; Neto, I.R.; Silva, L.K.D.; da Silva, I.L. Ergonomic adequacy of university tablet armchairs for male and female: A multigroup item response theory analysis. J. Saf. Sustain. 2024, 1, 223–233. [Google Scholar] [CrossRef]
- Yun, W.; Zhang, X.; Li, Z.; Liu, H.; Han, M. Knowledge modeling: A survey of processes and techniques. Int. J. Intell. Syst. 2021, 36, 1686–1720. [Google Scholar] [CrossRef]
- Haslinda, A.; Sarinah, A. A review of knowledge management models. J. Int. Soc. Res. 2009, 2, 9. [Google Scholar]
- Pan, J.Z.; Razniewski, S.; Kalo, J.C.; Singhania, S.; Chen, J.; Dietze, S.; Jabeen, H.; Omeliyanenko, J.; Zhang, W.; Lissandrini, M.; et al. Large language models and knowledge graphs: Opportunities and challenges. arXiv 2023, arXiv:2308.06374. [Google Scholar] [CrossRef]
- Abellanosa, A.D.; Pereira, E.; Lefsrud, L.; Mohamed, Y. Integrating Knowledge Management and Large Language Models to Advance Construction Job Hazard Analysis: A Systematic Review and Conceptual Framework. J. Saf. Sustain. 2025; in press, corrected proof. [Google Scholar]
- Chandak, P.; Huang, K.; Zitnik, M. Building a knowledge graph to enable precision medicine. Sci. Data 2023, 10, 67. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, R.; Gao, Q.; Liang, T.; Wang, X.; Li, S.; Lei, B.; Gao, K. A Review of Applying Large Language Models in Healthcare. IEEE Access 2024, 13, 6878–6892. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Petzold, L. Are Large Language Models Ready for Healthcare? A Comparative Study on Clinical Language Understanding. arXiv 2023, arXiv:2304.05368. [Google Scholar] [CrossRef]
- Chang, E.; Mostafa, J. The use of SNOMED CT, 2013–2020: A literature review. J. Am. Med. Inform. Assoc. 2021, 28, 2017–2026. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, J.; Su, J.; Shen, D.; Tan, C. Recognizing names in biomedical texts: A machine learning approach. Bioinformatics 2004, 20, 1178–1190. [Google Scholar] [CrossRef] [PubMed]
- Köhler, S.; Vasilevsky, N.A.; Engelstad, M.; Foster, E.; McMurry, J.; Aymé, S.; Baynam, G.; Bello, S.M.; Boerkoel, C.F.; Boycott, K.M.; et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017, 45, D865–D876. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.N.; Köhler, S.; Bauer, S.; Seelow, D.; Horn, D.; Mundlos, S. The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008, 83, 610–615. [Google Scholar] [CrossRef]
- Jahan, I.; Laskar, M.T.R.; Peng, C.; Huang, J.X. A comprehensive evaluation of large language models on benchmark biomedical text processing tasks. Comput. Biol. Med. 2024, 171, 108189. [Google Scholar] [CrossRef] [PubMed]
- Do, T.S.; Hier, D.B.; Obafemi-Ajayi, T. Mapping Biomedical Ontology Terms to IDs: Effect of Domain Prevalence on Prediction Accuracy. In Proceedings of the 2025 IEEE Conference on Artificial Intelligence (CAI), Santa Clara, CA, USA, 5–7 May 2025; IEEE: New York, NY, USA, 2025; pp. 1–6. [Google Scholar]
- Kandpal, N.; Deng, H.; Roberts, A.; Wallace, E.; Raffel, C. Large language models struggle to learn long-tail knowledge. In Proceedings of the International Conference on Machine Learning, Honolulu, HI, USA, 23–29 July 2023; PMLR: Cambridge, MA, USA, 2023; pp. 15696–15707. [Google Scholar]
- Wu, E.; Wu, K.; Zou, J. FineTuneBench: How well do commercial fine-tuning APIs infuse knowledge into LLMs? arXiv 2024, arXiv:2411.05059. [Google Scholar] [CrossRef]
- Braga, M. Personalized Large Language Models through Parameter Efficient Fine-Tuning Techniques. In Proceedings of the 47th International ACM SIGIR Conference on Research and Development in Information Retrieval, Washington, DC, USA, 14–18 July 2024. [Google Scholar] [CrossRef]
- Tinn, R.; Cheng, H.; Gu, Y.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Fine-tuning large neural language models for biomedical natural language processing. Patterns 2023, 4, 100729. [Google Scholar] [CrossRef]
- Ding, N.; Qin, Y.; Yang, G.; Wei, F.; Yang, Z.; Su, Y.; Hu, S.; Chen, Y.; Chan, C.M.; Chen, W.; et al. Parameter-efficient fine-tuning of large-scale pre-trained language models. Nat. Mach. Intell. 2023, 5, 220–235. [Google Scholar] [CrossRef]
- Wang, C.; Yan, J.; Zhang, W.; Huang, J. Towards Better Parameter-Efficient Fine-Tuning for Large Language Models: A Position Paper. arXiv 2023, arXiv:2311.13126. [Google Scholar] [CrossRef]
- Wu, E.; Wu, K.; Zou, J. Limitations of Learning New and Updated Medical Knowledge with Commercial Fine-Tuning Large Language Models. NEJM AI 2025, 2, AIcs2401155. [Google Scholar] [CrossRef]
- Mecklenburg, N.; Lin, Y.; Li, X.; Holstein, D.; Nunes, L.; Malvar, S.; Silva, B.; Chandra, R.; Aski, V.; Yannam, P.K.R.; et al. Injecting New Knowledge Into Large Language Models Via Supervised Fine-Tuning. arXiv 2024, arXiv:2404.00213. [Google Scholar] [CrossRef]
- Chu, T.; Zhai, Y.; Yang, J.; Tong, S.; Xie, S.; Schuurmans, D.; Le, Q.V.; Levine, S.; Ma, Y. SFT memorizes, RL generalizes: A comparative study of foundation model post-training. arXiv 2025, arXiv:2501.17161. [Google Scholar]
- Pan, X.; Hahami, E.; Zhang, Z.; Sompolinsky, H. Memorization and Knowledge Injection in Gated LLMs. arXiv 2025, arXiv:2504.21239. [Google Scholar] [CrossRef]
- Pletenev, S.; Marina, M.; Moskovskiy, D.; Konovalov, V.; Braslavski, P.; Panchenko, A.; Salnikov, M. How Much Knowledge Can You Pack into a LoRA Adapter without Harming LLM? arXiv 2025, arXiv:2502.14502. [Google Scholar] [CrossRef]
- Orgad, H.; Toker, M.; Gekhman, Z.; Reichart, R.; Szpektor, I.; Kotek, H.; Belinkov, Y. LLMS Know More Than They Show: On The Intrinsic Representation of LLM Hallucinations. arXiv 2025, arXiv:2410.02707. [Google Scholar]
- Gekhman, Z.; Yona, G.; Aharoni, R.; Eyal, M.; Feder, A.; Reichart, R.; Herzig, J. Does Fine-Tuning LLMs on New Knowledge Encourage Hallucinations? In Proceedings of the 2024 Conference on Empirical Methods in Natural Language Processing, Miami, FL, USA, 12–16 November 2024; pp. 7765–7784. [Google Scholar] [CrossRef]
- Gekhman, Z.; David, E.B.; Orgad, H.; Ofek, E.; Belinkov, Y.; Szpektor, I.; Herzig, J.; Reichart, R. Inside-out: Hidden factual knowledge in LLMs. arXiv 2025, arXiv:2503.15299. [Google Scholar]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM. org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef]
- Maiella, S.; Rath, A.; Angin, C.; Mousson, F.; Kremp, O. Orphanet and its consortium: Where to find expert-validated information on rare diseases. Rev. Neurol. 2013, 169, S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Sequeira, E. PubMed Central (PMC): An Archive for Literature from Life Sciences Journals. In The NCBI Handbook [Internet]; McEntyre, J., Ostell, J., Eds.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2002; Chapter 9. Available online: https://www.ncbi.nlm.nih.gov/sites/books/NBK21087/pdf/Bookshelf_NBK21087.pdf (accessed on 3 September 2025).
- Hier, D.B.; Carrithers, M.A.; Platt, S.K.; Nguyen, A.; Giannopoulos, I.; Obafemi-Ajayi, T. Preprocessing of Physician Notes by LLMs Improves Clinical Concept Extraction Without Information Loss. Information 2025, 16, 446. [Google Scholar] [CrossRef]
- Grattafiori, A.; Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Vaughan, A.; et al. The llama 3 herd of models. arXiv 2024, arXiv:2407.21783. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30, pp. 5998–6008. [Google Scholar]
- Hu, E.J.; Shen, Y.; Wallis, P.; Allen-Zhu, Z.; Li, Y.; Wang, S.; Wang, L.; Chen, W. Lora: Low-rank adaptation of large language models. ICLR 2022, 1, 3. [Google Scholar]
- Luo, Y.; Yang, Z.; Meng, F.; Li, Y.; Zhou, J.; Zhang, Y. An empirical study of catastrophic forgetting in large language models during continual fine-tuning. arXiv 2023, arXiv:2308.08747. [Google Scholar] [CrossRef]
- Kalajdzievski, D. Scaling laws for forgetting when fine-tuning large language models. arXiv 2024, arXiv:2401.05605. [Google Scholar] [CrossRef]
- Wang, A.; Liu, C.; Yang, J.; Weng, C. Fine-tuning large language models for rare disease concept normalization. J. Am. Med. Inform. Assoc. 2024, 31, 2076–2083. [Google Scholar] [CrossRef] [PubMed]
- Dettmers, T.; Pagnoni, A.; Holtzman, A.; Zettlemoyer, L. Qlora: Efficient finetuning of quantized llms. Adv. Neural Inf. Process. Syst. 2023, 36, 10088–10115. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hier, D.B.; Platt, S.K.; Nguyen, A. Prior Knowledge Shapes Success When Large Language Models Are Fine-Tuned for Biomedical Term Normalization. Information 2025, 16, 776. https://doi.org/10.3390/info16090776
Hier DB, Platt SK, Nguyen A. Prior Knowledge Shapes Success When Large Language Models Are Fine-Tuned for Biomedical Term Normalization. Information. 2025; 16(9):776. https://doi.org/10.3390/info16090776
Chicago/Turabian StyleHier, Daniel B., Steven K. Platt, and Anh Nguyen. 2025. "Prior Knowledge Shapes Success When Large Language Models Are Fine-Tuned for Biomedical Term Normalization" Information 16, no. 9: 776. https://doi.org/10.3390/info16090776
APA StyleHier, D. B., Platt, S. K., & Nguyen, A. (2025). Prior Knowledge Shapes Success When Large Language Models Are Fine-Tuned for Biomedical Term Normalization. Information, 16(9), 776. https://doi.org/10.3390/info16090776







