LLMs in Action: Robust Metrics for Evaluating Automated Ontology Annotation Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Creating Ontology Embeddings
- Walk length: the length of random walks was varied from 10 to 100 steps to capture different levels of contextual information for each node.
- Number of walks: the number of walks per node was adjusted from 5 to 30 to ensure sufficient sampling of network neighborhoods.
- Embedding dimensions: various dimensionalities were tested, including 64, 128, 256, 512, and 1024, to evaluate the impact of dimensionality on embedding quality.
- P and Q: These return (P) and in–out (Q) hyperparameters, which influence the probability of revisiting previously visited nodes, were tuned within the range of 0.2 to 2. These parameters balance the weighting between local and global context in the embeddings.
- Workers = 10
- Dimensions = 512
- Num_walks = 30
- Walk_length = 70
- P = 0
- Q = 1.4
2.2. Semantic Similarity Metrics
- Jaccard similarity: Jaccard semantic similarity evaluates the similarity between concepts within an ontology using the Jaccard Index, a measure traditionally applied to set similarity. In ontology-driven applications, the Jaccard Index can be adapted to capture semantic similarities based on shared features, relationships, or annotations.
- Cosine similarity using hierarchical embedding: hierarchical embedding with Node2Vec combines the idea of generating node embeddings for a graph with hierarchical graph structures, enabling the model to capture relationships and features at multiple levels of the hierarchy.
- Cosine similarity using LLM PubMed embeddings: LLM PubMed embeddings refer to vector representations of biomedical text derived from PubMed articles using large language models (LLMs). These embeddings capture the semantic meaning of biomedical terms, phrases, or documents, enabling advanced downstream applications in healthcare, life sciences, and biomedical research.
- Cosine similarity using MiniLM6 embeddings: MiniLM6 embeddings are lightweight, high-performance vector representations derived from MiniLM v6, a distilled version of a transformer-based language model. MiniLM6 focuses on delivering powerful semantic embeddings with reduced computational overhead, making it efficient for large-scale and real-time applications.
2.3. Dataset for Evaluating Semantic Similarity
2.4. Performance Evaluation
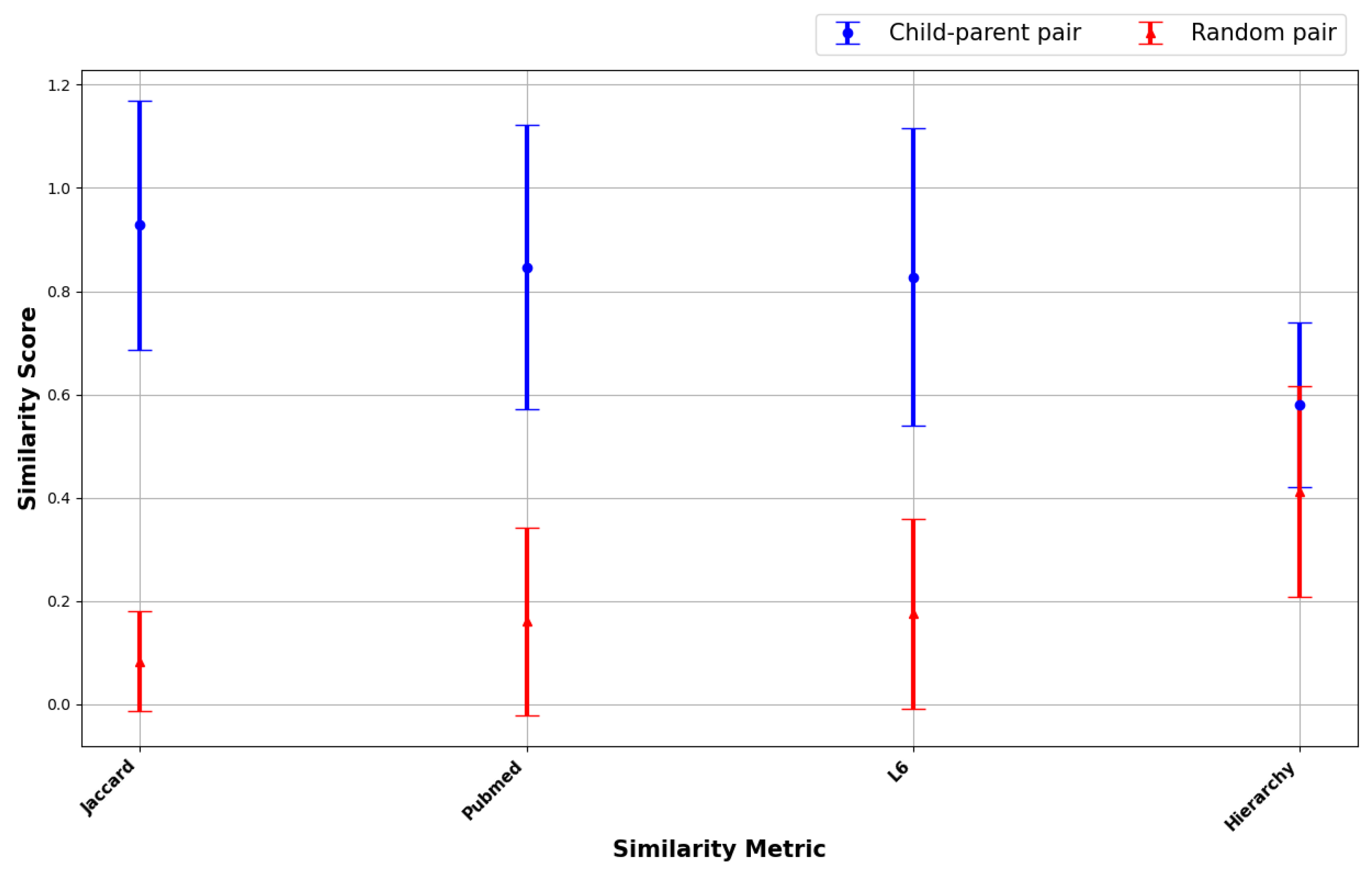
- Ability to distinguish between real similarity and noise: Different semantic similarity metrics were applied to the signal and noise datasets to observe the metrics’ ability to discriminate between the two. The metric with the largest disparity in similarity scores between the signal and noise datasets can be said to effectively distinguish meaningful relationships from random associations.
- Machine learning evaluation of semantic similarity metrics: we used scores from the four core, as well as the 11 hybrid metrics above, as features in machine learning models to predict whether the similarity was from a random pair or a child–parent pair.The goal of these models was to evaluate the predictive ability of different similarity metrics (or combinations thereof) to distinguish between child–parent and random node pairs.The dataset was split into training (80%) and testing (20%) sets, and we trained a linear regression model to predict relationships between nodes. Additionally, we used logistic regression to classify node pairs and evaluate model accuracy. Our primary focus was to determine whether combining hierarchical and LLM embeddings led to better classification performance.
3. Results
3.1. Ability to Distinguish Between Real Similarity and Noise
3.2. Machine Learning Evaluation of Semantic Similarity Metrics
4. Discussion
5. Conclusions
6. Future Directions
- Development of Next-Generation Semantic Similarity Metrics Existing similarity measures exhibit sensitivity to annotation depth and hierarchical structures, leading to inconsistencies in similarity assessments. To address these limitations, future work should explore the following:
- Context-aware semantic metrics: develop hybrid similarity measures that dynamically adjust based on term frequency, ontology structure, and contextual embeddings.
- Graph neural networks (GNNs) for semantic similarity: utilize GNNs to encode hierarchical relationships while incorporating domain-specific text embeddings, ensuring robust cross-domain applicability.
- Uncertainty-aware similarity metrics: introduce probabilistic methods that account for annotation uncertainty, particularly in incomplete or evolving ontologies.
- Real-Time Integration of Automated Ontology Annotation Pipelines To bridge the gap between NLP-based annotation systems and real-world biomedical applications, future research should focus on streaming annotation pipelines, federated learning for distributed annotation, and explainable AI (XAI) for ontology annotation. This requires the implementation of real-time text mining frameworks that continuously ingest scientific literature and dynamically update ontology annotations. Additionally, federated learning should be leveraged to train NLP models across multiple institutions while preserving data privacy, thereby enhancing annotation scalability. Finally, transparent annotation models with built-in interpretability modules must be developed to provide justifications for assigned ontology terms, fostering trust and reliability in automated annotation systems.
- Leveraging Advanced Neural Architectures for Ontology Embeddings Recent advances in deep learning models present promising opportunities for enhancing ontology-based text mining. One key direction is contrastive learning for ontology representations, where transformer-based models such as BioBERT and PubMedGPT are trained using contrastive loss to refine ontology embeddings by aligning contextual and structural information. Additionally, multi-modal ontology learning aims to improve annotation accuracy by integrating textual definitions, biological pathways, and experimental data, such as gene expression, into multimodal neural models. Another critical avenue is self-supervised pretraining on ontology graphs, which leverages techniques like masked node prediction and node contrastive learning to enhance representations of under-annotated ontology nodes, ensuring more robust and comprehensive knowledge extraction.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Smith, B.; Williams, J.; Steffen, S.K. The ontology of the gene ontology. AMIA Annu. Symp. Proc. 2003, 2003, 609–613. [Google Scholar] [PubMed]
- Lesteven, S.; Derriere, S.; Dubois, P.; Genova, F.; Preite Martinez, A.; Hernandez, N.; Mothe, J.; Napoli, A.; Toussaint, Y. Ontologies for Astronomy. In Library and Information Services in Astronomy V; Astronomical Society of the Pacific: San Francisco, CA, USA, 2007; Volume 377, p. 193. [Google Scholar]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Both, V. From Functional Similarity Among Gene Products to Dependence Relations Among Gene Ontology Terms. In Proceedings of the Second conference on Standards and Ontologies for Functional Genomics (SOFG 2), Philadelphia, PA, USA, 23–26 October 2004; p. 53. [Google Scholar]
- Bodenreider, O.; Smith, B.; Kumar, A.; Burgun, A. Investigating subsumption in SNOMED CT: An exploration into large description logic-based biomedical terminologies. Artif. Intell. Med. 2007, 39, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Consortium, T.G.O. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Dessimoz, C.; Škunca, N. The Gene Ontology Handbook; Humana Press: Totowa, NJ, USA, 2017. [Google Scholar]
- Robinson, P.N.; Köhler, S.; Bauer, S.; Seelow, D.; Horn, D.; Mundlos, S. The Human Phenotype Ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008, 83, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Cui, C.; Zhang, J.; Yang, R.; Liu, B. Predicting gene ontology annotations using domain-specific word embeddings. PLoS Comput. Biol. 2021, 17, e1009056. [Google Scholar]
- Radivojac, P.; Clark, W.T.; Oron, T.R.; Schnoes, A.M.; Wittkop, T.; Sokolov, A.; Graim, K.; Funk, C.; Verspoor, K.; Ben-Hur, A.; et al. A large-scale evaluation of computational protein function prediction. Nat. Methods 2013, 10, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Havrilla, J.M.; Fang, L.; Chen, Y.; Peng, J.; Liu, C.; Wu, C.; Sarmady, M.; Botas, P.; Isla, J.; et al. Phen2Gene: Rapid phenotype-driven gene prioritization for rare diseases. NAR Genom. Bioinform. 2020, 2, lqaa032. [Google Scholar] [CrossRef] [PubMed]
- Groth, P.; Pavlova, N.; Kalev, I.; Tonov, S.; Georgiev, G.; Pohjalainen, E. Automated assignment of biomedical annotations using statistical and machine learning methods. Nat. Biotechnol. 2010, 28, 977–982. [Google Scholar]
- Baumgartner, W.A.; Cohen, K.B.; Fox, L.; Acquaah-Mensah, G.; Hunter, L. Manual curation is not sufficient for annotation of genomic data. Bioinformatics 2008, 24, 1846–1852. [Google Scholar]
- Manda, P.; SayedAhmed, S.; Mohanty, S.D. Automated Ontology-Based Annotation of Scientific Literature Using Deep Learning. In Proceedings of the SBD ’20: International Workshop on Semantic Big Data, New York, NY, USA, 14–19 June 2020. [Google Scholar] [CrossRef]
- Devkota, P.; Mohanty, S.D.; Manda, P. A Gated Recurrent Unit based architecture for recognizing ontology concepts from biological literature. BioData Min. 2022, 15, 22. [Google Scholar] [CrossRef] [PubMed]
- Devkota, P.; Mohanty, S.; Manda, P. Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature [Ontology-Powered Boosting for Improved Recognition of Ontology Concepts from Biological Literature]. In Proceedings of the 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023), Lisbon, Portugal, 16–18 February 2023; Volume 3. [Google Scholar]
- Devkota, P.; Mohanty, S.; Manda, P. Knowledge of the Ancestors: Intelligent Ontology-aware Annotation of Biological Literature using Semantic Similarity. In Proceedings of the International Conference on Biomedical Ontology, Ann Arbor, MI, USA, 25–28 September 2022. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2019, arXiv:1810.04805. [Google Scholar]
- Brown, T.B.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2020, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Jiang, Y.; Bergquist, T.R.; Lee, A.J.; Kacsoh, B.Z.; Crocker, A.W.; Gillis, J. The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef]
- Pesquita, C.; Faria, D.; Falcao, A.O.; Lord, P.; Couto, F.M. Semantic similarity in biomedical ontologies. PLoS Comput. Biol. 2009, 5, e1000443. [Google Scholar] [CrossRef] [PubMed]
- Resnik, P. Semantic similarity in a taxonomy: An information-based measure and its application to problems of ambiguity in natural language. J. Artif. Intell. Res. 1999, 11, 95–130. [Google Scholar] [CrossRef]
- Lin, D. An information-theoretic definition of similarity. In Proceedings of the 15th International Conference on Machine Learning (ICML), Madison, WI, USA, 24–27 July 1998; pp. 296–304. [Google Scholar]
- Jiang, J.J.; Conrath, D.W. Semantic similarity based on corpus statistics and lexical taxonomy. In Proceedings of the 10th Research on Computational Linguistics International Conference, Taipei, Taiwan, 19–22 August 1997; pp. 19–33. [Google Scholar]
- Wang, J.; Zhang, Z.; Li, J. A new method to measure the semantic similarity in gene ontology. BMC Bioinform. 2007, 8, 44. [Google Scholar]
- Manda, P.; Vision, T.J. On the Statistical Sensitivity of Semantic Similarity Metrics. In Proceedings of the 9th International Conference on Biological Ontology (ICBO 2018), Corvallis, OR, USA, 7–10 August 2018. [Google Scholar]
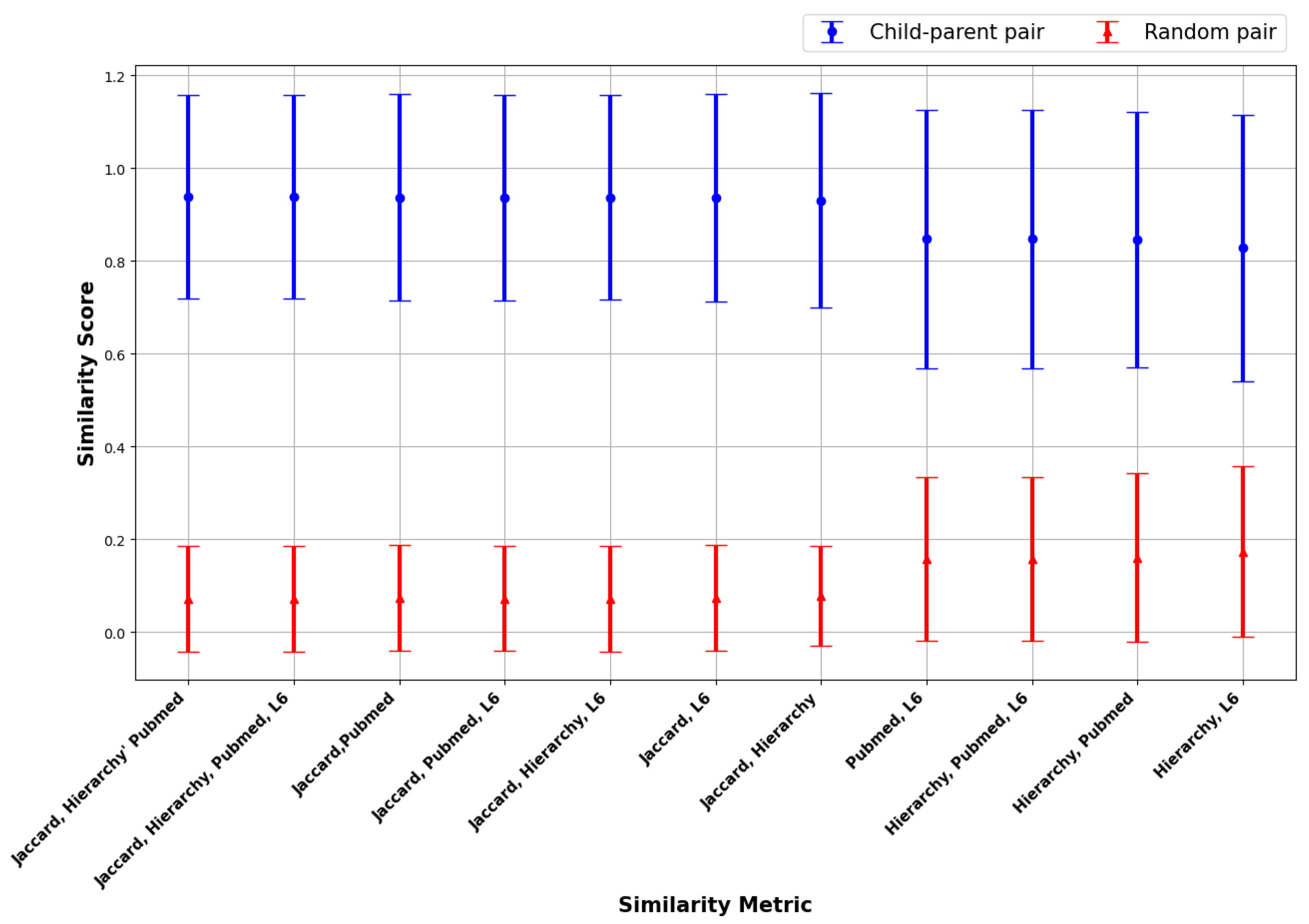
| Jaccard & Hierarchy |
| Jaccard & Pubmed |
| Jaccard & L6 |
| Hierarchy & Pubmed |
| Hierarchy & L6 |
| Pubmed & L6 |
| Jaccard, Hierarchy & Pubmed |
| Jaccard, Hierarchy & L6 |
| Jaccard, Pubmed & L6 |
| Hierarchy, Pubmed & L6 |
| Jaccard, Hierarchy, Pubmed & L6 |
| Predictive Metrics | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|
| Jaccard, Hierarchy, L6 | 0.98 | 0.98 | 0.97 | 0.97 |
| Jaccard | 0.97 | 0.97 | 0.96 | 0.97 |
| Jaccard, Hierarchy | 0.97 | 0.98 | 0.97 | 0.97 |
| Jaccard, Pubmed | 0.97 | 0.97 | 0.97 | 0.97 |
| Jaccard, L6 | 0.97 | 0.97 | 0.96 | 0.97 |
| Jaccard, Hierarchy, Pubmed | 0.97 | 0.98 | 0.97 | 0.97 |
| Jaccard, Hierarchy, Pubmed, L6 | 0.97 | 0.98 | 0.97 | 0.97 |
| Jaccard, Pubmed, L6 | 0.97 | 0.97 | 0.97 | 0.97 |
| Hierarchy, Pubmed, L6 | 0.91 | 0.90 | 0.90 | 0.90 |
| Pubmed | 0.90 | 0.90 | 0.89 | 0.90 |
| Hierarchy, Pubmed | 0.90 | 0.90 | 0.89 | 0.90 |
| Hierarchy, L6 | 0.90 | 0.90 | 0.89 | 0.90 |
| Pubmed, L6 | 0.90 | 0.90 | 0.90 | 0.90 |
| L6 | 0.89 | 0.89 | 0.88 | 0.89 |
| Hierarchy | 0.68 | 0.64 | 0.80 | 0.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noori, A.; Devkota, P.; Mohanty, S.D.; Manda, P. LLMs in Action: Robust Metrics for Evaluating Automated Ontology Annotation Systems. Information 2025, 16, 225. https://doi.org/10.3390/info16030225
Noori A, Devkota P, Mohanty SD, Manda P. LLMs in Action: Robust Metrics for Evaluating Automated Ontology Annotation Systems. Information. 2025; 16(3):225. https://doi.org/10.3390/info16030225
Chicago/Turabian StyleNoori, Ali, Pratik Devkota, Somya D. Mohanty, and Prashanti Manda. 2025. "LLMs in Action: Robust Metrics for Evaluating Automated Ontology Annotation Systems" Information 16, no. 3: 225. https://doi.org/10.3390/info16030225
APA StyleNoori, A., Devkota, P., Mohanty, S. D., & Manda, P. (2025). LLMs in Action: Robust Metrics for Evaluating Automated Ontology Annotation Systems. Information, 16(3), 225. https://doi.org/10.3390/info16030225







