Automated Fungal Identification with Deep Learning on Time-Lapse Images
Abstract
1. Introduction
2. Materials and Methods
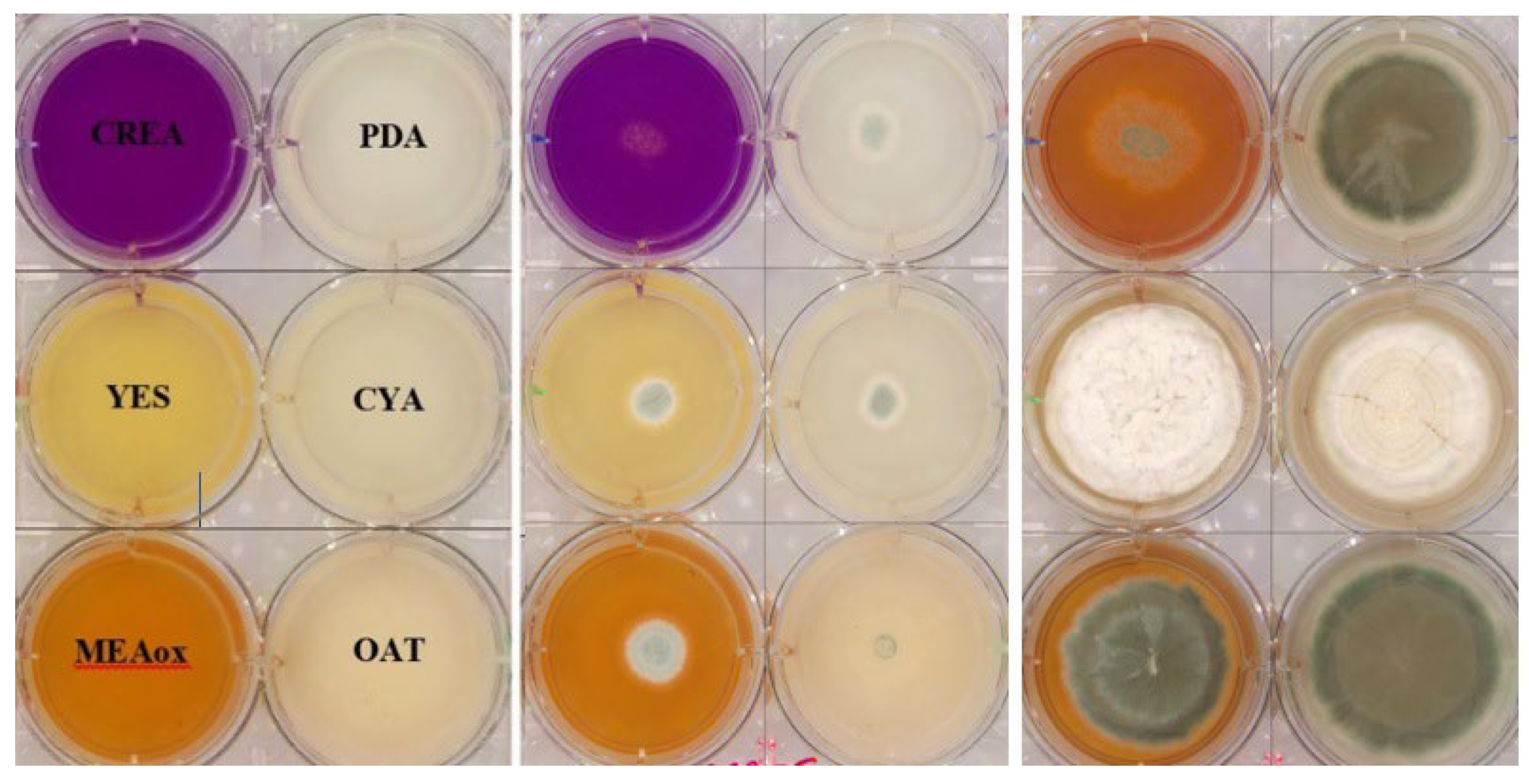
2.1. Preparation and Inoculation of 6-Well Plates for Image Capture
2.2. Method
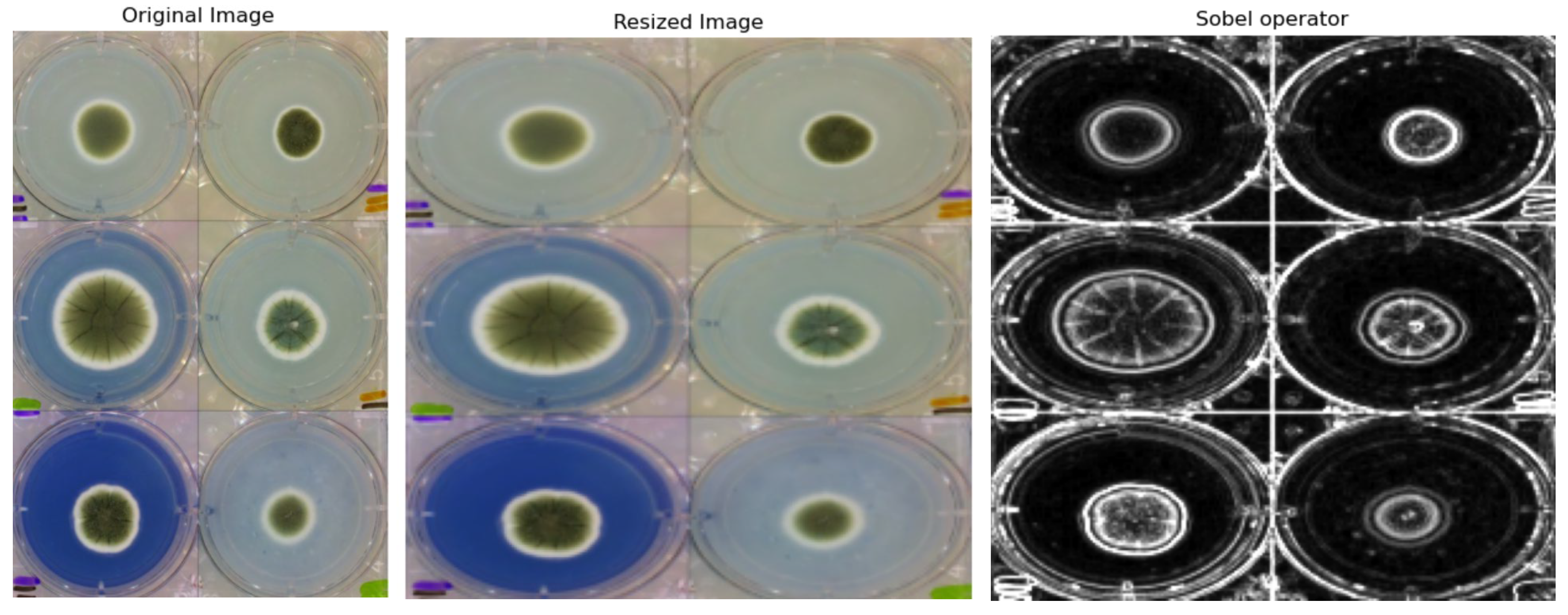
2.2.1. Preprocessing
2.2.2. Data Augmentation
2.2.3. Deep Learning Architectures for Fungal Classification
2.2.4. Model Training and Hyperparameters
3. Results
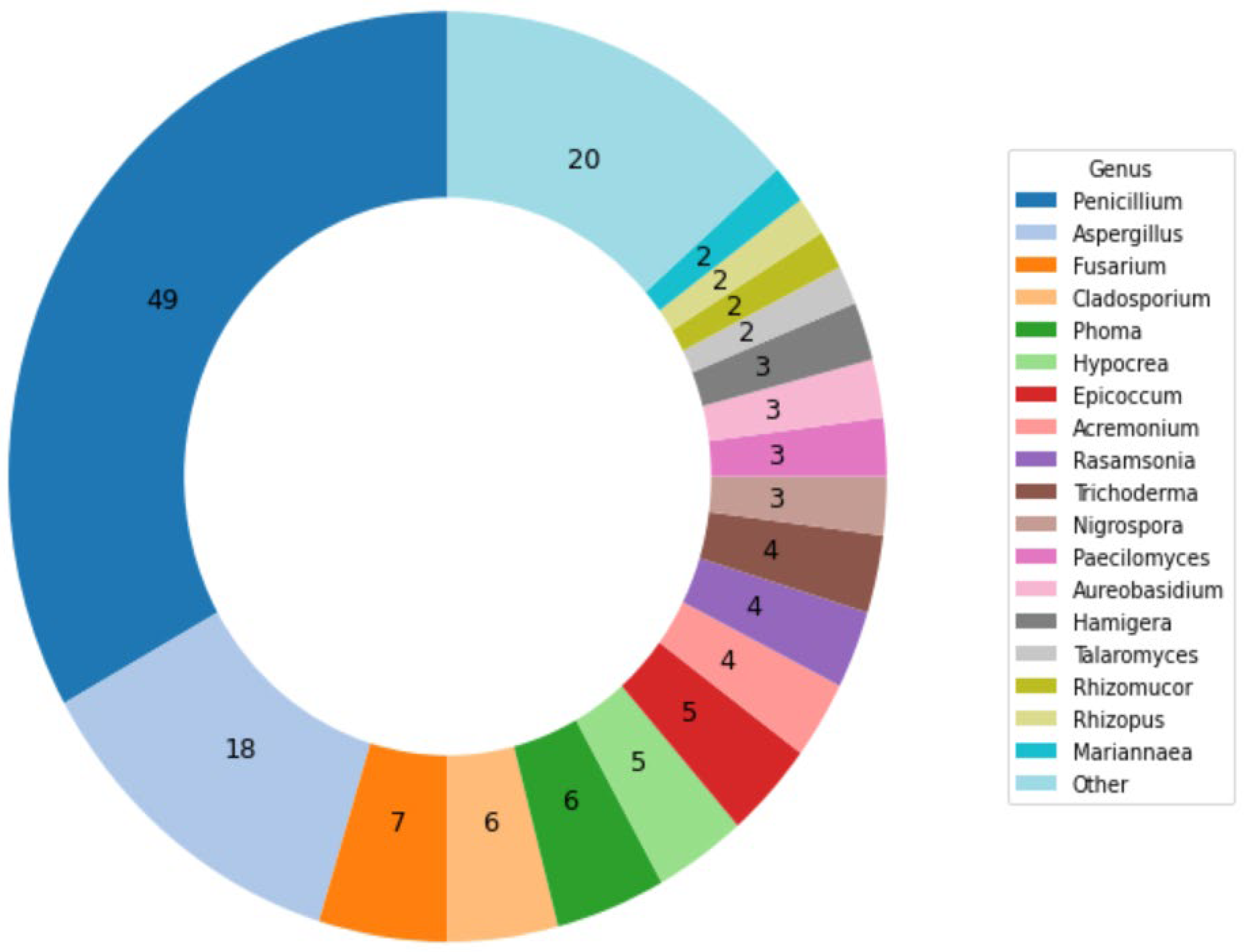
3.1. Cultivations and Images
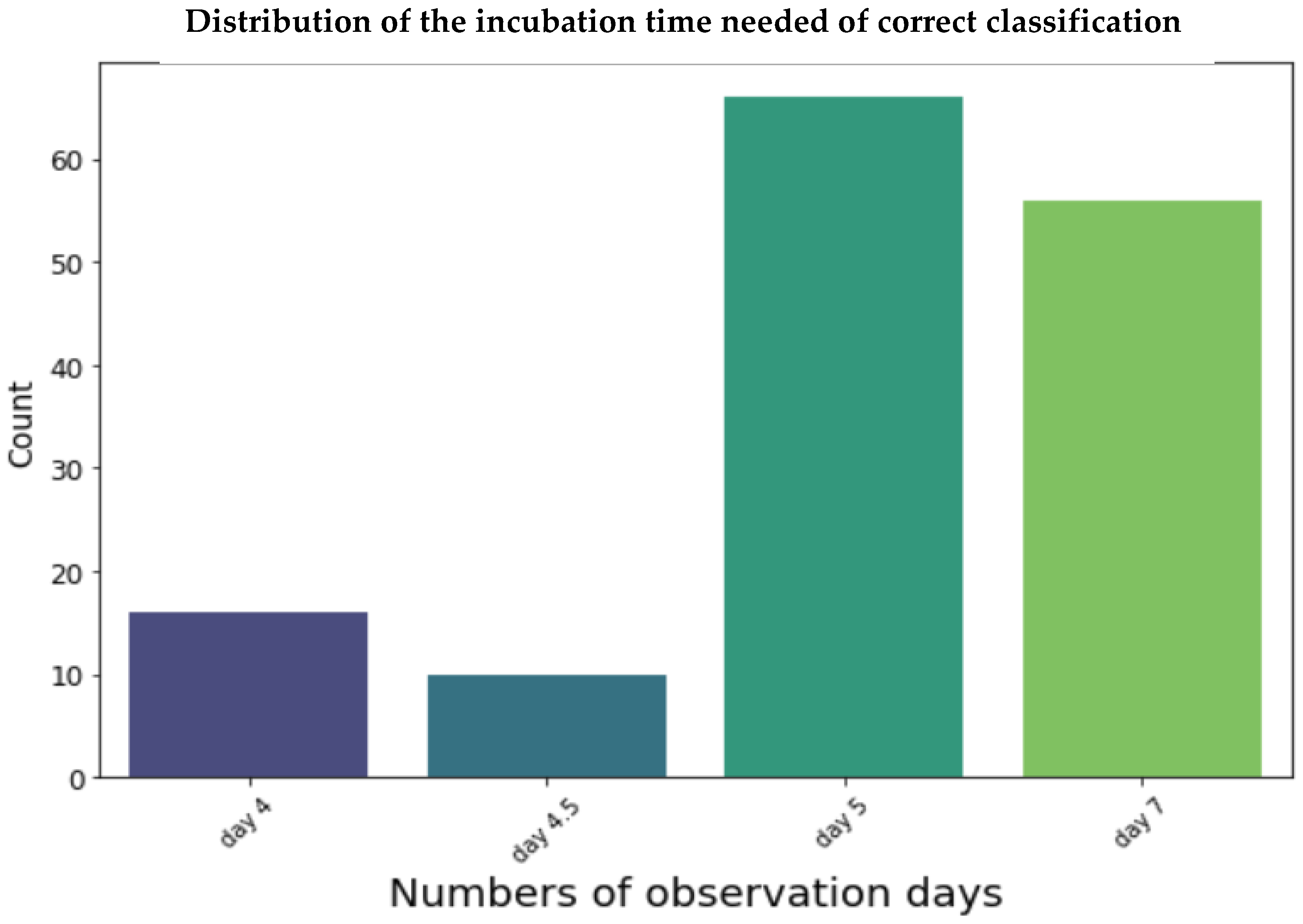
3.2. Evaluation and Performance of Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5. [Google Scholar] [CrossRef] [PubMed]
- Money, N.P. Fungal Diversity. In The Fungi, 3rd ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 1–36. [Google Scholar] [CrossRef]
- Mendonça, A.; Santos, H.; Franco-Duarte, R.; Sampaio, P. Fungal infections diagnosis—Past, present and future. Res. Microbiol. 2022, 173, 103915. [Google Scholar] [CrossRef] [PubMed]
- Bhunjun, C.S.; Niskanen, T.; Suwannarach, N.; Wannathes, N.; Chen, Y.J.; McKenzie, E.H.; Maharachchikumbura, S.S.; Buyck, B.; Zhao, C.L.; Fan, Y.G.; et al. The numbers of fungi: Are the most speciose genera truly diverse? Fungal Divers. 2022, 114, 387–462. [Google Scholar] [CrossRef]
- Aboul-Ella, H.; Hamed, R.; Abo-Elyazeed, H. Recent trends in rapid diagnostic techniques for dermatophytosis. Int. J. Vet. Sci. Med. 2020, 8, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Jana, C.; Raus, M.; Sedlářová, M.; Šebela, M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol. Adv. 2014, 32, 230–241. [Google Scholar]
- Desingu, K.; Bhaskar, A.; Palaniappan, M.; Chodisetty, E.A.; Bharathi, H. Classification of Fungi Species: A Deep Learning Based Image Feature Extraction and Gradient Boosting Ensemble Approach. 2022. Available online: http://ceur-ws.org (accessed on 13 September 2024).
- Yin, H.; Yi, W.; Hu, D. Computer vision and machine learning applied in the mushroom industry: A critical review. Comput. Electron. Agric. 2022, 198, 107015. [Google Scholar] [CrossRef]
- Mansourvar, M.; Funk, J.; Petersen, S.D.; Tavakoli, S.; Hoof, J.B.; Corcoles, D.L.; Pittroff, S.M.; Jelsbak, L.; Jensen, N.B.; Ding, L.; et al. Automatic classification of fungal-fungal interactions using deep leaning models. Comput. Struct. Biotechnol. J. 2024, 23, 4222–4231. [Google Scholar] [CrossRef]
- Tahir, M.W.; Zaidi, N.A.; Rao, A.A.; Blank, R.; Vellekoop, M.J.; Lang, W. A fungus spores dataset and a convolutional neural network based approach for fungus detection. IEEE Trans. Nanobiosci. 2018, 17, 281–290. [Google Scholar] [CrossRef]
- Marandi, B.; Deep, M.K. Advancements in Machine Learning for Detection and Prediction of Infectious and Parasitic Diseases: A Comprehensive Investigation. Machine Learning. Int. J. Adv. Multidiscip. Sci. Res. 2024, 7, 82–97. [Google Scholar]
- Aldogan, K.Y.; Kayan, C.E.; Gumus, A. Intensity and phase stacked analysis of a Φ-OTDR system using deep transfer learning and recurrent neural networks. Appl. Opt. 2023, 62, 1753–1764. [Google Scholar] [CrossRef]
- Kristensen, K.; Ward, L.M.; Mogensen, M.L.; Cichosz, S.L. Using image processing and automated classification models to classify microscopic gram stain images. Comput. Methods Programs Biomed. Update 2023, 3, 100091. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, H.; Ye, T.; Juhas, M. Deep Learning for Imaging and Detection of Microorganisms. Trends Microbiol. 2021, 29, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Picek, L.; Šulc, M.; Matas, J.; Heilmann-Clausen, J.; Jeppesen, T.S.; Lind, E. Automatic Fungi Recognition: Deep Learning Meets Mycology. Sensors 2022, 22, 633. [Google Scholar] [CrossRef] [PubMed]
- Gaikwad, S.S.; Rumma, S.S.; Hangarge, M. Fungi Classification using Convolution Neural Network. Turk. J. Comput. Math. Educ. 2021, 12, 4563–4569. [Google Scholar]
- Picek, L.; Šulc, M.; Matas, J.; Jeppesen, T.S.; Heilmann-Clausen, J.; Læssøe, T.; Frøslev, T. Danish Fungi 2020—Not Just Another Image Recognition Dataset. In Proceedings of the 2022 IEEE/CVF Winter Conference on Applications of Computer Vision, WACV 2022, Waikoloa, HI, USA, 3–8 January 2022; pp. 3281–3291. [Google Scholar] [CrossRef]
- Koo, T.; Kim, M.H.; Jue, M.S. Automated detection of superficial fungal infections from microscopic images through a regional convolutional neural network. PLoS ONE 2021, 16, e0256290. [Google Scholar] [CrossRef]
- Gao, W.; Li, M.; Wu, R.; Du, W.; Zhang, S.; Yin, S.; Chen, Z.; Huang, H. The design and application of an automated microscope developed based on deep learning for fungal detection in dermatology. Mycoses 2021, 64, 245–251. [Google Scholar] [CrossRef]
- Rahman, M.A.; Clinch, M.; Reynolds, J.; Dangott, B.; Villegas, D.M.M.; Nassar, A.; Hata, D.J.; Akkus, Z. Classification of fungal genera from microscopic images using artificial intelligence. J. Pathol. Inform. 2023, 14, 100314. [Google Scholar] [CrossRef]
- Cinar, I.; Taspinar, Y.S.; Taspinar, Y.S. Detection of Fungal Infections from Microscopic Fungal Images Using Deep Learning Techniques. Int. Conf. Adv. Technol. 2023. [Google Scholar] [CrossRef]
- Gümüş, A. Classification of Microscopic Fungi Images Using Vision Transformers for Enhanced Detection of Fungal Infections. Turk. J. Nat. Sci. 2024, 13, 152–160. [Google Scholar] [CrossRef]
- Ikechukwu, A.V.; Murali, S.; Deepu, R.; Shivamurthy, R.C. ResNet-50 vs VGG-19 vs training from scratch: A comparative analysis of the segmentation and classification of Pneumonia from chest X-ray images. Glob. Transit. Proc. 2021, 2, 375–381. [Google Scholar] [CrossRef]
- Alomar, K.; Aysel, H.I.; Cai, X. Data Augmentation in Classification and Segmentation: A Survey and New Strategies. J. Imaging 2023, 9, 46. [Google Scholar] [CrossRef] [PubMed]
- Barshooi, A.H.; Amirkhani, A. A novel data augmentation based on Gabor filter and convolutional deep learning for improving the classification of COVID-19 chest X-Ray images. Biomed. Signal Process. Control 2022, 72, 103326. [Google Scholar] [CrossRef]
- Shabbir, A.; Ali, N.; Ahmed, J.; Zafar, B.; Rasheed, A.; Sajid, M.; Ahmed, A.; Dar, S.H. Satellite and Scene Image Classification Based on Transfer Learning and Fine Tuning of ResNet50. Math. Probl. Eng. 2021, 2021, 5843816. [Google Scholar] [CrossRef]
- Nandhini, S.; Ashokkumar, K. An automatic plant leaf disease identification using DenseNet-121 architecture with a mutation-based henry gas solubility optimization algorithm. Neural Comput. Appl. 2022, 34, 5513–5534. [Google Scholar] [CrossRef]
- Arulananth, T.S.; Prakash, S.W.; Ayyasamy, R.K.; Kavitha, V.P.; Kuppusamy, P.G.; Chinnasamy, P. Classification of Paediatric Pneumonia Using Modified DenseNet-121 Deep-Learning Model. IEEE Access 2024, 12, 35716–35727. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, Z.; Cai, X.; Zhu, T. The application of improved densenet algorithm in accurate image recognition. Sci. Rep. 2024, 14, 8645. [Google Scholar] [CrossRef]
- Thisanke, H.; Deshan, C.; Chamith, K.; Seneviratne, S.; Vidanaarachchi, R.; Herath, D. Semantic segmentation using Vision Transformers: A survey. Eng. Appl. Artif. Intell. 2023, 126, 106669. [Google Scholar] [CrossRef]
- Yang, J.; Luo, K.Z.; Li, J.; Deng, C.; Guibas, L.; Krishnan, D.; Weinberger, K.Q.; Tian, Y.; Wang, Y. Denoising Vision Transformers. arXiv 2024, arXiv:2401.02957. [Google Scholar]
- Yunusa, H.; Qin, S.; Chukkol, A.H.A.; Yusuf, A.A.; Bello, I.; Lawan, A. Exploring the Synergies of Hybrid CNNs and ViTs Architectures for Computer Vision: A Survey. arXiv 2024, arXiv:2402.02941. [Google Scholar]
- Ahn, K.; Zhang, Z.; Kook, Y.; Dai, Y. Understanding Adam Optimizer via Online Learning of Updates: Adam is FTRL in Disguise. arXiv 2024, arXiv:2402.01567. [Google Scholar]
- Rainio, O.; Teuho, J.; Klén, R. Evaluation metrics and statistical tests for machine learning. Sci. Rep. 2024, 14, 6086. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Qin, Y.; Zhao, C. Rapid detection of adulterated lamb meat using near infrared and electronic nose: A F1-score-MRE data fusion approach. Food Chem. 2024, 439, 138123. [Google Scholar] [CrossRef] [PubMed]




| Dataset | Number of Images |
|---|---|
| Train set size (70%) | 10,027 |
| Test set size | 4827 |
| Validation set size | 11,597 |
| Classification Class Name | Number of Images |
|---|---|
| Penicillium svalbardense | 218 |
| Epicoccum nigrum | 217 |
| Rhizomucor pusillus | 216 |
| Penicillium onobense | 212 |
| Nigrospora oryzae | 198 |
| Aspergillus uvarum | 198 |
| Penicillium restrictum | 197 |
| Penicillium rotoruae | 197 |
| Paecilomyces maximus | 197 |
| Penicillium canescens | 184 |
| Phoma pomorum | 182 |
| Penicillium wotroi | 179 |
| Mariannaea elegans | 177 |
| Penicillium scabrosum | 175 |
| Penicillium ochrochloron | 174 |
| Aspergillus flavus | 174 |
| Fusarium tricinctum | 174 |
| Penicillium glabrum | 173 |
| Purpureocillium lilacinum | 173 |
| Penicillium fagi | 170 |
| Hypocrea pulvinata | 169 |
| Penicillium janczewskii | 167 |
| Hamigera avellanea | 165 |
| Rasamsonia piperina | 163 |
| Penicillium olsonii | 121 |
| Others (86 classes with 121 each) | 10,406 |
| Total | 26,451 |
| Architecture | Accuracy | Precision | Recall | F1 Score |
|---|---|---|---|---|
| Resnet | 76.75% | 89.35% | 89.75% | 88.54% |
| DenseNet121 | 86.77% | 93.08% | 91.10% | 89.20% |
| ViT-16 | 92.64% | 95.77% | 96.35% | 93.84% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansourvar, M.; Charylo, K.R.; Frandsen, R.J.N.; Brewer, S.S.; Hoof, J.B. Automated Fungal Identification with Deep Learning on Time-Lapse Images. Information 2025, 16, 109. https://doi.org/10.3390/info16020109
Mansourvar M, Charylo KR, Frandsen RJN, Brewer SS, Hoof JB. Automated Fungal Identification with Deep Learning on Time-Lapse Images. Information. 2025; 16(2):109. https://doi.org/10.3390/info16020109
Chicago/Turabian StyleMansourvar, Marjan, Karol Rafal Charylo, Rasmus John Normand Frandsen, Steen Smidth Brewer, and Jakob Blæsbjerg Hoof. 2025. "Automated Fungal Identification with Deep Learning on Time-Lapse Images" Information 16, no. 2: 109. https://doi.org/10.3390/info16020109
APA StyleMansourvar, M., Charylo, K. R., Frandsen, R. J. N., Brewer, S. S., & Hoof, J. B. (2025). Automated Fungal Identification with Deep Learning on Time-Lapse Images. Information, 16(2), 109. https://doi.org/10.3390/info16020109







