Structure Learning and Hyperparameter Optimization Using an Automated Machine Learning (AutoML) Pipeline
Abstract
1. Introduction
2. Related Studies and the Current Contribution
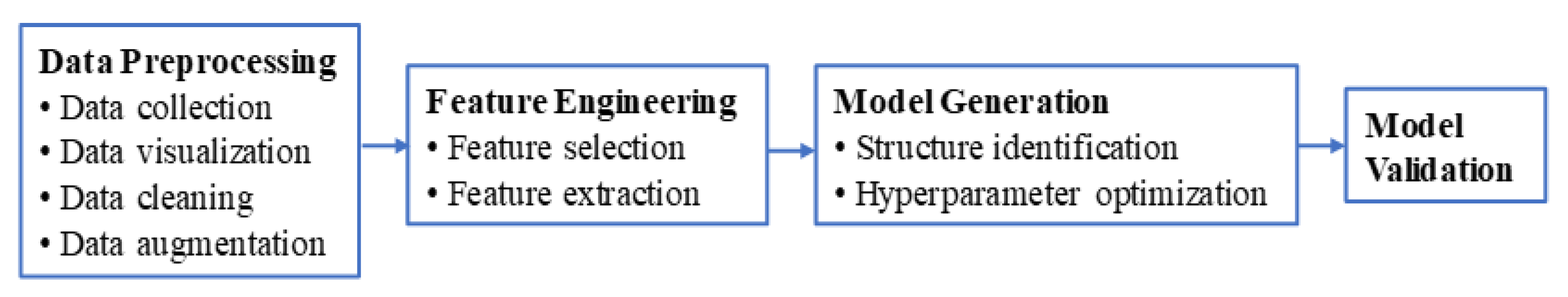
3. The Proposed Pipeline
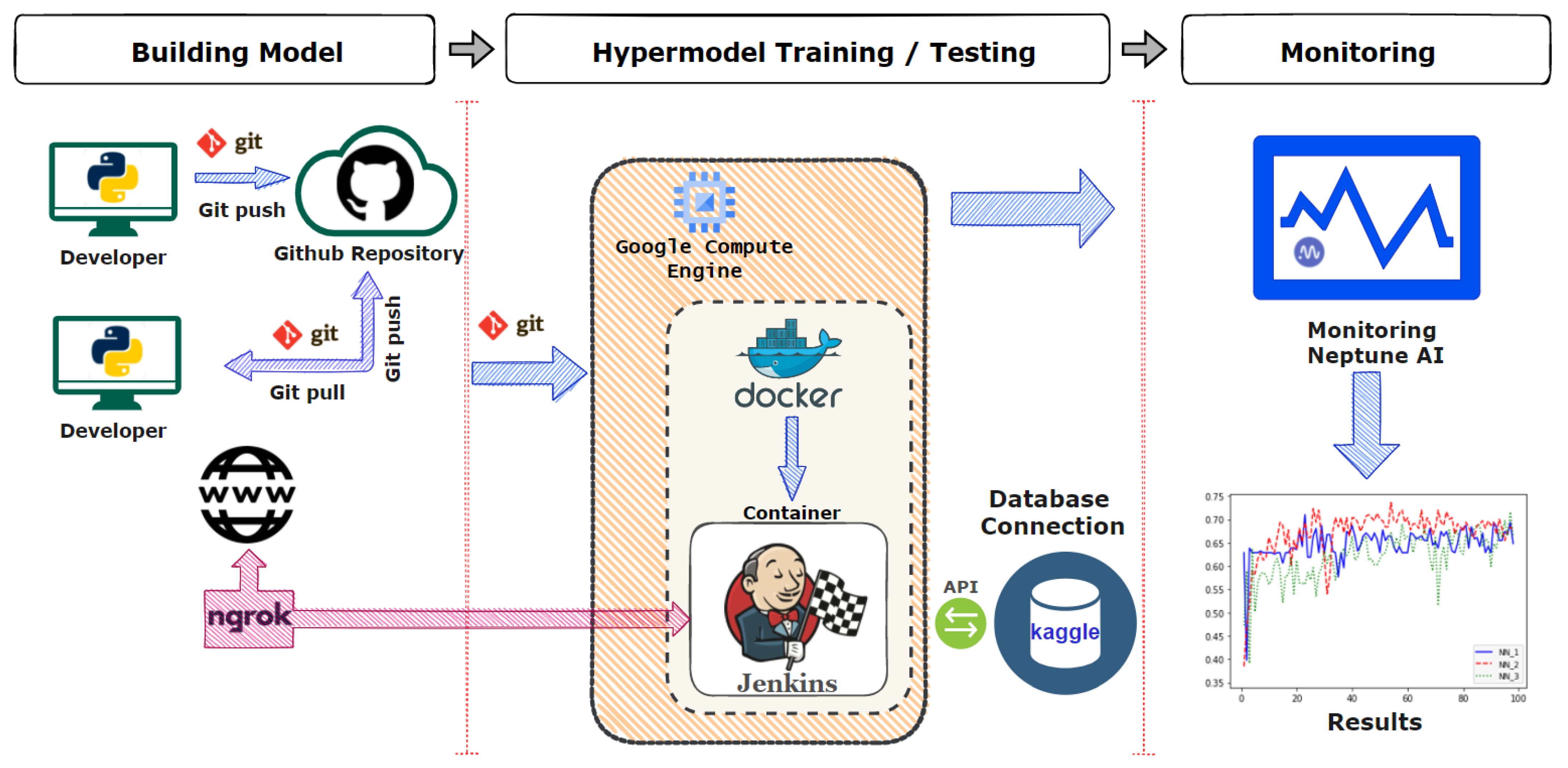
3.1. The Overall Operational Structure of the Pipeline
3.2. Technologies Used in the Pipeline
3.2.1. Git Version Control System
3.2.2. Jenkins Automation Server
3.2.3. Google Cloud Virtual Machines
3.2.4. Ngrok API
3.2.5. Kaggle API
3.2.6. Docker Platform
3.2.7. Monitoring through Neptune AI
- Easy integration with popular libraries and tools (e.g., TensorFlow, Keras, PyTorch, Scikit-Learn, etc.)
- A web-based interface for tracking and organizing experiments and projects
- Collaboration features that allow multiple people to work on the same project
- A platform for sharing and reproducing results
- Flexibility in handling complex projects that deal with different experiments and the proper data/results tracking
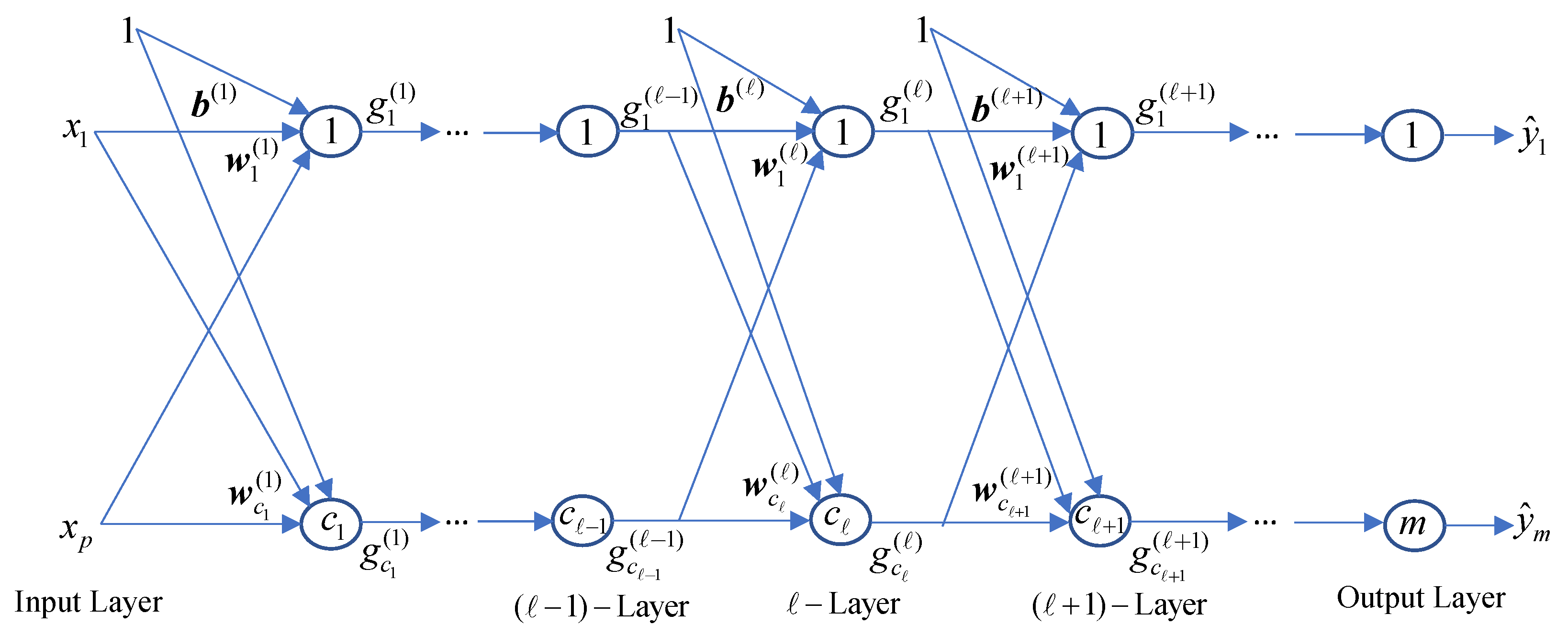
3.2.8. Polynomial Neural Network
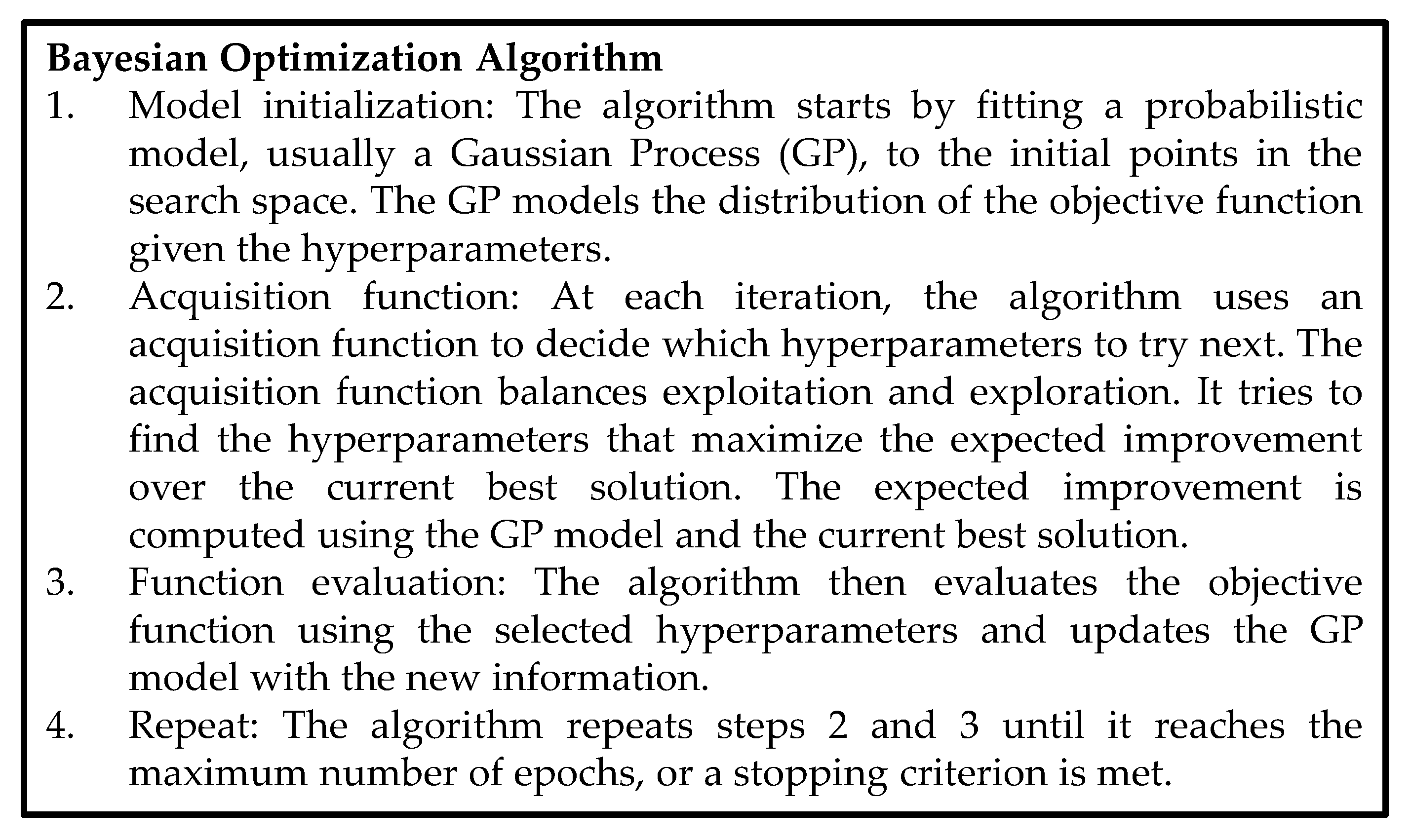
3.2.9. Hyperparameter Tuning
- the learning rate parameter
- the number of layers
- the number of nodes in each layer
- the type of activation function in each layer (the activation function types are given in Equations (4)–(6))
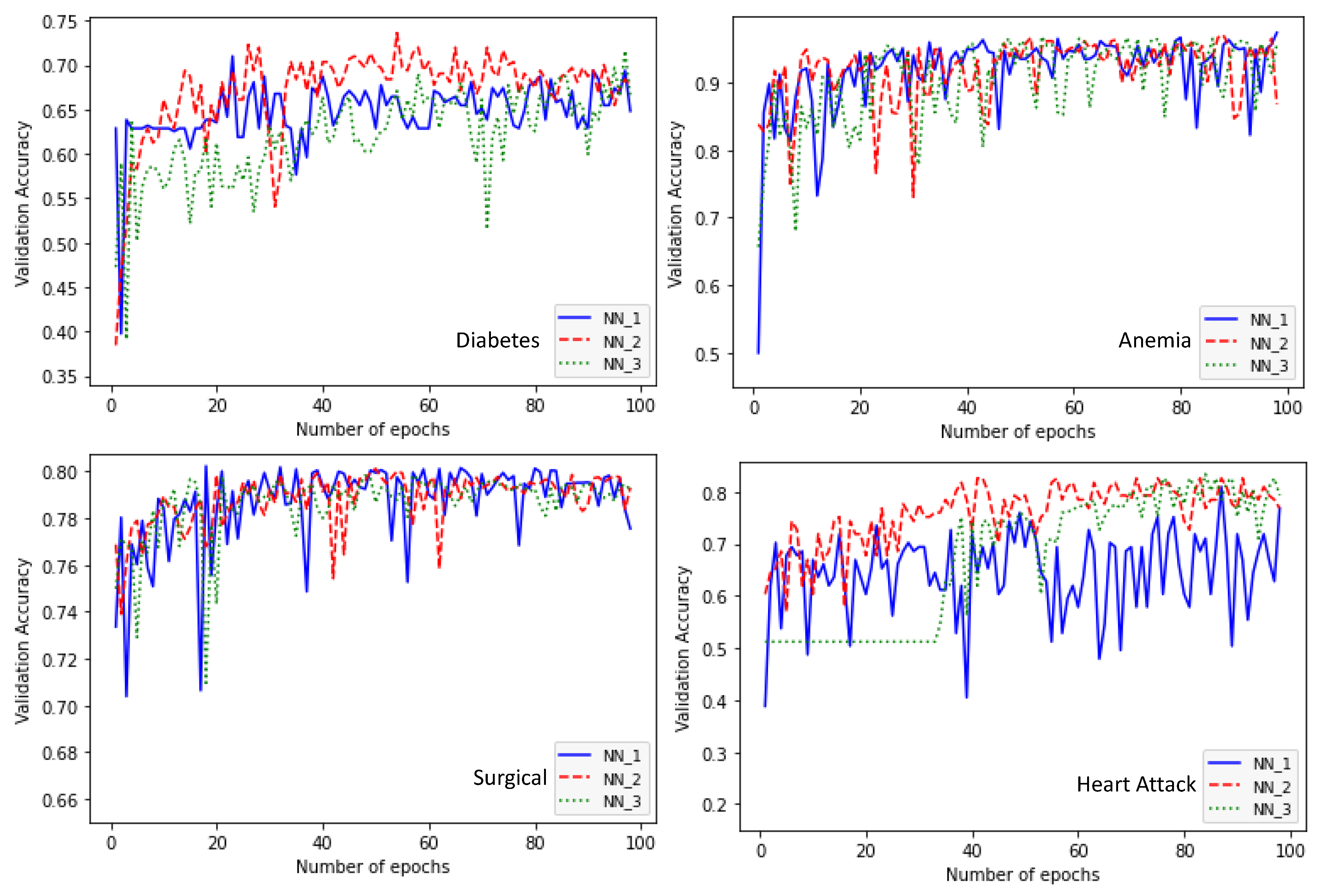
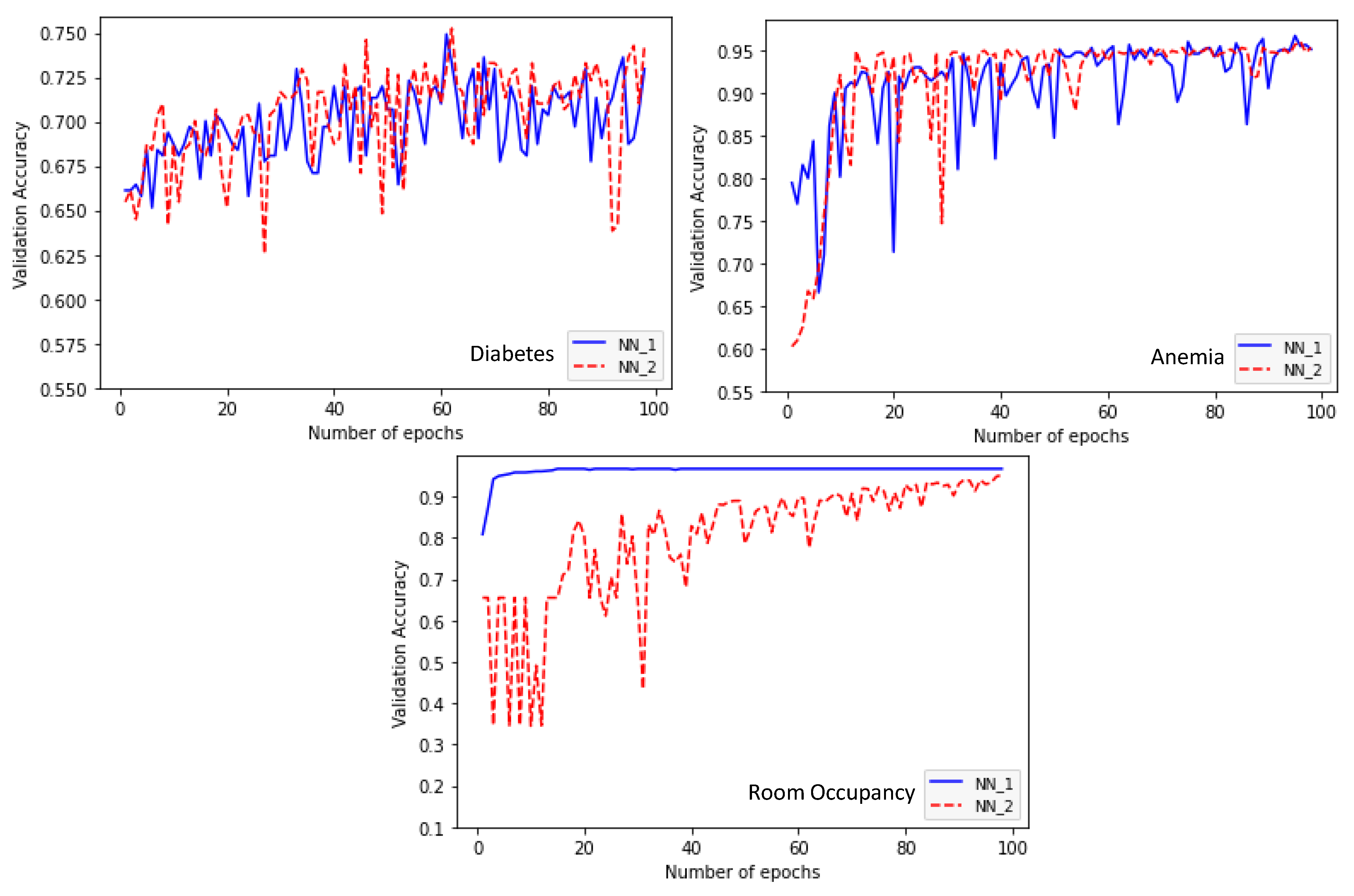
4. Experimental Study
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.; Blum, M.; Hutter, F. Efficient and robust automated machine learning. In Proceedings of the Annual Conference on Neural Information Processing Systems 2015 (Advances in Neural Information Processing Systems 28), Montreal, QC, Canada, 7–12 December 2015; pp. 2962–2970. [Google Scholar]
- Liu, H.; Simonyan, K.; Yang, Y.; Anderson, A.; Zisserman, A. DARTS: Differentiable architecture search. In Proceedings of the 7th International Conference on Learning Representations (ICLR 2019), New Orleans, LA, USA, 6–9 May 2019; pp. 1–13. [Google Scholar]
- Zoph, B.; Le, Q.V. Neural architecture search with reinforcement learning. In Proceedings of the 5th International Conference on Learning Representations (ICLR 2016), Toulon, France, 24–26 April 2017; pp. 1–16. [Google Scholar]
- He, X.; Zhao, K.; Chu, X. AutoML: A survey of the state-of-the-art. Knowl.-Based Syst. 2021, 212, 106622. [Google Scholar] [CrossRef]
- Cheng, S.; Chen, J.; Anastasiou, C.; Angeli, P.; Matar, O.K.; Guo, Y.-K.; Pain, C.C.; Arcucci, R. Generalised latent assimilation in heterogeneous reduced spaces with machine learning surrogate models. J. Sci. Comput. 2023, 94, 11. [Google Scholar] [CrossRef]
- Cheng, S.; Prentice, I.C.; Huang, Y.; Jin, Y.; Guo, Y.K.; Arcucci, R. Data-driven surrogate model with latent data assimilation: Application to wildfire forecasting. J. Comput. Phys. 2022, 464, 111302. [Google Scholar] [CrossRef]
- Zoller, M.; Huber, M.F. Benchmark and survey of automated machine learning frameworks. arXiv 2019, arXiv:1904.12054. [Google Scholar] [CrossRef]
- Karmaker, S.K.; Hassan, M.M.; Smith, M.J.; Xu, L.; Zhai, C.-X.; Veeramachaneni, K. AutoML to date and beyond: Challenges and opportunities. ACM Comput. Surv. 2021, 54, 175. [Google Scholar]
- Nagarajah, T.; Poravi, G. A review on automated machine learning (AutoML) systems. In Proceedings of the 5th IEEE International Conference for Convergence in Technology (I2CT), Bombay, India, 29–31 March 2019; pp. 1–6. [Google Scholar]
- Kotthoff, L.; Thornton, C.; Hoos, H.H.; Hutter, F.; Leyton-Brown, K. Auto-WEKA 2.0: Automatic model selection and hyperparameter optimization in WEKA. J. Mach. Learn. Res. 2017, 18, 1–5. [Google Scholar]
- Autokeras. Available online: https://autokeras.com/ (accessed on 15 October 2022).
- Zimmer, L.; Lindauer, M.; Hutter, F. Auto-pytorch tabular: Multiidelity metalearning for efficient and robust autodl. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3079–3090. [Google Scholar] [CrossRef]
- Feurer, M.; Eggensperger, K.; Falkner, S.; Lindauer, M.; Hutter, F. Auto-sklearn 2.0: Hands-free automl via meta-learning. arXiv 2020, arXiv:2007.04074. [Google Scholar]
- Khan, M.A.; Iqbal, N.; Imran; Jamil, H.; Kim, D.-H. An optimized ensemble prediction model using AutoML based on soft voting classifier for network intrusion detection. J. Netw. Comput. Appl. 2023, 212, 103560. [Google Scholar] [CrossRef]
- Pecnik, L.; Fister, I.; Fister, I., Jr. NiaAML2: An improved AutoML using nature-inspired algorithms. Lect. Notes Comput. Sci. 2021, 12690, 243–252. [Google Scholar]
- Ferreira, L.; Pilastri, A.; Martins, C.M.; Pires, P.M.; Cortez, P. A comparison of AutoML tools for machine learning, deep learning and XGBoost. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN’ 21), Shenzhen, China, 18–22 July 2021; pp. 1–8. [Google Scholar]
- Renza, D.; Cardenas, E.A.; Jaramillo, C.M.; Weber, S.S.; Martinez, E. Landslide susceptibility model by means of remote sensing images and AutoML. Commun. Comput. Inf. Sci. 2021, 1431, 25–37. [Google Scholar]
- Opara, E.; Wimmer, H.; Rebman, C.M., Jr. Auto-ML cyber security data analysis using Google, Azure and IBM Cloud Platforms. In Proceedings of the International Conference on Electrical, Computer and Energy Technologies (ICECET 2022), Prague, Czech Republic, 20–22 July 2022; pp. 1–10. [Google Scholar]
- Yan, C.; Zhang, Y.; Zhang, Q.; Yang, Y.; Jiang, X.; Yang, Y.; Wang, B. Privacy-preserving online AutoML for domain-specific face detection. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 4124–4134. [Google Scholar]
- Singh, D.; Pant, P.K.; Pant, H.; Dobhal, D.C. Robust automated machine learning (AutoML) system for early stage hepatic disease detection. Lect. Notes Data Eng. Commun. Technol. 2021, 57, 65–76. [Google Scholar]
- Mukherjee, S.; Rao, Y.S. Auto-ML Web-application for automated machine learning algorithm training and evaluation. In Proceedings of the 7th IEEE International Conference for Convergence in Technology (I2CT), Pune, India, 7–9 April 2022; pp. 1–6. [Google Scholar]
- Javeri, I.Y.; Toutiaee, M.; Arpinar, I.B.; Miller, J.A. Improving neural networks for time-series forecasting using data augmentation and AutoML. arXiv 2021, arXiv:2103.01992. [Google Scholar]
- Symeonidis, G.; Nerantzis, E.; Kazakis, A.; Papakostas, G.A. MLOps—Definitions, Tools and Challenges. In Proceedings of the 12th IEEE Annual Computing and Communication Workshop and Conference (CCWC 2022), Las Vegas, NV, USA, 26–29 January 2022; pp. 453–460. [Google Scholar]
- Gijsbers, P.; LeDell, E.; Thomas, J.; Poirier, S.; Bischl, B.; Vanschoren, J. An open source AutoML benchmark. arXiv 2019, arXiv:1907.00909. [Google Scholar]
- Patibandla, R.S.M.L.; Srinivas, V.S.; Mohanty, S.N.; Pattanaik, C.R. Automatic machine learning: An exploratory review. In Proceedings of the 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Noida, India, 3–4 September 2021; pp. 1–9. [Google Scholar]
- Stamoulis, D.; Ding, R.; Wang, D.; Lymberopoulos, D.; Priyantha, B.; Liu, J.; Marculescu, D. Single-path mobile AutoML: Efficient ConvNet design and NAS hyperparameter optimization. IEEE J. Sel. Top. Signal 2020, 14, 609–622. [Google Scholar] [CrossRef]
- Cai, H.; Lin, J.; Lin, Y.; Liu, Z.; Wang, K.; Wang, T.; Zhu, L.; Han, S. AutoML for architecting efficient and specialized neural networks. IEEE Micro 2019, 40, 75–82. [Google Scholar] [CrossRef]
- Kreuzberger, D.; Kühl, N.; Hirschl, S. Machine Learning Operations (MLOps): Overview, Definition, and Architecture. arXiv 2022, arXiv:2205.02302. [Google Scholar] [CrossRef]
- Hewage, N.; Meedeniya, D. Machine Learning Operations: A Survey on MLOps Tool Support. arXiv 2022, arXiv:2202.10169. [Google Scholar]
- Treveil, M.; Omont, N.; Stenac, C.; Lefevre, K.; Phan, D.; Zentici, J.; Lavoillotte, A.; Miyazaki, M.; Heidmann, L. Introducing MLOps: How to Scale Machine Learning in the Enterprise; O’Reilly Media: Sebastopol, ON, Canada, 2021. [Google Scholar]
- Subramanya, R.; Sierla, S.; Vyatkin, V. From DevOps to MLOps: Overview and Application to Electricity Market Forecasting. Appl. Sci. 2022, 12, 9851. [Google Scholar] [CrossRef]
- Granlund, T.; Kopponen, A.; Stirbu, V.; Myllyaho, L.; Mikkonen, T. MLOps challenges in multi-organization setup: Experiences from two real-world cases. In Proceedings of the 1st Workshop on AI Engineering—Software Engineering for AI (WAIN’21), Virtual Conference, Madrid, Spain, 30–31 May 2021; pp. 82–88. [Google Scholar]
- Makinen, S.; Skogstrom, H.; Laaksonen, E.; Mikkonen, T. Who needs MLOps: What data scientists seek to accomplish and how can MLOps help? In Proceedings of the 1st Workshop on AI Engineering—Software Engineering for AI (WAIN’21), Virtual Conference, Madrid, Spain, 30–31 May 2021; pp. 109–112. [Google Scholar]
- Humble, J.; Farley, D. Continuous Delivery: Reliable Software Releases through Build, Test, and Deployment Automation; Pearson Education Inc.: Boston, MA, USA, 2011. [Google Scholar]
- Garg, S.; Pundir, P.; Rathee, G.; Gupta, P.K.; Garg, S.; Ahlawat, S. On continuous integration/continuous delivery for automated deployment of machine learning models using MLOps. In Proceedings of the 4th IEEE International Conference on Artificial Intelligence and Knowledge Engineering (AIKE), Laguna Hills, CA, USA, 1–3 December 2021; pp. 25–28. [Google Scholar]
- Karlas, B.; Interlandi, M.; Renggli, C.; Wu, W.; Zhang, C.; Mukunthu, D.; Babu, I.; Edward, J.; Lauren, C.; Xu, A.; et al. Building continuous integration services for machine learning. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Long Beach, CA, USA, 6–10 July 2020; pp. 2407–2415. [Google Scholar]
- Durbha, K.S.; Amuru, S. AutoML models for wireless signals classification and their effectiveness against adversarial attacks. In Proceedings of the 14th International Conference on COMmunication Systems & NETworks (COMSNETS’ 22), Bangalore, India, 4–8 January 2022; pp. 265–269. [Google Scholar]
- Goodfellow, I.; Benzio, Y.; Courville, A. Deep Learning; MIT Press Ltd.: Boston, MA, USA, 2016. [Google Scholar]
- Kurian, J.J.; Dix, M.; Amihai, I.; Ceusters, G.; Prabhune, A. BOAT: A Bayesian optimization AutoML time-series framework for industrial applications. In Proceedings of the 7th IEEE International Conference on Big Data Computing Service and Applications (BigDataService’ 21), Oxford, UK, 23–26 August 2021; pp. 17–24. [Google Scholar]
- Esmaeili, A.; Ghorrati, Z.; Matson, E.T. Hierarchical collaborative hyperparameter tuning. Lect. Notes Artif. Intell. 2022, 13616, 127–139. [Google Scholar]
- Bardenet, R.; Brendel, M.; Kegl, B. Collaborative hyperparameter tuning. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; pp. II-199–II-207. [Google Scholar]
- Filippou, K.; Aifantis, G.; Mavrikos, E.; Tsekouras, G. Deep feedforward neural network classifier with polynomial layer and shared weights. In Proceedings of the 4th International Conference on Advances in Signal Processing and Artificial Intelligence (ASPAI 2022), Corfu, Greece, 19–21 October 2022. [Google Scholar]
- Tsekouras, G.E.; Trygonis, V.; Maniatopoulos, A.; Rigos, A.; Chatzipavlis, A.; Tsimikas, J.; Mitianoudis, N.; Velegrakis, A.F. A Hermite neural network incorporating artificial bee colony optimization to model shoreline realignment at a reef-fronted beach. Neurocomputing 2018, 280, 32–45. [Google Scholar] [CrossRef]
- Git. Available online: https://git-scm.com/ (accessed on 12 November 2022).
- Docker. Available online: https://www.docker.com/ (accessed on 12 November 2022).
- Jenkins. Available online: https://jenkins.io/ (accessed on 12 November 2022).
- Kaggle. Available online: https://www.kaggle.com/ (accessed on 18 November 2022).
- Neptune AI. Available online: https://neptune.ai/ (accessed on 15 November 2022).
- Bird, C.; Rigby, P.C.; Barr, E.T.; Hamilton, D.J.; German, D.M.; Devanbu, P. The promises and perils of mining git. In Proceedings of the 6th IEEE International Working Conference on Mining Software Repositories, Vancouver, BC, Canada, 16–17 May 2009; pp. 1–10. [Google Scholar]
- Zolkifli, N.N.; Ngah, A.; Deraman, A. Version control system: A review. Procedia Comput. Sci. 2018, 135, 408–415. [Google Scholar] [CrossRef]
- Shahin, M.; Babar, M.A.; Zhu, L. Continuous integration, delivery and deployment: A systematic review on approaches, tools, challenges and practices. IEEE Access 2017, 5, 3909–3943. [Google Scholar] [CrossRef]
- Zhao, Y.; Serebrenik, A.; Zhou, Y.; Filkov, V.; Vasilescu, B. The impact of continuous integration on other software development practices: A large-scale empirical study. In Proceedings of the 32nd IEEE/ACM International Conference on Automated Software Engineering (ASE), Urbana, IL, USA, 30 October–3 November 2017; pp. 60–71. [Google Scholar]
- Google Cloud Platform. Available online: https://cloud.google.com/ (accessed on 13 November 2022).
- Ngrok. Available online: https://ngrok.com/ (accessed on 21 November 2022).
- Uslu, C. What is Kaggle? Available online: https://www.datacamp.com/blog/what-is-kaggle (accessed on 18 November 2022).
- Anderson, C. Docker [software engineering]. IEEE Softw. 2015, 32, 102-c3. [Google Scholar] [CrossRef]
- Kwon, S.; Lee, J.H. Divds: Docker image vulnerability diagnostic system. IEEE Access 2020, 8, 42666–42673. [Google Scholar] [CrossRef]
- Jaramillo, D.; Nguyen, D.V.; Smart, R. Leveraging microservices architecture by using Docker technology. In Proceedings of the IEEE SoutheastCon 2016, Norfolk, VA, USA, 30 March–3 April 2016; pp. 1–5. [Google Scholar]
- Bui, T. Analysis of docker security. arXiv 2015, arXiv:1501.02967. [Google Scholar]
- Feurer, M.; Hutter, F. Hyperparameter Optimization. In Automated Machine Learning: Methods, Systems, Challenges; Hutter, F., Kotthoff, L., Vanschoren, J., Eds.; Springer Nature: Cham, Switzerland, 2019; pp. 3–33. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Montgomery, D. Design and Analysis of Experiments; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Brochu, E.; Cora, V.M.; de Freitas, N. A tutorial on Bayesian optimization of expensive cost, with application to active user modeling and hierarchical reinforcement learning. arXiv 2010, arXiv:1012.2599. [Google Scholar]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian optimization of machine learning algorithms. arXiv 2012, arXiv:1206.2944. [Google Scholar]
- BayesianOptimization Tuner. Available online: https://keras.io/api/keras_tuner/tuners/bayesian/ (accessed on 3 December 2022).



| Component/Resource | Google Cloud Platform |
|---|---|
| CPU | 2 vCPU (Intel® Xeon® E5-2696V4 Processor Base frequency 2.2 (GHz)) |
| CPU platform | Intel Broadwell 5 |
| Memory | 8 GB RAM (4 GB RAM per CPU) |
| Operating System | Debian 11.6 (bullseye) |
| Storage | SSD Balanced persistent disk, 80 GB |
| No | Data Set | Number of Instances | Number of Inputs |
|---|---|---|---|
| 1 | Diabetes | 767 | 8 |
| 2 | Surgical | 14,634 | 24 |
| 3 | Anemia | 1420 | 5 |
| 4 | Heart Attack | 3584 | 13 |
| 5 | Room Occupancy | 2664 | 5 |
| 6 | Blood Transfusion | 747 | 4 |
| 7 | BankNote Authentication | 1371 | 4 |
| 8 | Ionosphere | 349 | 34 |
| 9 | Brain Tumor | 35 | 7465 |
| 10 | Phishing Website | 1352 | 9 |
| Hyperparameter | Domain of Values |
|---|---|
| Learning rate | {0.0001, 0.001, 0.01} |
| Number of layers | {3, 4, 5} |
| Number of neurons | [5, 30] with step 5 |
| Type of activation function | {“ReLU”, “Tanh”, “2nd order Hermite polynomial (H2)”} |
| Argument | Value |
|---|---|
| max_trials | 100 |
| num_initial_points | 2 |
| alpha | 0.001 |
| beta | 2.6 |
| seed | random.seed |
| Data Set | NN_1 | NN_2 | NN_3 | Total Hyper-Tuning Time (in Minutes) |
|---|---|---|---|---|
| Diabetes | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: Tanh-15 nodes Layer3: Hermite-30 nodes Layer4: ReLU-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.6938 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-15 nodes Layer2: ReLU-30 nodes Layer3: Hermite-5 nodes Layer4: Tanh-30 nodes Layer5: Hermite-30 nodes Accuracy: 0.7003 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-15 nodes Layer2: ReLU-30 nodes Layer3: Hermite-5 nodes Layer4: Tanh-30 nodes Layer5: Hermite-30 nodes Accuracy: 0.7166 | 41 |
| Anemia | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-25 nodes Layer3: ReLU-25 nodes Layer4: ReLU-5 nodes Layer5: ReLU-20 nodes Accuracy: 0.9735 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-20 nodes Layer3: ReLU-20 nodes Layer4: ReLU-5 nodes Layer5: Tanh-5 nodes Accuracy: 0.9647 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-25 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: Hermite-5 nodes Accuracy: 0.9612 | 47.59 |
| Data Set | NN_1 | NN_2 | NN_3 | Total Hyper-Tuning Time (in Minutes) |
|---|---|---|---|---|
| Surgical | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-5 nodes Layer3: ReLU-10 nodes Layer4: ReLU-20 nodes Layer5: Tanh-30 nodes Accuracy: 0.7982 | learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-20 nodes Layer3: ReLU-15 nodes Layer4: ReLU-30 nodes Layer5: Hermite-5 nodes Accuracy: 0.7931 | learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-5 nodes Layer3: ReLU-10 nodes Layer4: ReLU-25 nodes Layer5: ReLU-25 nodes Accuracy: 0.7948 | 237.42 |
| Heart Attack | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: ReLU-30 nodes Layer3: Hermite-5 nodes Layer4: Hermite-30 nodes Layer5: Tanh-5 nodes Accuracy: 0.7685 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: ReLU-30 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: Hermite/30 nodes Accuracy: 0.8264 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: ReLU-30 nodes Layer3: ReLU- 30 nodes Layer4: ReLU-5 nodes Layer5: Hermite-30 nodes Accuracy: 0.8264 | 29.57 |
| Room Occupancy | Learning rate: 0.01 Number of layers: 5 Layer1: Hermite-30 nodes Layer2: ReLU-10 nodes Layer3: Hermite-20 nodes Layer4: ReLU-5 nodes Layer5: Tanh-5 nodes Accuracy: 0.9762 | Learning rate: 0.0001 Number of layers: 5 Layer1: Hermite-5 nodes Layer2: ReLU-30 nodes Layer3: Hermite-30 nodes Layer4: ReLU-5 nodes Layer5: Hermite-5 nodes Accuracy: 0.9762 | Learning rate: 0.0001 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: ReLU-10 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.9762 | 51.96 |
| Blood Transfusion | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: Hermite-30 nodes Layer3: ReLU-30 nodes Layer4: Hermite-30 nodes Layer5: Hermite-30 nodes Accuracy: 0.7224 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: Hermite-5 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: Hermite-5 nodes Accuracy: 0.7425 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-5 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: Hermite-5 nodes Accuracy: 0.7492 | 35.75 |
| Banknote Authentication | Learning rate: 0.0001 Number of layers: 4 Layer1: Tanh-20 nodes Layer2: Tanh-20 nodes Layer3: ReLU-30 nodes Layer4: ReLU-15 nodes Accuracy: 0.9963 | Learning rate: 0.01 Number of layers: 5 Layer1: Tanh-30 nodes Layer2: ReLU-15 nodes Layer3: ReLU- 30 nodes Layer4: ReLU-30 nodes Layer5: ReLU-15 nodes Accuracy: 0.9981 | Learning rate: 0.01 Number of layers: 5 Layer1: Hermitte-30 nodes Layer2: Tanh-20 nodes Layer3: ReLU-30 nodes Layer4: ReLU-30 nodes Layer5: ReLU-30 nodes Accuracy: 0.9945 | 56.13 |
| Ionosphere | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-20 nodes Layer2: Tanh-30 nodes Layer3: Tanh-5 nodes Layer4: ReLU-5 nodes Layer5: ReLU-15 nodes Accuracy: 0.9426 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-10 nodes Layer2: ReLU-30 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: ReLU-20 nodes Accuracy: 0.8929 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: ReLU-15 nodes Layer3: ReLU-15 nodes Layer4: ReLU-5 nodes Layer5: Hermite-5 nodes Accuracy: 0.9143 | 34.76 |
| Data Set | NN_1 | NN_2 | NN_3 | Total Hyper-Tuning Time (in Minutes) |
|---|---|---|---|---|
| Brain Tumor | Learning rate: 0.01 Number of layers: 5 Layer1: Hermite-5 nodes Layer2: Hermite-30 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.7857 | Learning rate: 0.01 Number of layers: 5 Layer1: Hermite-5 nodes Layer2: Hermite-30 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: Hermite-25 nodes Accuracy: 0.9286 | Learning rate: 0.01 Number of layers: 5 Layer1: Hermite-30 nodes Layer2: Hermit-30 nodes Layer3: ReLU-30 nodes Layer4: ReLU-5 nodes Layer5: ReLU-25 nodes Accuracy: 0.9286 | 34.96 |
| Phishing Website | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: ReLU-30 nodes Layer3: ReLU-25 nodes Layer4: Tanh-5 nodes Layer5: Tanh-5 nodes Accuracy: 0.4233 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-25 nodes Layer2: ReLU-30 nodes Layer3: ReLU-20 nodes Layer4: Tanh-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.4344 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-25 nodes Layer2: Tanh-30 nodes Layer3: ReLU-25 nodes Layer4: Tanh-5 nodes Layer5: Tanh-5 nodes Accuracy: 0.4370 | 49.50 |
| Data Set | NN_1 (Use of Hyperspace A) | NN_2 (Use of Hyperspace B) | Mean Hyper-Tuning Time (in Minutes) |
|---|---|---|---|
| Diabetes | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-15 nodes Layer2: ReLU-20 nodes Layer3: ReLU-30 nodes Layer4: Hermite-5 nodes Layer5: ReLU-30 nodes Accuracy: 0.7361 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-15 nodes Layer2: ReLU-20 nodes Layer3: ReLU-30 nodes Layer4: ReLU-30 nodes Layer5: ReLU-30 nodes Accuracy: 0.7427 | 42.57 |
| Anemia | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-10 nodes Layer2: ReLU-30 nodes Layer3: Hermite-5 nodes Layer4: ReLU-10 nodes Layer5: ReLU-30 nodes Accuracy: 0.9665 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: ReLU-5 nodes Layer3: ReLU-15 nodes Layer4: ReLU-5 nodes Layer5: ReLU-10 nodes Accuracy: 0.9595 | 56.08 |
| Room Occupancy | Learning rate: 0.0001 Number of layers: 5 Layer1: Hermite-5 nodes Layer2: ReLU-30 nodes Layer3: ReLU-5 nodes Layer4: ReLU-30 nodes Layer5: ReLU-5 nodes Accuracy: 0.9675 | Learning rate: 0.0001 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: ReLU-25 nodes Layer3: ReLU-30 nodes Layer4: ReLU-30 nodes Layer5: ReLU-5 nodes Accuracy: 0.9525 | 73.85 |
| Data Set | NN_1 (Use of Hyperspace A) | NN_2 (Use of Hyperspace B) | Mean Hyper-Tuning Time (in Minutes) |
|---|---|---|---|
| Banknote Authentication | Learning rate: 0.0001 Number of layers: 3 Layer1: ReLU-20 nodes Layer2: Hermite-10 nodes Layer3: Tanh-5 nodes Accuracy: 0.9982 | Learning rate: 0.01 Number of layers: 3 Layer1: ReLU-20 nodes Layer2: ReLU-10 nodes Layer3: Tanh-5 nodes Accuracy: 0.9944 | 58.48 |
| Ionosphere | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 Layer2: ReLU-30 Layer3: Hermite-5 Layer4: ReLU-5 Layer5: ReLU-20 Accuracy: 0.9214 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: ReLU-30 nodes Layer3: Tanh-5 nodes Layer4: ReLU-5 nodes Layer5: ReLU-20 nodes Accuracy: 0.9071 | 38.96 |
| Data Set | NN_1 | NN_2 | NN_3 | Total Hyper-Tuning Time (in Minutes) |
|---|---|---|---|---|
| Diabetes | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-10 nodes Layer2: Hermite-15 nodes Layer3: Tanh-15 nodes Layer4: ReLU-30 nodes Layer5: Tanh-30 nodes Accuracy: 0.6818 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: ReLU-15 nodes Layer3: Tanh-30 nodes Layer4: Hermite-10 nodes Layer5: Tanh-10 nodes Accuracy: 0.6628 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-15 nodes Layer2: Hermite-15 nodes Layer3: Tanh-30 nodes Layer4: Hermite-30 nodes Layer5: Tanh-10 nodes Accuracy: 0.6958 | 42.61 |
| Anemia | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-10 nodes Layer2: Hermite-20 nodes Layer3: ReLU-25 nodes Layer4: ReLU-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.9484 | Learning rate: 0.001 Number of layers: 5 Layer1: ReLU-30 nodes Layer2: Hermite-10 nodes Layer3: ReLU-25 nodes Layer4: ReLU-30 nodes Layer5: Hermite-5 nodes Accuracy: 0.9528 | Learning rate: 0.01 Number of layers: 5 Layer1: Hermite-10 nodes Layer2: Hermite-20 nodes Layer3: ReLU-30 nodes Layer4: Hermite-30 nodes Layer5: Hermite-30 nodes Accuracy: 0.9390 | 48.09 |
| Data Set | NN_1 (Use of Hyperspace A) | NN_2 (Use of Hyperspace B) | Mean Hyper-Tuning Time (in Minutes) |
|---|---|---|---|
| Diabetes | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-15 nodes Layer2: ReLU-30 nodes Layer3: Hermite-5 nodes Layer4: ReLU-5 nodes Layer5: ReLU-30 nodes Accuracy: 0.7272 | Learning rate: 0.01 Number of layers: 3 Layer1: Tanh-30 nodes Layer2: Tanh-30 nodes Layer3: Tanh-5 nodes Accuracy: 0.6963 | 46.08 |
| Anemia | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: ReLU-30 nodes Layer3: Hermite-5 nodes Layer4: ReLU-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.9613 | Learning rate: 0.01 Number of layers: 5 Layer1: ReLU-5 nodes Layer2: ReLU-30 nodes Layer3: ReLU-15 nodes Layer4: ReLU-5 nodes Layer5: ReLU-5 nodes Accuracy: 0.9417 | 52.63 |
| Stage | Process | Time Difference (Minutes) | Total Time Difference (Minutes) | Time Difference per Dataset (Minutes) |
|---|---|---|---|---|
| 1 | 1.1 Collaboration between team’s members | >>40 min per day | >>40 min per day | ---- |
| 2 | 2.1 Execution to open the appropriate software | 9 | >89 | >9 |
| 2.2 Dataset search and download | 60 | |||
| 2.3 Creation of folders and subfolders | 30 | |||
| 2.4 Hyperparameter optimization | 0 | |||
| 2.5 Create Jenkin jobs (the respective time is subtracted from the total time) | 10 (subtracted) | |||
| 3 | 3.1 Save tracked data values in a file and open manually another script to read the values and produce the figures | 200 | 550 | 55 |
| 3.2 Track a different variable (for example different loss function) | 50 | |||
| 3.3 Share results and figures between team’s members | 300 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippou, K.; Aifantis, G.; Papakostas, G.A.; Tsekouras, G.E. Structure Learning and Hyperparameter Optimization Using an Automated Machine Learning (AutoML) Pipeline. Information 2023, 14, 232. https://doi.org/10.3390/info14040232
Filippou K, Aifantis G, Papakostas GA, Tsekouras GE. Structure Learning and Hyperparameter Optimization Using an Automated Machine Learning (AutoML) Pipeline. Information. 2023; 14(4):232. https://doi.org/10.3390/info14040232
Chicago/Turabian StyleFilippou, Konstantinos, George Aifantis, George A. Papakostas, and George E. Tsekouras. 2023. "Structure Learning and Hyperparameter Optimization Using an Automated Machine Learning (AutoML) Pipeline" Information 14, no. 4: 232. https://doi.org/10.3390/info14040232
APA StyleFilippou, K., Aifantis, G., Papakostas, G. A., & Tsekouras, G. E. (2023). Structure Learning and Hyperparameter Optimization Using an Automated Machine Learning (AutoML) Pipeline. Information, 14(4), 232. https://doi.org/10.3390/info14040232








