AutoML with Bayesian Optimizations for Big Data Management
Abstract
1. Introduction
- Hyperparameter tuning: This involves automatically searching for the best combination of hyperparameters for a given machine-learning model. This can be done using techniques such as grid search, random search, or Bayesian optimization.
- Feature selection and engineering: AutoML can be used to automatically select the most relevant features for a given dataset and to perform feature engineering tasks such as scaling, normalization, and dimensionality reduction.
- Model selection: AutoML can be used to automatically select the best machine learning model for a given dataset. This can be done by comparing the performance of different models on the dataset, or by using techniques such as ensembling or stacking to combine the predictions of multiple models.
- Automated Deployment: AutoML can be used to automate the process of deploying machine learning models into production. This can include tasks such as model versioning, monitoring, and scaling.
- Model selection: It is necessary to first use a model to identify the class of hypothesis spaces from which the final hypothesis will be selected. The model of choice is typically embedded implicitly in the class of hypothesis spaces of a learner L. Automating this procedure is challenging. The model is chosen, in practice, by experts who have a thorough grasp of the problem at hand.
- Hyperparameter search: Optimizing a vector in the hyperparameter space of the learner L representing a hypothesis space . A naïve approach to do this is to systematically try configurations using a grid search or a random search over . To evaluate the quality of a given , L is usually trained on a training dataset using . This yields a hypothesis that is evaluated using a validation dataset . The goal of hyperparameter optimization is to minimize the loss of on , i.e., to find an approximation:
- Training or parameter search: Let w be a vector in the parameter space , describing a hypothesis given a hyperparameter configuration . The goal of parameter search is to find an approximation of the hypothesis , with being the empirical loss of on a given training dataset according to some loss function ℓ. Depending on the learner L, various kinds of optimization methods are used to find this minimum, e.g., Bayesian optimization, quadratic programming or, if is computable, gradient descent. The quality l of is measured by the loss on a validation or test dataset, i.e.,
2. Related Work
2.1. Automated Machine Learning in Industry
2.2. Feature Engineering and Selection
2.3. Meta-Learning
2.4. Neural Architecture Search (NAS)
2.5. The CASH Problem
2.6. Optimization Techniques
2.7. Tiny Machine Learning
2.8. AutoML
3. Hyperparameter Optimization
- Number T of evaluations of l: During optimization multiple hyperparameter configurations will be evaluated using l. T is usually fixed when using a grid search or a random search. After evaluating T configurations, the best one is chosen. Those naïve approaches assume that is independent of for all pairs . We will see that this strong assumption of independence is not necessarily true which in turn allows us to reduce T.
- Training dataset size S: The performance of a given configuration is computed by training the learner on which is expensive for big datasets. By training on S instead of datapoints the evaluation can be sped up.
- Number of training iterations E: Training is frequently an iterative process, e.g., gradient descent, depending on the learner. The training phase of hyperparameter optimization might end before convergence.
3.1. FABOLAS
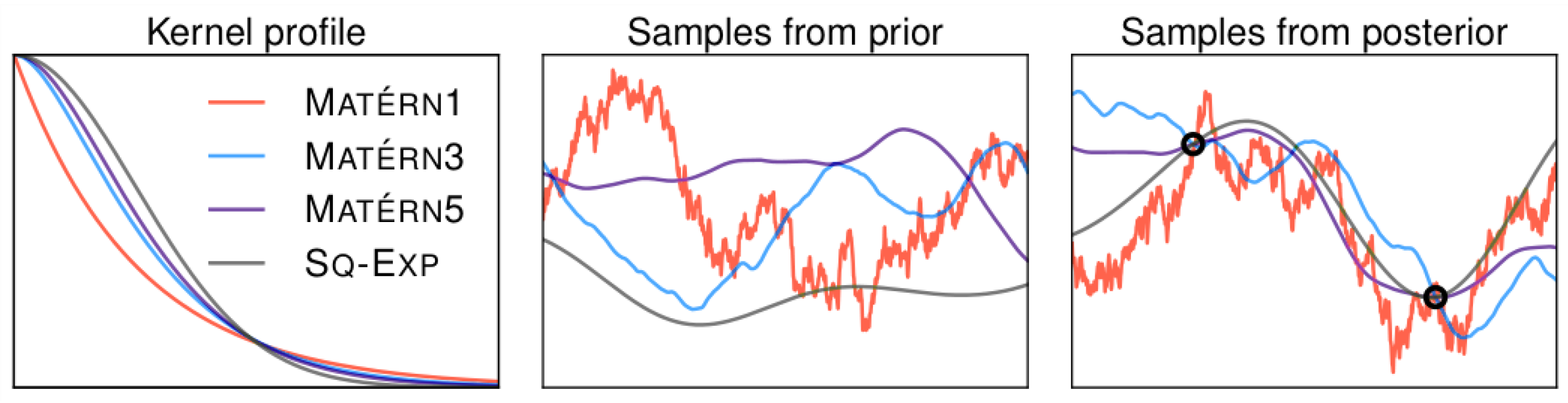
- The validation loss l is modeled as a Gaussian process (GP) f based on the assumption that two configurations and will perform similarly if they are similar according to some kernel . The Gaussian process f is used as a surrogate to estimate the expected value and variance of l given . Using Bayesian optimization l will be sampled at promising positions to iteratively improve f. Hyperparameter configurations that are expected to perform worse than the current optimum will not be sampled. This effectively reduces T.
- The optimizer is given an additional degree of freedom by modeling the training dataset size S as an additional hyperparameter of f. When trained on the whole dataset, this enables projecting the value of l while only probing smaller sections, thereby reducing the size of S.
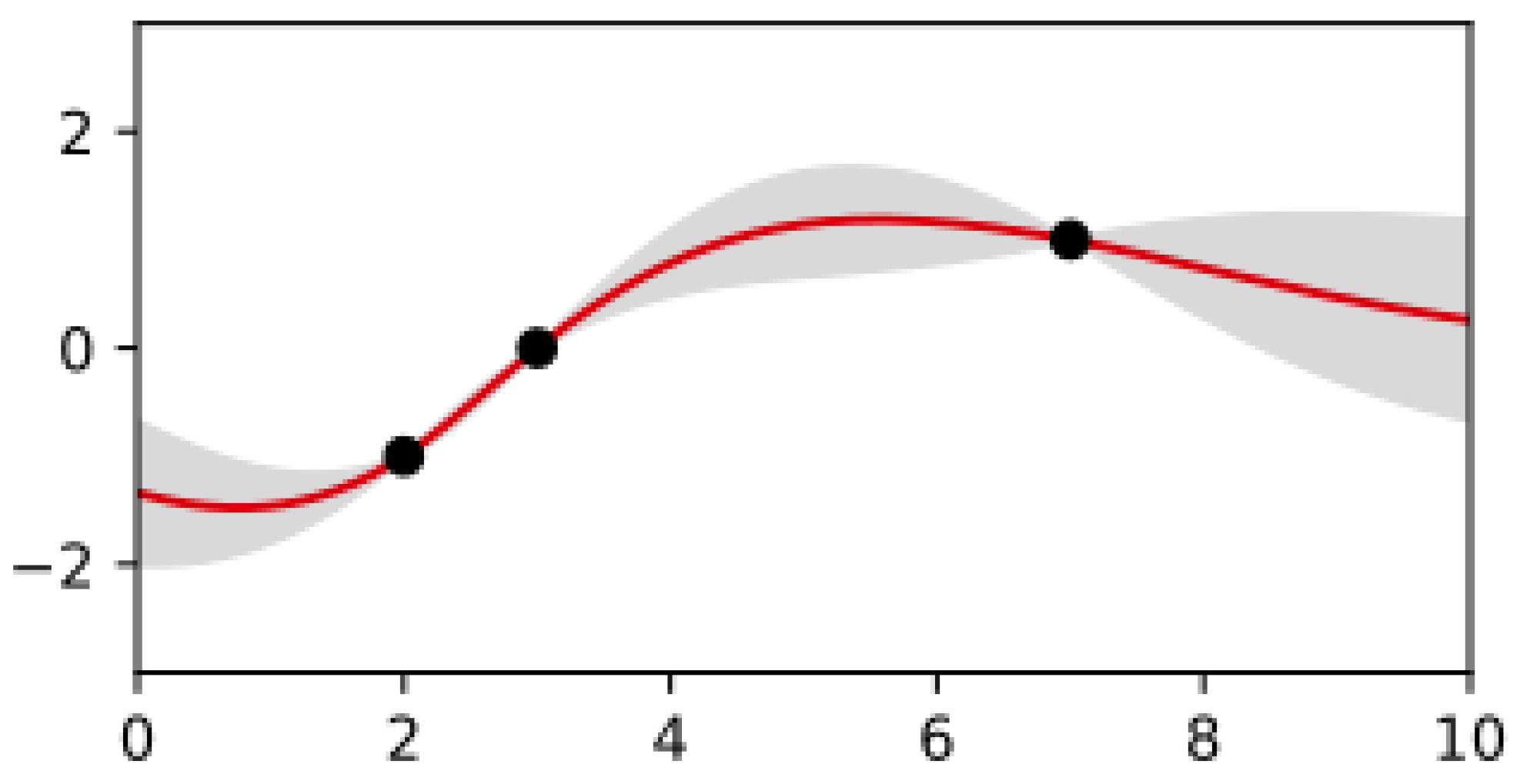
3.1.1. Gaussian Processes
3.1.2. Bayesian Optimization
3.2. Simulation Interface and Datasets
3.2.1. Simulation Interface
3.2.2. Datasets
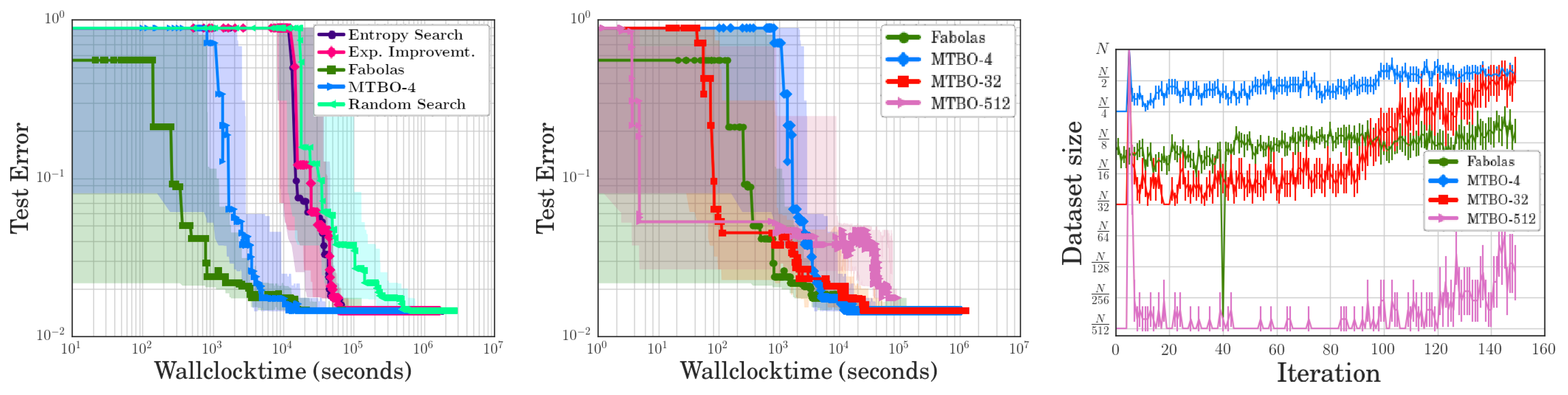
3.2.3. Evaluation
- Random Search: Simple random hyperparameter search. Each configuration is evaluated on the full dataset.
- Entropy Search & Expected Improvement: Bayesian optimization methods always evaluate the full dataset. Expected Improvement uses an acquisition function that simply samples at the current expected optimum. Entropy Search uses an acquisition function similar to the one used by Fabolas but without the cost model.
- MTBO-N (Multi-Task Bayesian Optimization [61]): Like Fabolas but restricts samples to two sizes , i.e., either a small subsample or the entire dataset is used. Multiple values for N are evaluated: 4, 32, and 512.
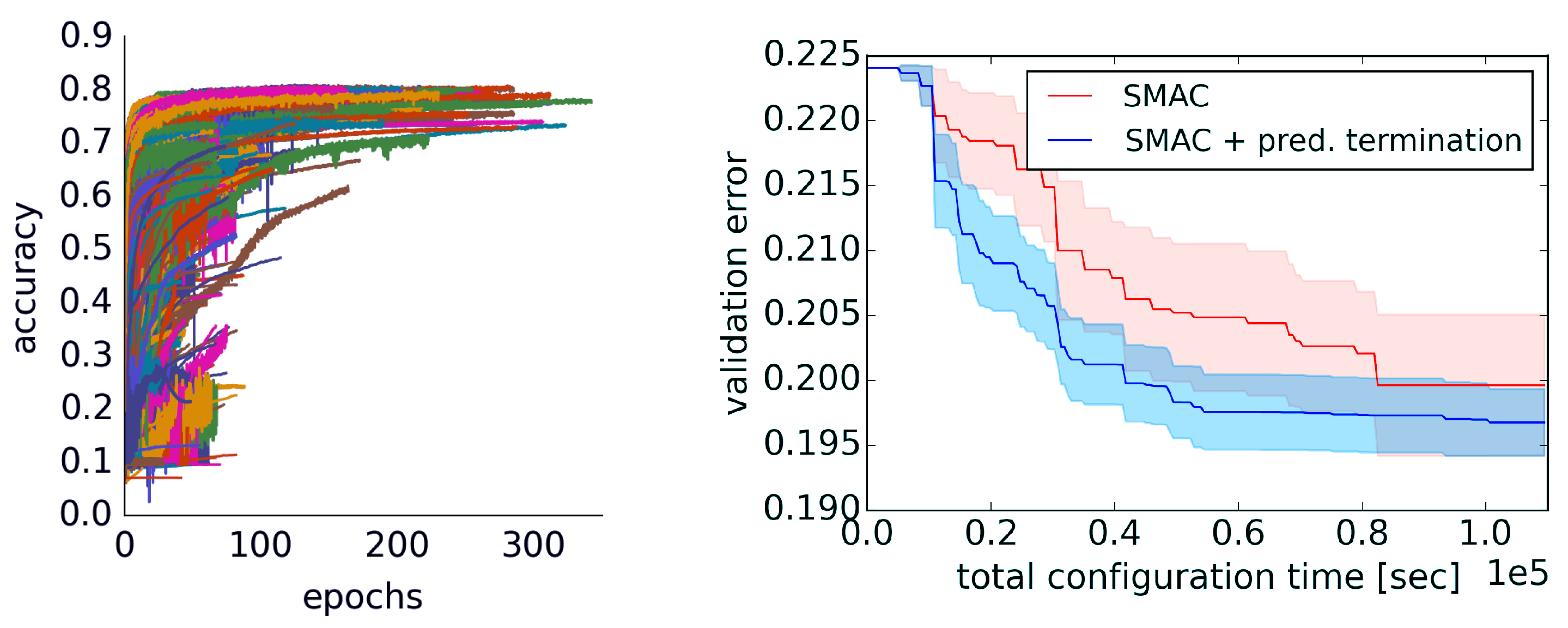
3.3. Learning Curve Extrapolation
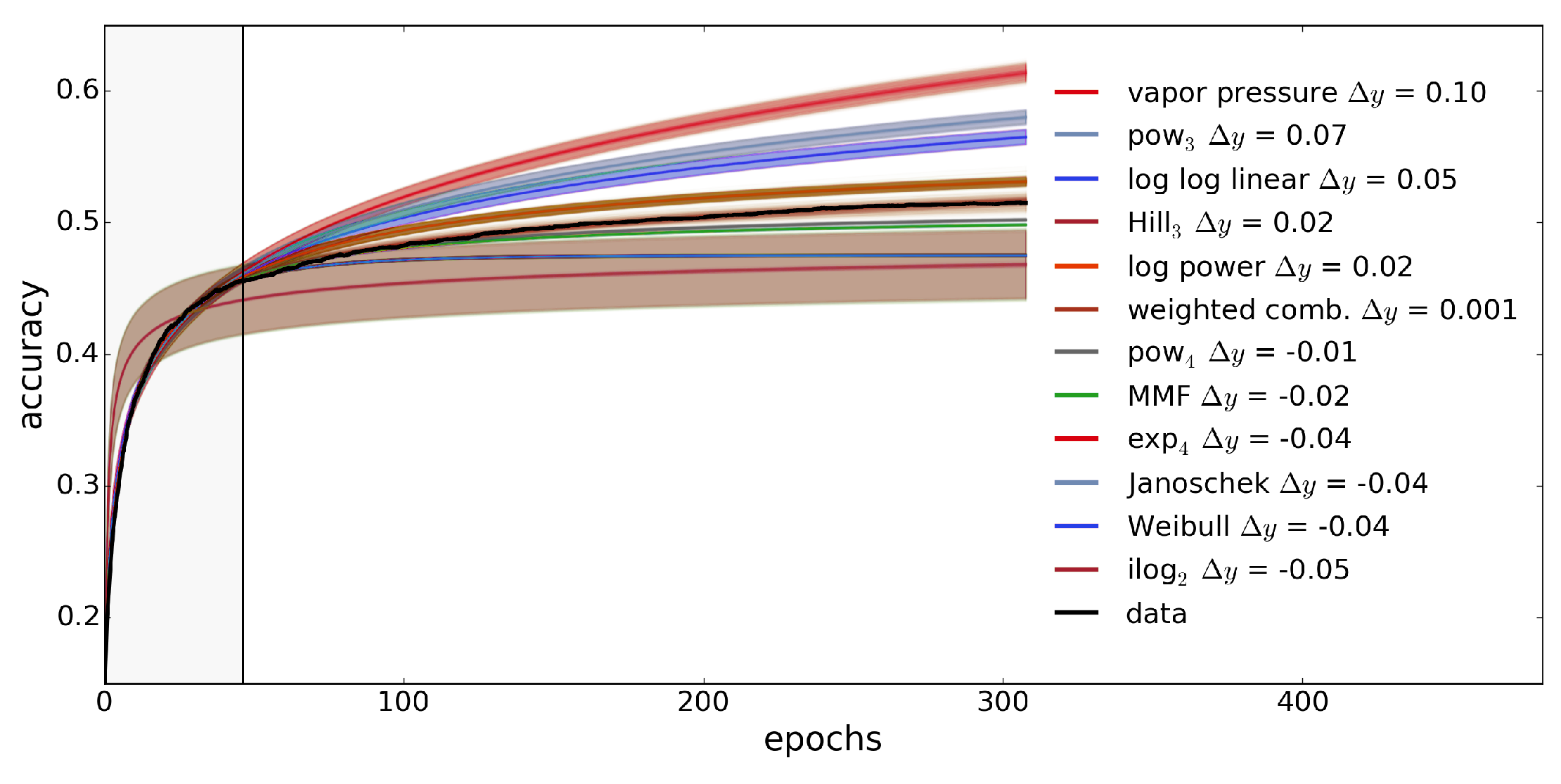
3.3.1. Extrapolation Method
| Algorithm 1: Extrapolation Method. |
|
3.3.2. Evaluation
3.4. Fine-Tuning
| Algorithm 2: Extrapolation Method Optimized. |
|
| Algorithm 3: Extrapolation Method Optimized (Parallel). |
|
| Algorithm 4: Gradient Descent. |
|
| Algorithm 5: Adaptive Stochastic Gradient Descent. |
|
| Listing 1: PySpark Linear Regression Cross-Validation. |
| import pyspark.ml.tuning.CrossValidator import pyspark.ml.evaluation.RegressionEvaluator import pyspark.ml.regression.LinearRegression import pyspark.ml.feature.VectorAssembler |
| df = spark.read.csv("path/to/data.csv", header=True) assembler = VectorAssembler(inputCols=["col1", "col2", "col3"], outputCol="features") |
| lr = LinearRegression() |
| paramGrid = ParamGridBuilder() .addGrid(lr.regParam, [0.1, 0.01, 0.001]) .addGrid(lr.fitIntercept, [False, True]) .build() |
| cv = CrossValidator(estimator = lr, estimatorParamMaps = paramGrid, evaluator = RegressionEvaluator(), numFolds = 5) |
| cvModel = cv.fit(df) |
| Listing 2: Random Grid Search for Logistic Regression. |
| import pyspark.ml.tuning.RandomGridSearch |
| model = LogisticRegression() |
| paramGrid = RandomGridSearch(model.regParam, [0.1, 0.01, 0.001]) .add(model.elasticNetParam, [0.0, 0.5, 1.0]) |
| evaluator = BinaryClassificationEvaluator() |
| cv = CrossValidator(estimator = model, estimatorParamMaps = paramGrid, evaluator = evaluator, numFolds = 5), cvModel = cv.fit(train_data) |
| Listing 3: Bayesian Optimization for Logistic Regression. |
| from pyspark.ml.tuning import BayesianOptimization |
| model = LogisticRegression() |
| paramSpace = {‘regParam’: (0.1,0.01),‘elasticNetParam’: (0.0,1.0)} |
| evaluator = BinaryClassificationEvaluator() |
| bo = BayesianOptimization(estimator = model, paramSpace = paramSpace, evaluator = evaluator, maxIter = 10) |
| boModel = bo.fit(train_data) |
| return boModel |
4. Optimizing Training
- A general-purpose approach that fuses bootstrapping with subsampling.
- A technique that iteratively chooses the best subsample size for gradient descent.
- Weighting the samples will enhance the quality of the logistic regression subsampling.
- Through k-means clustering, accelerating the training of SVMs.
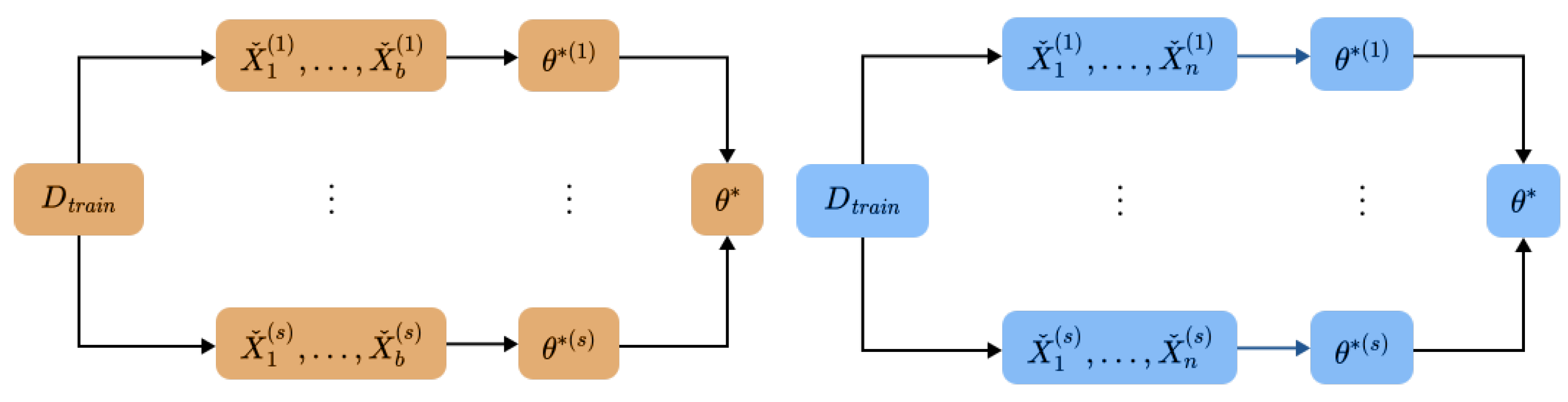
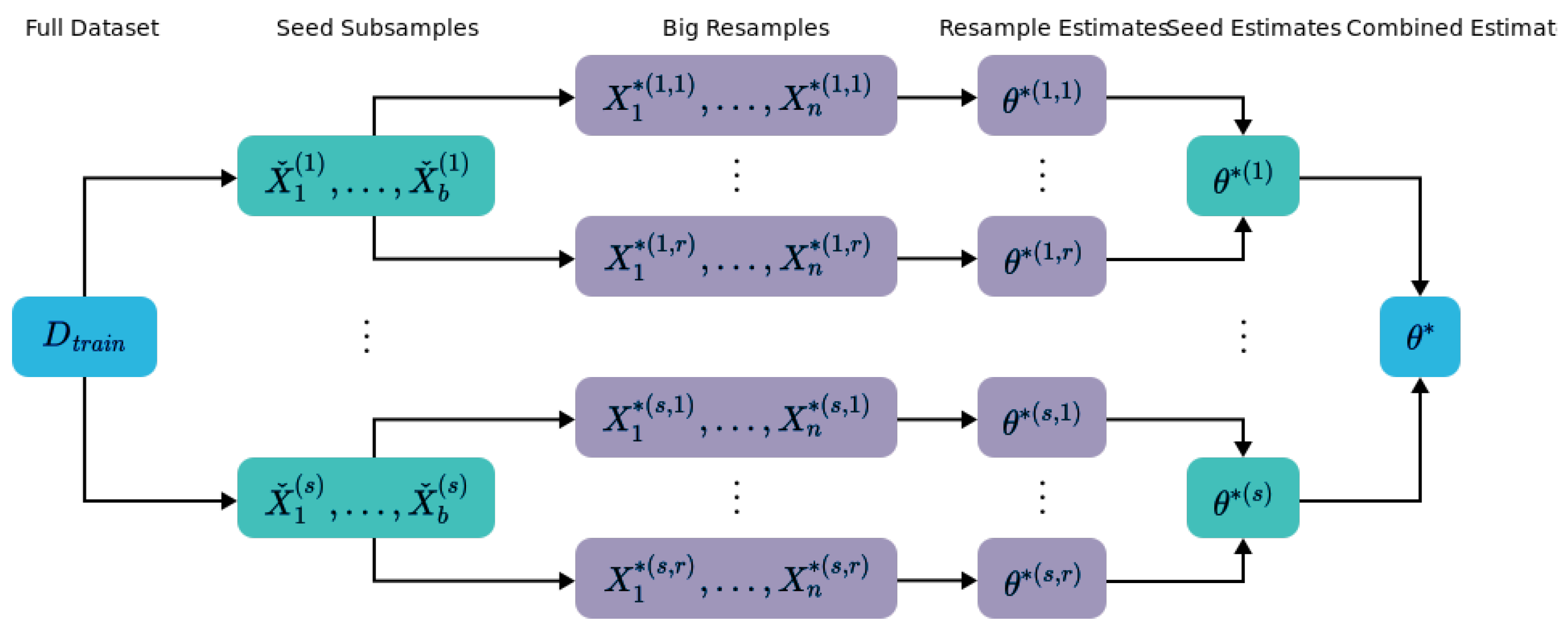
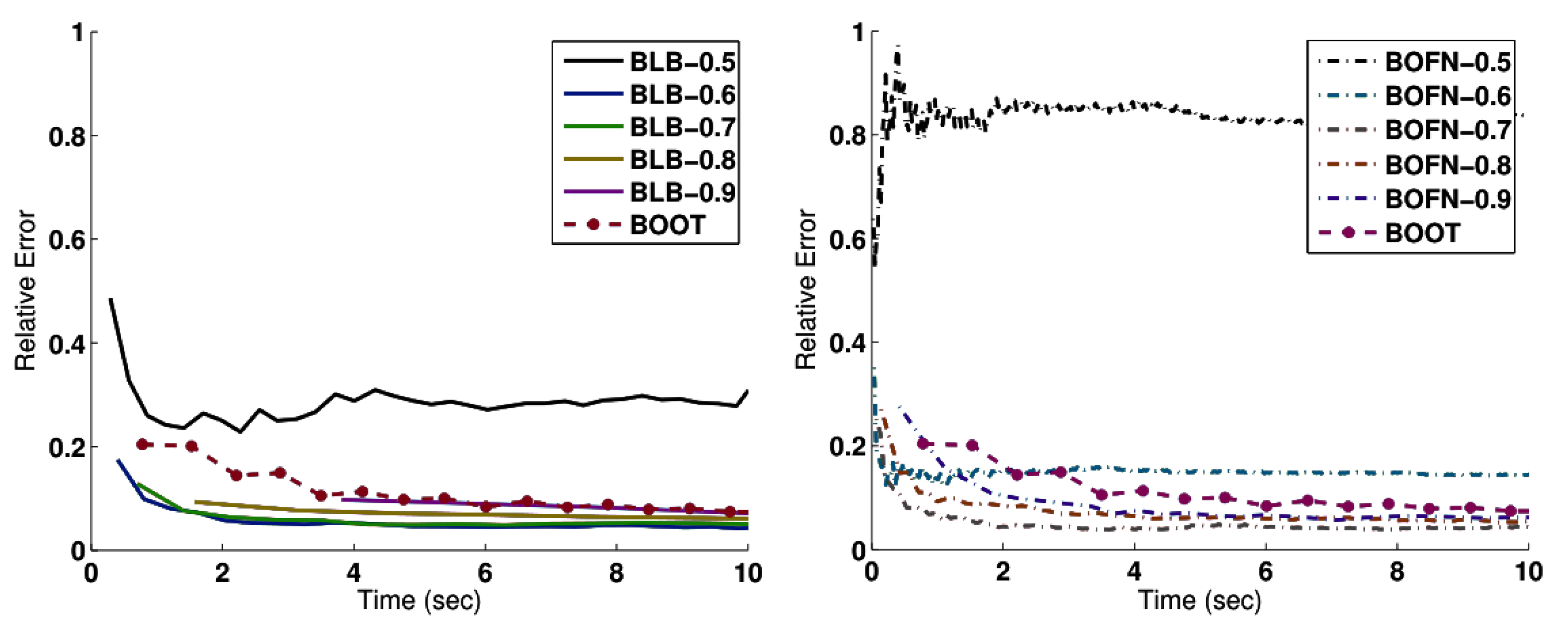
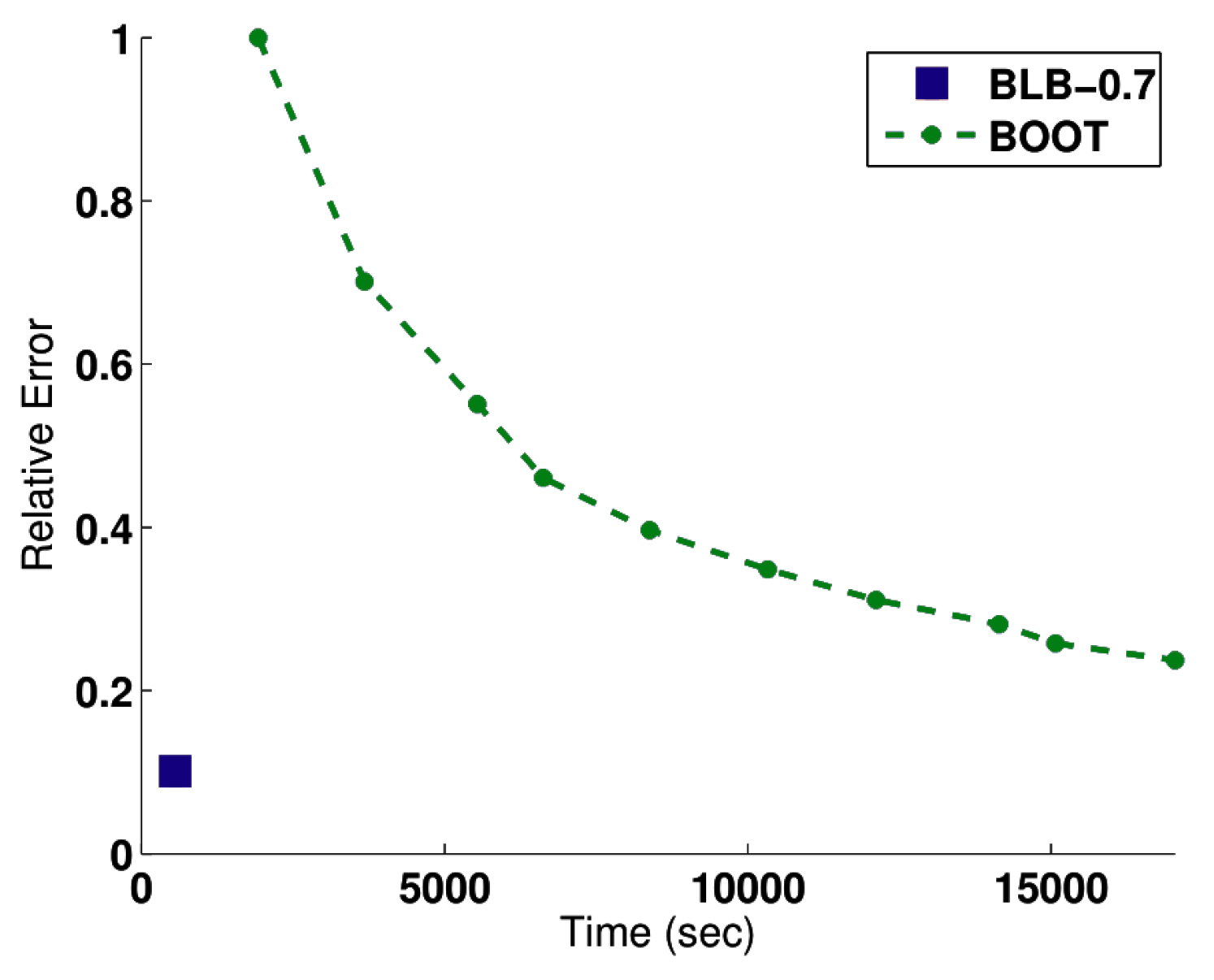
4.1. Bag of Little Bootstraps
4.1.1. Intuition
4.1.2. Evaluation
4.2. Subsample Size Selection for Gradient Descent
- In the stochastic approximation regime small samples, typically , are used. This causes fast but noisy steps.
- In the batch regime large samples are used, typically with . Steps are expensive to compute but more reliable.
4.2.1. Size Selection Method
4.2.2. Evaluation
4.3. Subsampling for Logistic Regression
4.3.1. Case Control
4.3.2. Local Case Control
4.3.3. OSMAC
4.3.4. Evaluation
- mzNormal: Uses a multivariate normal distribution with mean 0 and . Contains a roughly equal amount of positive and negative samples.
- nzNormal: Uses a multivariate normal distribution with mean . About 95% of the samples are positive.
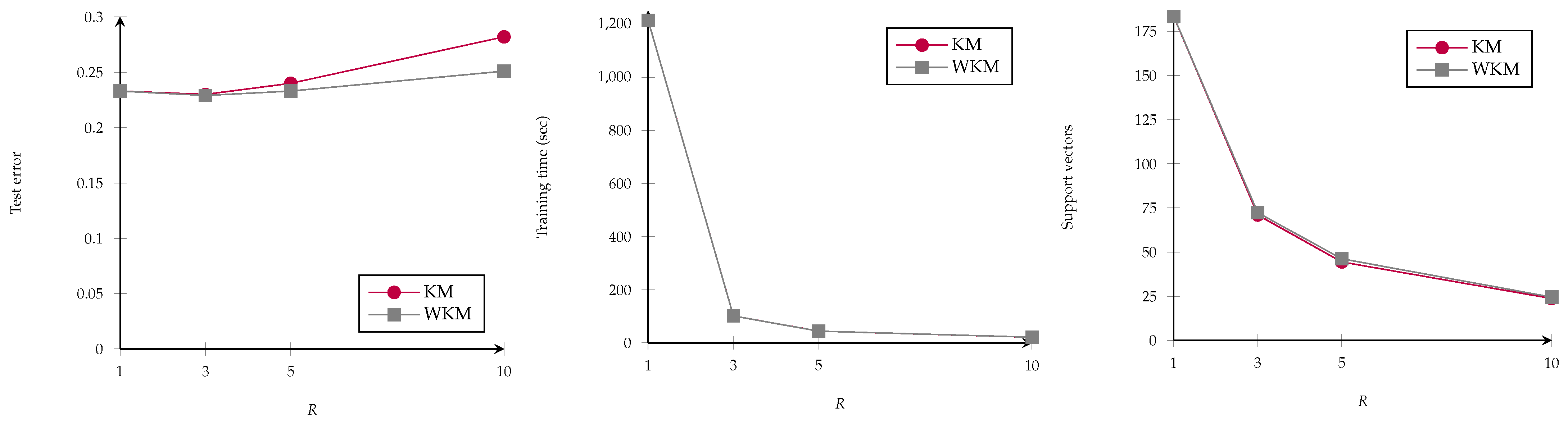
4.4. Clustering for SVMs
- Group the training samples into k clusters with centers where k should be determined via hyperparameter optimization.
- Check for each cluster whether all associated data points belong to the same class, i.e., . If yes, all datapoints in are removed from and replaced by . If not, they are kept in the dataset. The intuition behind this is that clusters with points from multiple classes might be near the decision boundary so they are kept to serve as potential support vectors.
- Finally standard SVM training is performed on the reduced training dataset.
4.4.1. Evaluation
5. Discussion
6. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AutoML | Automated Machine Learning |
| SVM | Support Vector Machine |
| MCMC | Markov Chain Monte Carlo |
| NAS | Neural Architecture Search |
| LFE | Learning Feature Engineering |
| CNN | Convolutional Neural Network |
| NASH | Neural Architecture Search by Hillclimbing |
| AMC | Model Compression and Acceleration |
| SMAC | Sequential model-based Algorithm Configuration |
| Fabolas | Fast Bayesian Optimization of Machine Learning Hyperparameters on Large Datasets |
| RV | Random Variables |
| GP | Gaussian Process |
| SE kernel | Squared Exponential kernel |
| SQ-EXP | Squared Exponential |
| UCB | Upper Confidence Bound |
| MTBO | Multi-Task Bayesian Optimization |
| DNNs | Deep Neural Networks |
| SGD | Stochastic Gradient Descent |
| HMC | Hamiltonian Monte Carlo |
| ELBO | Evidence Lower Bound |
| ADAM | Adaptive Moment Estimation Optimizer |
| BLB | Bag of Little Bootstraps |
| BOFN | B out of N Bootstrapping |
| BOOT | Bootstrapping |
| NCG | Newton Conjugate Gradient |
| MSE | Mean Squared Error |
| NCG | Nonlinear Conjugate Gradient |
| CC | Case-Control |
| LCC | Local Case-Control |
| OSMAC | Optimal Subsampling Motivated by the A-Optimality Criterion |
| KM-SVM | K-means support vector machine |
| WKM-SVM | Weighted K-means support vector machine |
| HMM | Hidden Markov Models |
References
- Kang, J.S.; Kang, J.; Kim, J.J.; Jeon, K.W.; Chung, H.J.; Park, B.H. Neural Architecture Search Survey: A Computer Vision Perspective. Sensors 2023, 23, 1713. [Google Scholar] [CrossRef]
- Baymurzina, D.; Golikov, E.; Burtsev, M. A review of neural architecture search. Neurocomputing 2022, 474, 82–93. [Google Scholar] [CrossRef]
- Lindauer, M.; Hutter, F. Best Practices for Scientific Research on Neural Architecture Search. J. Mach. Learn. Res. 2020, 21, 9820–9837. [Google Scholar]
- Jin, H.; Song, Q.; Hu, X. Auto-Keras: An Efficient Neural Architecture Search System. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (KDD ’19), Anchorage, AK, USA, 4–8 August 2019; Association for Computing Machinery: New York, NY, USA, 2019; pp. 1946–1956. [Google Scholar] [CrossRef]
- Figueiredo, E.; Park, G.; Farrar, C.R.; Worden, K.; Figueiras, J. Machine learning algorithms for damage detection under operational and environmental variability. Struct. Health Monit. 2011, 10, 559–572. [Google Scholar] [CrossRef]
- Susto, G.A.; Schirru, A.; Pampuri, S.; McLoone, S.; Beghi, A. Machine learning for predictive maintenance: A multiple classifier approach. IEEE Trans. Ind. Inform. 2014, 11, 812–820. [Google Scholar] [CrossRef]
- Li, H.; Parikh, D.; He, Q.; Qian, B.; Li, Z.; Fang, D.; Hampapur, A. Improving rail network velocity: A machine learning approach to predictive maintenance. Transp. Res. Part Emerg. Technol. 2014, 45, 17–26. [Google Scholar] [CrossRef]
- Stühler, E.; Braune, S.; Lionetto, F.; Heer, Y.; Jules, E.; Westermann, C.; Bergmann, A.; van Hövell, P. Framework for personalized prediction of treatment response in relapsing remitting multiple sclerosis. BMC Med. Res. Methodol. 2020, 20, 24. [Google Scholar] [CrossRef]
- Handzic, M.; Tjandrawibawa, F.; Yeo, J. How neural networks can help loan officers to make better informed application decisions. Informing Sci. 2003, 6, 97–109. [Google Scholar]
- Viaene, S.; Dedene, G.; Derrig, R.A. Auto claim fraud detection using Bayesian learning neural networks. Expert Syst. Appl. 2005, 29, 653–666. [Google Scholar] [CrossRef]
- Pérez, J.M.; Muguerza, J.; Arbelaitz, O.; Gurrutxaga, I.; Martín, J.I. Consolidated tree classifier learning in a car insurance fraud detection domain with class imbalance. In Proceedings of the International Conference on Pattern Recognition and Image Analysis, Bath, UK, 23–25 August 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 381–389. [Google Scholar]
- Tsoumakas, G. A survey of machine learning techniques for food sales prediction. Artif. Intell. Rev. 2019, 52, 441–447. [Google Scholar] [CrossRef]
- Karras, C.; Karras, A.; Tsolis, D.; Avlonitis, M.; Sioutas, S. A Hybrid Ensemble Deep Learning Approach for Emotion Classification. In Proceedings of the 2022 IEEE International Conference on Big Data (Big Data), Atlanta, GA, USA, 17–20 December 2022; pp. 3881–3890. [Google Scholar] [CrossRef]
- Li, L.; Jamieson, K.; DeSalvo, G.; Rostamizadeh, A.; Talwalkar, A. Hyperband: A novel bandit-based approach to hyperparameter optimization. J. Mach. Learn. Res. 2017, 18, 6765–6816. [Google Scholar]
- Duan, J.; Zeng, Z.; Oprea, A.; Vasudevan, S. Automated generation and selection of interpretable features for enterprise security. In Proceedings of the 2018 IEEE International Conference on Big Data (Big Data), Seattle, WA, USA, 10–13 December 2018; pp. 1258–1265. [Google Scholar]
- Andrychowicz, M.; Denil, M.; Gómez, S.; Hoffman, M.W.; Pfau, D.; Schaul, T.; Shillingford, B.; de Freitas, N. Learning to learn by gradient descent by gradient descent. In Advances in Neural Information Processing Systems; Lee, D., Sugiyama, M., Luxburg, U., Guyon, I., Garnett, R., Eds.; Curran Associates, Inc.: New York, NY, USA, 2016; Volume 29. [Google Scholar]
- Zoph, B.; Le, Q.V. Neural architecture search with reinforcement learning. arXiv 2016, arXiv:1611.01578. [Google Scholar]
- Feurer, M.; Klein, A.; Eggensperger, K.; Springenberg, J.; Blum, M.; Hutter, F. Efficient and robust automated machine learning. Adv. Neural Inf. Process. Syst. 2015, 28. [Google Scholar]
- Gaudel, R.; Sebag, M. Feature selection as a one-player game. In Proceedings of the International Conference on Machine Learning, Haifa, Israel, 21–25 June 2010; pp. 359–366. [Google Scholar]
- Katz, G.; Shin, E.C.R.; Song, D. Explorekit: Automatic feature generation and selection. In Proceedings of the 2016 IEEE 16th International Conference on Data Mining (ICDM), Barcelona, Spain, 12–15 December 2016; pp. 979–984. [Google Scholar]
- Nargesian, F.; Samulowitz, H.; Khurana, U.; Khalil, E.B.; Turaga, D.S. Learning Feature Engineering for Classification. In Proceedings of the IJCAI, Melbourne, Australia, 19–25 August 2017; pp. 2529–2535. [Google Scholar]
- Kaul, A.; Maheshwary, S.; Pudi, V. Autolearn—Automated feature generation and selection. In Proceedings of the 2017 IEEE International Conference on data mining (ICDM), New Orleans, LA, USA, 18–21 November 2017; pp. 217–226. [Google Scholar]
- Meinshausen, N.; Bühlmann, P. Stability selection. J. R. Stat. Soc. Ser. (Stat. Methodol.) 2010, 72, 417–473. [Google Scholar] [CrossRef]
- Pfahringer, B.; Bensusan, H.; Giraud-Carrier, C.G. Meta-Learning by Landmarking Various Learning Algorithms. In Proceedings of the ICML, Stanford, CA, USA, 29 June–2 July 2000; pp. 743–750. [Google Scholar]
- Klein, A.; Falkner, S.; Springenberg, J.T.; Hutter, F. Learning Curve Prediction with Bayesian Neural Networks. In Proceedings of the ICLR, Toulon, France, 24–26 April 2017. [Google Scholar]
- Eggensperger, K.; Lindauer, M.; Hutter, F. Neural networks for predicting algorithm runtime distributions. arXiv 2017, arXiv:1709.07615. [Google Scholar]
- Brazdil, P.B.; Soares, C. A comparison of ranking methods for classification algorithm selection. In Proceedings of the European Conference on Machine Learning, Barcelona, Spain, 31 May–2 June 2000; Springer: Berlin/Heidelberg, Germany, 2000; pp. 63–75. [Google Scholar]
- Andrychowicz, M.; Denil, M.; Gomez, S.; Hoffman, M.W.; Pfau, D.; Schaul, T.; Shillingford, B.; De Freitas, N. Learning to learn by gradient descent by gradient descent. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Graves, A. Long short-term memory. In Supervised Sequence Labelling with Recurrent Neural Networks; Springer: Berlin/Heidelberg, Germany, 2012; pp. 37–45. [Google Scholar]
- Chen, Y.; Hoffman, M.W.; Colmenarejo, S.G.; Denil, M.; Lillicrap, T.P.; Botvinick, M.; Freitas, N. Learning to learn without gradient descent by gradient descent. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 748–756. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Elsken, T.; Metzen, J.H.; Hutter, F. Simple and efficient architecture search for convolutional neural networks. arXiv 2017, arXiv:1711.04528. [Google Scholar]
- Real, E.; Moore, S.; Selle, A.; Saxena, S.; Suematsu, Y.L.; Tan, J.; Le, Q.V.; Kurakin, A. Large-scale evolution of image classifiers. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 2902–2911. [Google Scholar]
- He, Y.; Lin, J.; Liu, Z.; Wang, H.; Li, L.J.; Han, S. Amc: Automl for model compression and acceleration on mobile devices. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 784–800. [Google Scholar]
- Guyon, I.; Sun-Hosoya, L.; Boullé, M.; Escalante, H.J.; Escalera, S.; Liu, Z.; Jajetic, D.; Ray, B.; Saeed, M.; Sebag, M.; et al. Analysis of the automl challenge series. Autom. Mach. Learn. 2019, 177–219. [Google Scholar] [CrossRef]
- Brochu, E.; Cora, V.M.; De Freitas, N. A tutorial on Bayesian optimization of expensive cost functions, with application to active user modeling and hierarchical reinforcement learning. arXiv 2010, arXiv:1012.2599. [Google Scholar]
- Hutter, F.; Hoos, H.H.; Leyton-Brown, K. Sequential model-based optimization for general algorithm configuration. In Proceedings of the International Conference on Learning and Intelligent Optimization, Rome, Italy, 17–21 January 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 507–523. [Google Scholar]
- Feurer, M.; Springenberg, J.; Hutter, F. Initializing Bayesian Hyperparameter Optimization via Meta-Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015; Volume 29. [Google Scholar] [CrossRef]
- Jamieson, K.; Talwalkar, A. Non-stochastic best arm identification and hyperparameter optimization. In Proceedings of the Artificial Intelligence and Statistics, PMLR, Cadiz, Spain, 9–11 May 2016; pp. 240–248. [Google Scholar]
- Jaderberg, M.; Dalibard, V.; Osindero, S.; Czarnecki, W.M.; Donahue, J.; Razavi, A.; Vinyals, O.; Green, T.; Dunning, I.; Simonyan, K.; et al. Population based training of neural networks. arXiv 2017, arXiv:1711.09846. [Google Scholar]
- Maclaurin, D.; Duvenaud, D.; Adams, R. Gradient-based hyperparameter optimization through reversible learning. In Proceedings of the International Conference on Machine Learning, PMLR, Lille, France, 6–11 July 2015; pp. 2113–2122. [Google Scholar]
- Zacharia, A.; Zacharia, D.; Karras, A.; Karras, C.; Giannoukou, I.; Giotopoulos, K.C.; Sioutas, S. An Intelligent Microprocessor Integrating TinyML in Smart Hotels for Rapid Accident Prevention. In Proceedings of the 2022 7th South-East Europe Design Automation, Computer Engineering, Computer Networks and Social Media Conference (SEEDA-CECNSM), Ioannina, Greece, 23–25 September 2022; pp. 1–7. [Google Scholar] [CrossRef]
- Schizas, N.; Karras, A.; Karras, C.; Sioutas, S. TinyML for Ultra-Low Power AI and Large Scale IoT Deployments: A Systematic Review. Future Internet 2022, 14, 363. [Google Scholar] [CrossRef]
- Nagarajah, T.; Poravi, G. A Review on Automated Machine Learning (AutoML) Systems. In Proceedings of the 2019 IEEE 5th International Conference for Convergence in Technology (I2CT), Bombay, India, 29–31 March 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Bahri, M.; Salutari, F.; Putina, A.; Sozio, M. Automl: State of the art with a focus on anomaly detection, challenges, and research directions. Int. J. Data Sci. Anal. 2022, 14, 113–126. [Google Scholar] [CrossRef]
- Remeseiro, B.; Bolon-Canedo, V. A review of feature selection methods in medical applications. Comput. Biol. Med. 2019, 112, 103375. [Google Scholar] [CrossRef] [PubMed]
- Isabona, J.; Imoize, A.L.; Kim, Y. Machine Learning-Based Boosted Regression Ensemble Combined with Hyperparameter Tuning for Optimal Adaptive Learning. Sensors 2022, 22, 3776. [Google Scholar] [CrossRef]
- Guo, P.; Yang, D.; Hatamizadeh, A.; Xu, A.; Xu, Z.; Li, W.; Zhao, C.; Xu, D.; Harmon, S.; Turkbey, E.; et al. Auto-FedRL: Federated Hyperparameter Optimization for Multi-institutional Medical Image Segmentation. In Proceedings of the Computer Vision—ECCV 2022, Tel Aviv, Israel, 23–27 October 2022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2022; pp. 437–455. [Google Scholar]
- Li, Y.; Shen, Y.; Jiang, H.; Zhang, W.; Li, J.; Liu, J.; Zhang, C.; Cui, B. Hyper-Tune: Towards Efficient Hyper-parameter Tuning at Scale. arXiv 2022, arXiv:2201.06834. [Google Scholar] [CrossRef]
- Passos, D.; Mishra, P. A tutorial on automatic hyperparameter tuning of deep spectral modelling for regression and classification tasks. Chemom. Intell. Lab. Syst. 2022, 223, 104520. [Google Scholar] [CrossRef]
- Yu, T.; Zhu, H. Hyper-parameter optimization: A review of algorithms and applications. arXiv 2020, arXiv:2003.05689. [Google Scholar]
- Bischl, B.; Binder, M.; Lang, M.; Pielok, T.; Richter, J.; Coors, S.; Thomas, J.; Ullmann, T.; Becker, M.; Boulesteix, A.L.; et al. Hyperparameter optimization: Foundations, algorithms, best practices, and open challenges. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2021, 13, e1484. [Google Scholar] [CrossRef]
- Sipper, M. High Per Parameter: A Large-Scale Study of Hyperparameter Tuning for Machine Learning Algorithms. Algorithms 2022, 15, 315. [Google Scholar] [CrossRef]
- Giotopoulos, K.C.; Michalopoulos, D.; Karras, A.; Karras, C.; Sioutas, S. Modelling and Analysis of Neuro Fuzzy Employee Ranking System in the Public Sector. Algorithms 2023, 16, 151. [Google Scholar] [CrossRef]
- Klein, A.; Falkner, S.; Bartels, S.; Hennig, P.; Hutter, F. Fast Bayesian Optimization of Machine Learning Hyperparameters on Large Datasets. In Proceedings of the 20th International Conference on Artificial Intelligence and Statistics, Ft. Lauderdale, FL, USA, 20–22 April 2017; Singh, A., Zhu, J., Eds.; PMLR: Fort Lauderdale, FL, USA, 2017; Volume 54, pp. 528–536. [Google Scholar]
- Schön, S.; Kermarrec, G.; Kargoll, B.; Neumann, I.; Kosheleva, O.; Kreinovich, V. Why Student Distributions? Why Matern’s Covariance Model? A Symmetry-Based Explanation. In Econometrics for Financial Applications; Springer International Publishing: Berlin/Heidelberg, Germany, 2017; pp. 266–275. [Google Scholar] [CrossRef]
- Karras, C.; Karras, A.; Avlonitis, M.; Sioutas, S. An Overview of MCMC Methods: From Theory to Applications. In Proceedings of the Artificial Intelligence Applications and Innovations. AIAI 2022 IFIP WG 12.5 International Workshops, Crete, Greece, 17–20 June 2022; Maglogiannis, I., Iliadis, L., Macintyre, J., Cortez, P., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 319–332. [Google Scholar]
- Karras, C.; Karras, A.; Tsolis, D.; Giotopoulos, K.C.; Sioutas, S. Distributed Gibbs Sampling and LDA Modelling for Large Scale Big Data Management on PySpark. In Proceedings of the 2022 7th South-East Europe Design Automation, Computer Engineering, Computer Networks and Social Media Conference (SEEDA-CECNSM), Ioannina, Greece, 23–25 September 2022; pp. 1–8. [Google Scholar] [CrossRef]
- Karras, C.; Karras, A.; Avlonitis, M.; Giannoukou, I.; Sioutas, S. Maximum Likelihood Estimators on MCMC Sampling Algorithms for Decision Making. In Proceedings of the Artificial Intelligence Applications and Innovations. AIAI 2022 IFIP WG 12.5 International Workshops, Crete, Greece, 17–20 June 2022; Maglogiannis, I., Iliadis, L., Macintyre, J., Cortez, P., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 345–356. [Google Scholar]
- Swersky, K.; Snoek, J.; Adams, R.P. Multi-task Bayesian Optimization. In Advances in Neural Information Processing Systems; NIPS’13; Curran Associates Inc.: New York, NY, USA, 2013; pp. 2004–2012. [Google Scholar]
- Domhan, T.; Springenberg, J.T.; Hutter, F. Speeding Up Automatic Hyperparameter Optimization of Deep Neural Networks by Extrapolation of Learning Curves. In Proceedings of the 24th International Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015; pp. 3460–3468. [Google Scholar]
- Kleiner, A.; Talwalkar, A.; Sarkar, P.; Jordan, M.I. A Scalable Bootstrap for Massive Data. J. R. Stat. Soc. Ser. (Stat. Methodol.) 2014, 76, 795–816. [Google Scholar] [CrossRef]
- Norazan, M.; Habshah, M.; Imon, A.; Chen, S. Weighted bootstrap with probability in regression. In WSEAS International Conference. Proceedings. Mathematics and Computers in Science and Engineering; World Scientific and Engineering Academy and Society: South Wales, Australia, 2009; Volume 8, p. 16. [Google Scholar]
- Bickel, P.J.; Götze, F.; van Zwet, W.R. Resampling fewer than n observations: Gains, losses, and remedies for losses. Stat. Sin. 1997, 7, 1–31. [Google Scholar]
- Byrd, R.H.; Chin, G.M.; Nocedal, J.; Wu, Y. Sample size selection in optimization methods for machine learning. Math. Program. 2012, 134, 127–155. [Google Scholar] [CrossRef]
- Fithian, W.; Hastie, T. Local case-control sampling: Efficient subsampling in imbalanced data sets. Ann. Stat. 2014, 42, 1693. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. More efficient estimation for logistic regression with optimal subsamples. J. Mach. Learn. Res. 2019, 20, 1–59. [Google Scholar]
- Wang, H.; Zhu, R.; Ma, P. Optimal Subsampling for Large Sample Logistic Regression. J. Am. Stat. Assoc. 2018, 113, 829–844. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, M.B.; de Pádua Braga, A.; Braga, J.P. SVM-KM: Speeding SVMs learning with a priori cluster selection and k-means. In Proceedings of the Vol. 1. Sixth Brazilian Symposium on Neural Networks, Rio de Janeiro, Brazil, 25 November 2000; pp. 162–167. [Google Scholar]
- Lee, S.J.; Park, C.; Jhun, M.; Ko, J.Y. Support vector machine using K-means clustering. J. Korean Stat. Soc. 2007, 36, 175–182. [Google Scholar]
- Bang, S.; Jhun, M. Weighted Support Vector Machine Using k-Means Clustering. Commun. Stat.-Simul. Comput. 2014, 43, 2307–2324. [Google Scholar] [CrossRef]
- Leng, L.; Li, M.; Kim, C.; Bi, X. Dual-source discrimination power analysis for multi-instance contactless palmprint recognition. Multimed. Tools Appl. 2017, 76, 333–354. [Google Scholar] [CrossRef]
- Leng, L.; Li, M.; Teoh, A.B.J. Conjugate 2DPalmHash code for secure palm-print-vein verification. In Proceedings of the 2013 6th International congress on image and signal processing (CISP), Hangzhou, China, 16–18 December 2013; Volume 3, pp. 1705–1710. [Google Scholar]
- Leng, L.; Zhang, J. Palmhash code vs. palmphasor code. Neurocomputing 2013, 108, 1–12. [Google Scholar] [CrossRef]



| CPU | Memory | Programming Language | Operating System |
|---|---|---|---|
| i9-10850k | 32GB | Python 3.10 | Windows 11 |
| Dataset | Evaluation for Method | No. of Samples |
|---|---|---|
| CIFAR-10 | Fabolas | 60,000 |
| MNIST | Fabolas | 70,000 |
| Randomly Generated | BLB | 20,000 |
| Randomly Generated | OSMAC | 10,000 |
| PimaIndiansDiabetes2 | KM-SVM and WKM-SVM | 768 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karras, A.; Karras, C.; Schizas, N.; Avlonitis, M.; Sioutas, S. AutoML with Bayesian Optimizations for Big Data Management. Information 2023, 14, 223. https://doi.org/10.3390/info14040223
Karras A, Karras C, Schizas N, Avlonitis M, Sioutas S. AutoML with Bayesian Optimizations for Big Data Management. Information. 2023; 14(4):223. https://doi.org/10.3390/info14040223
Chicago/Turabian StyleKarras, Aristeidis, Christos Karras, Nikolaos Schizas, Markos Avlonitis, and Spyros Sioutas. 2023. "AutoML with Bayesian Optimizations for Big Data Management" Information 14, no. 4: 223. https://doi.org/10.3390/info14040223
APA StyleKarras, A., Karras, C., Schizas, N., Avlonitis, M., & Sioutas, S. (2023). AutoML with Bayesian Optimizations for Big Data Management. Information, 14(4), 223. https://doi.org/10.3390/info14040223









