An Improved Co-Training and Generative Adversarial Network (Diff-CoGAN) for Semi-Supervised Medical Image Segmentation
Abstract
1. Introduction
- (1)
- (2)
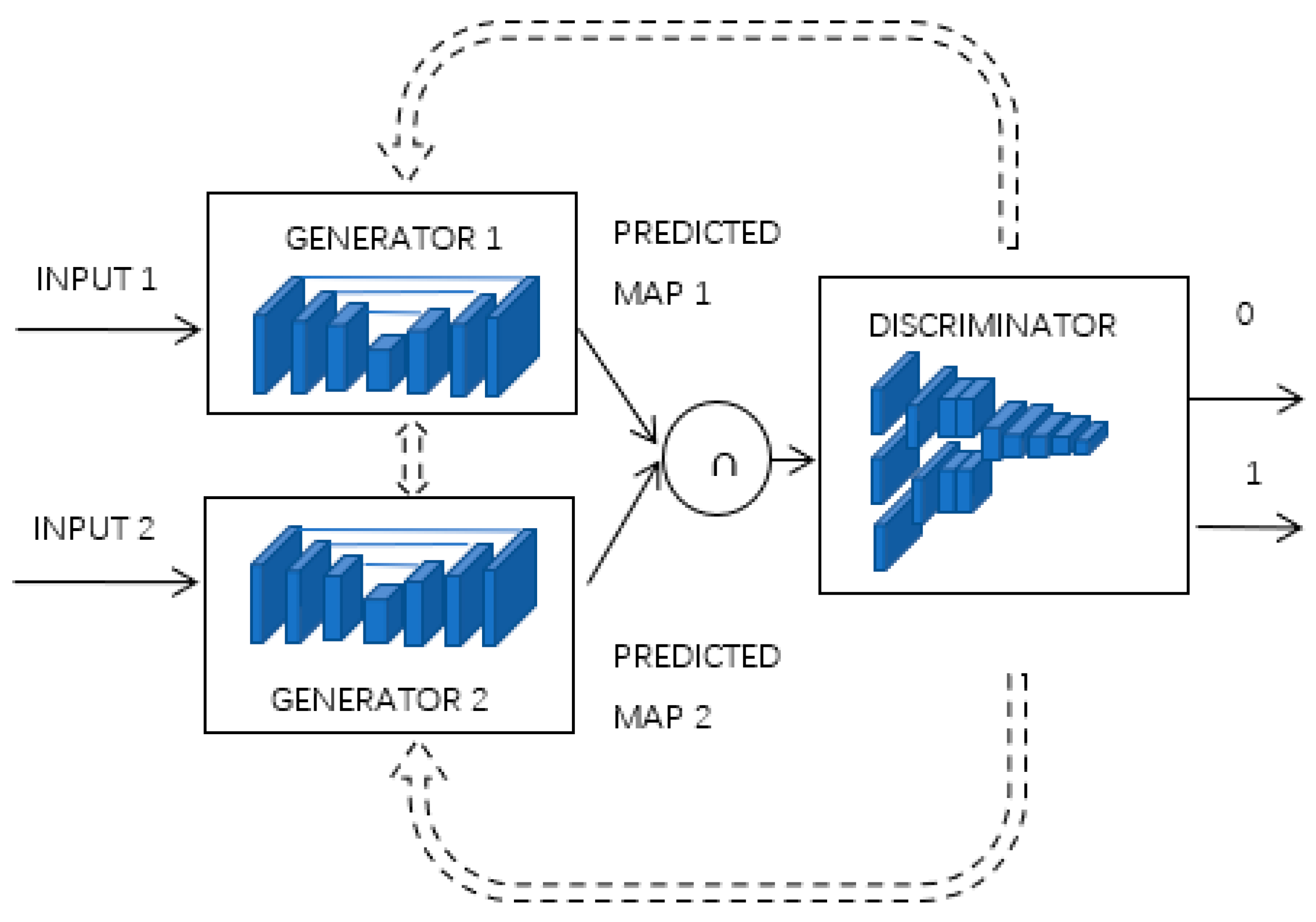
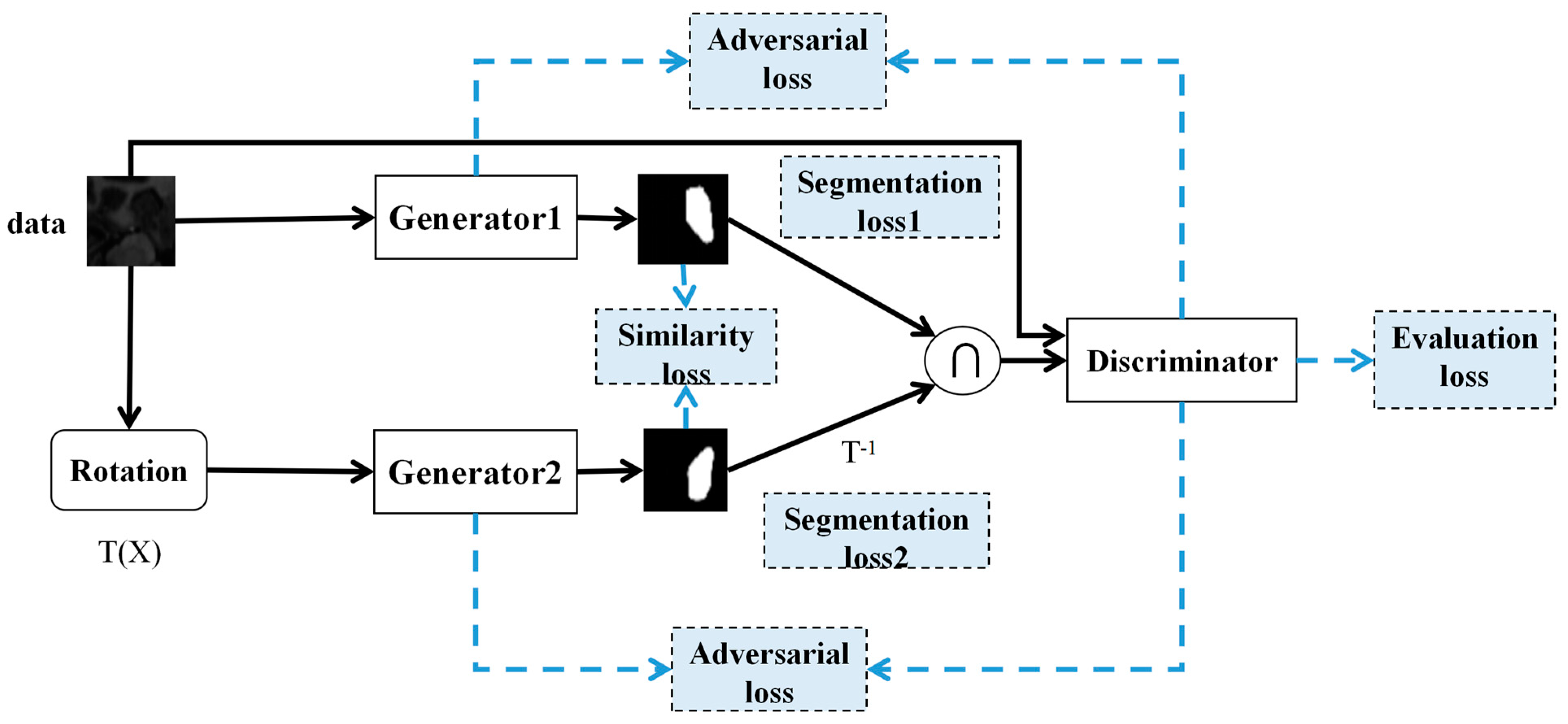
- In Diff-CoGAN, the two generators provide mutual segmentation information, which also supervises each other. In this paper, we introduced the intersection of two predicted maps with high confidence region produced by the outputs of the two generators as the input to the discriminator (see Section 2.3.3). Diff-CoGAN achieves higher segmentation accuracy through adversarial training because the discriminator can guide the generators to generate more accurate predicted maps, and the mutual information guidance of the two generators can also promote the improvement of segmentation performance.
- (3)
- The two generators adopt different networks (see Section 2.3.1 and Section 2.3.2) to increase the diversity of extracted features, consequently providing complementary information for the same data along with the strategy of co-training.
2. Materials and Methods
2.1. Overview of Diff-CoGAN Framework
2.2. Configurations of Diff-CoGAN Framework
2.3. The Network Design of Diff-CoGAN
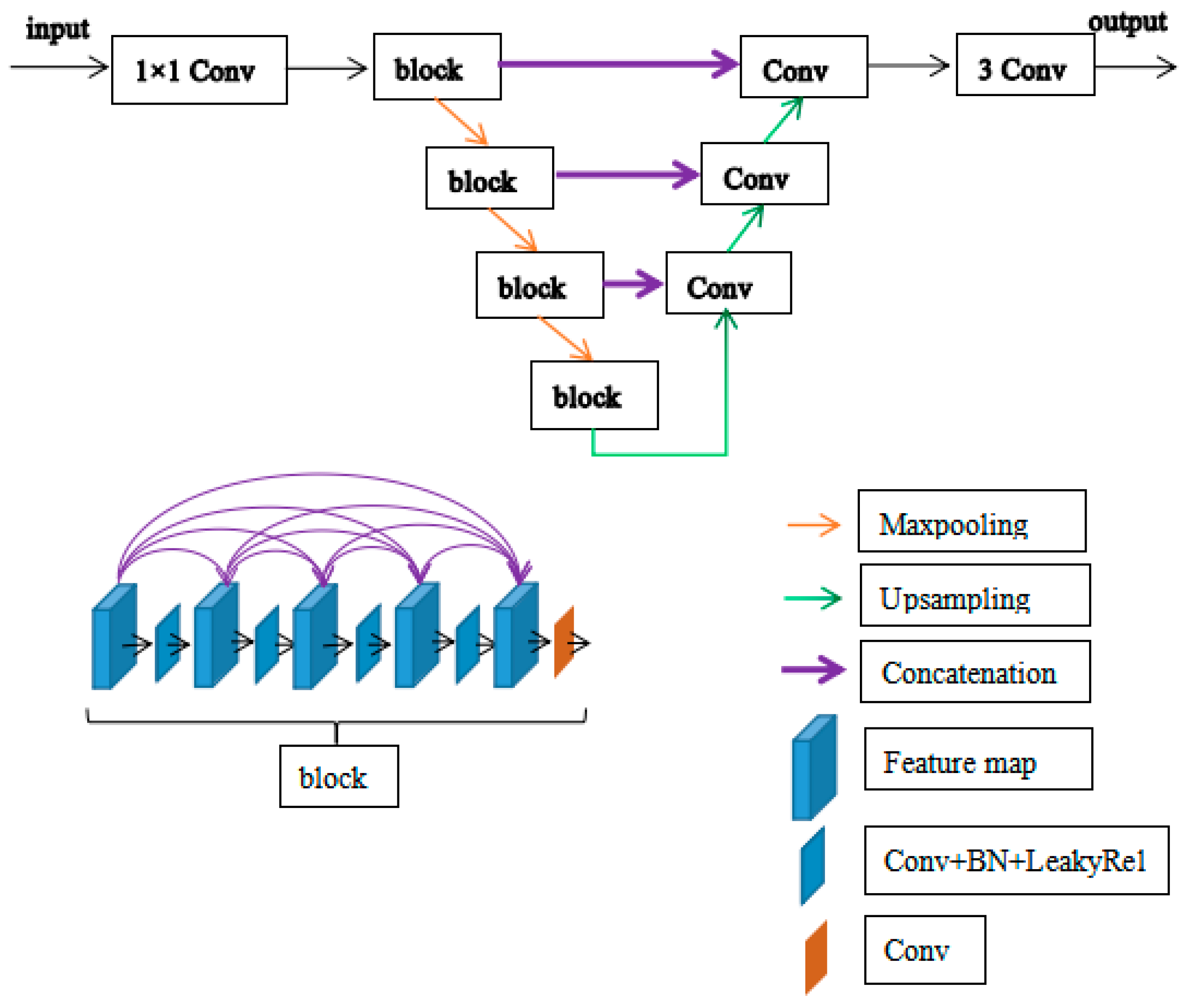
2.3.1. The Network of Generator 1
2.3.2. The Network of Generator 2
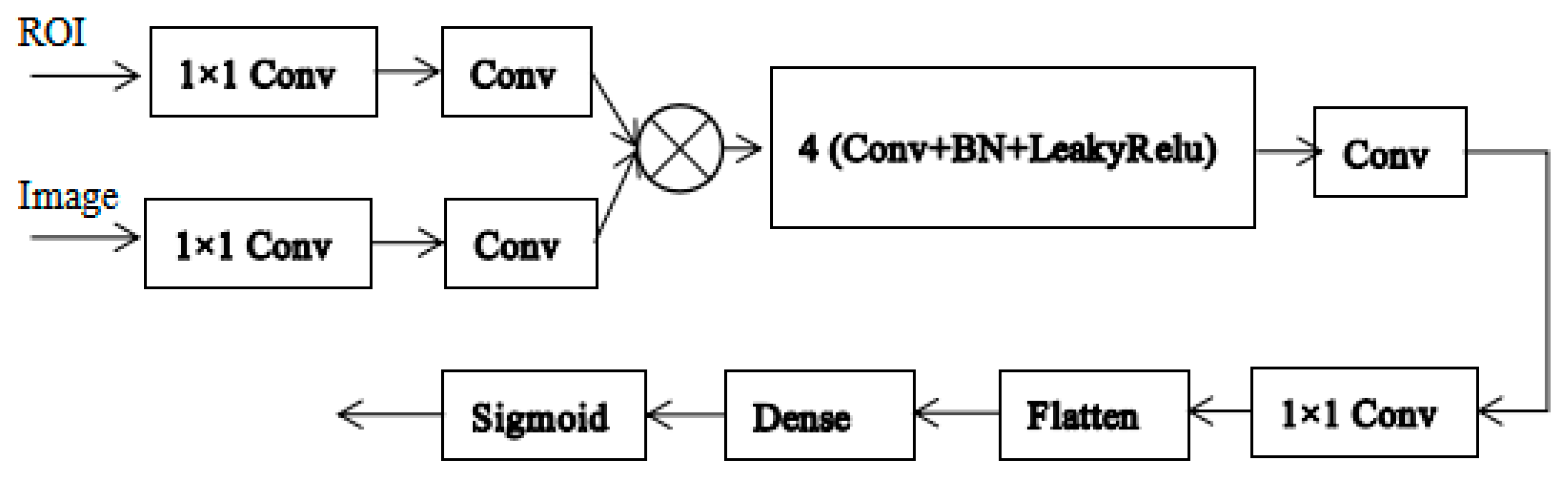
2.3.3. The Network of the Discriminator
2.4. Training Strategy of Diff-CoGAN Framework
2.4.1. Loss Functions
- = labeled data used by Generator 1
- = unlabeled data used by Generator 1
- = labeled data used by Generator 2
- = unlabeled data used by Generator 2
- = ground truth of the labeled data used by Generator 1
- = predicted map for the labeled data generated by Generator 1
- = predicted map for the unlabeled data generated by Generator 1
- = ground truth of the labeled data used by Generator 2
- = predicted map for the labeled data generated by Generator 2
- = predicted map for the unlabeled data generated by Generator 2
2.4.2. Training Settings
- (1)
- The first step was to initialize Generator 1 and Generator 2 using the labeled data only. In this step, the optimizer was stochastic gradient descent (SGD), the epoch was set to 50, and batch_size was set to 8. The training aim was to minimize the loss functions, and .
- (2)
- In the second step, Diff-CoGAN adopted a semi-supervised learning strategy. In the training process, the optimizer for the generators was SGD, and the learning rate was set to 0.01. For the discriminator, the optimizer was Root Mean Squared Propagation (RMSprop), and the learning rate was set to 0.001. The batch_size of the labeled data was set to 8, and the batch_size of the unlabeled data was set to 16. The epoch was set to 100. In the optimization process, the discriminator was trained to minimize , and the generators were trained to minimize . For the generators’ training, the labeled and unlabeled data were firstly used to minimize and , and was optimized using the labeled data only. In the training process, the generators were trained ten times and the discriminator was trained once.
- (1)
- The first step was to initialize Generator 1 and Generator 2 using the labeled data only. This step adopted the initialized generators in Diff-CoGAN.
- (2)
- The second step was to train GAN under semi-supervised learning. There were two generators to realize medical image segmentation in Diff-CoGAN. However, segmentation based on GAN was separated into two experiments: Semi-GAN (G1) which adopted the segmentation approach from [34] and Semi-GAN (G2) from [39]. Semi-GAN (G1) consisted of Generator 1 and the discriminator, and Semi-GAN (G2) consisted of Generator 2 and the discriminator. The optimizer of the generator was SGD, and the learning rate was set to 0.01. The optimizer of the discriminator was RMSprop, and the learning rate was set to 0.001. The batch_size of the labeled data was set to 8, and the batch_size of the unlabeled data was set to 16. The epoch was set to 100.
- (1)
- The first step was to initialize Generator 1 and Generator 2 using the labeled data only. This step adopted the initialized generators in Diff-CoGAN.
- (2)
- In the second step, the unlabeled data and labeled data were used to train the model together inspired by the work by [7]. In the training process, the optimizer for the generators was SGD, and the learning rate was set to 0.01. The batch_size of the labeled data was set to 8, and the batch_size of the unlabeled data was set to 16. The epoch was set to 100. For these three experiments, the Dropout layer and the Batch normalization layer were imported into the model’s structure to prevent overfitting.
3. Experiments and Analysis
3.1. Dataset
3.2. Experimental Setup
3.3. Performance Evaluation
3.4. Results
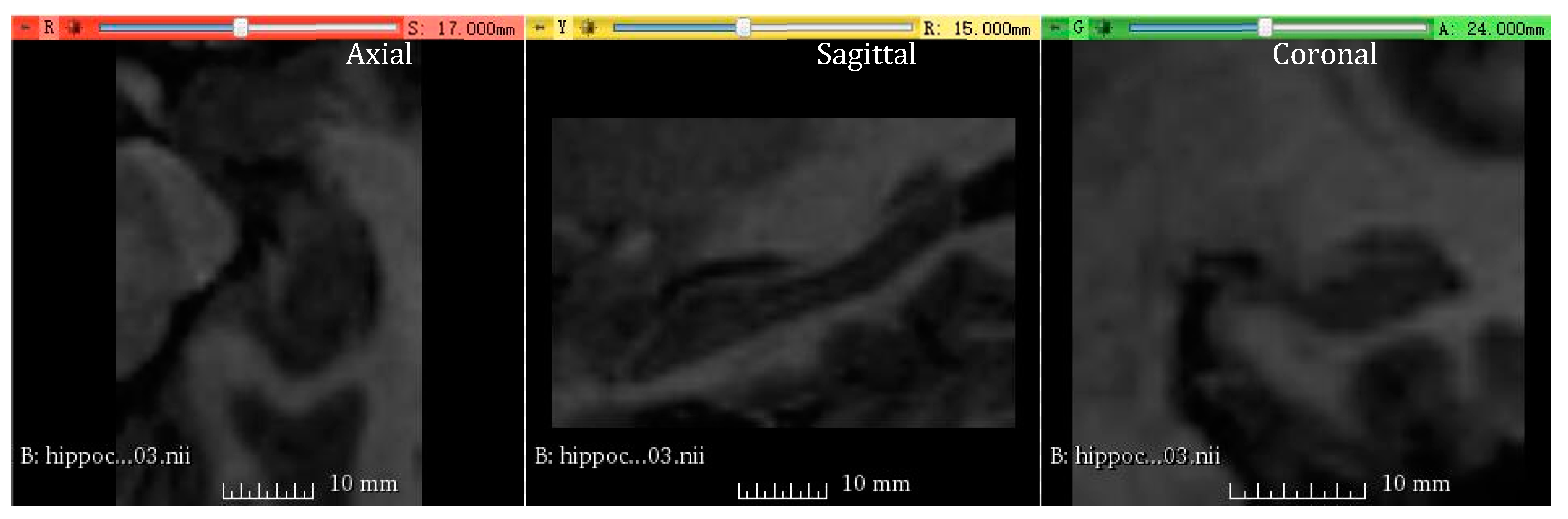
3.4.1. Medical Image Segmentation Using Hippocampus Dataset
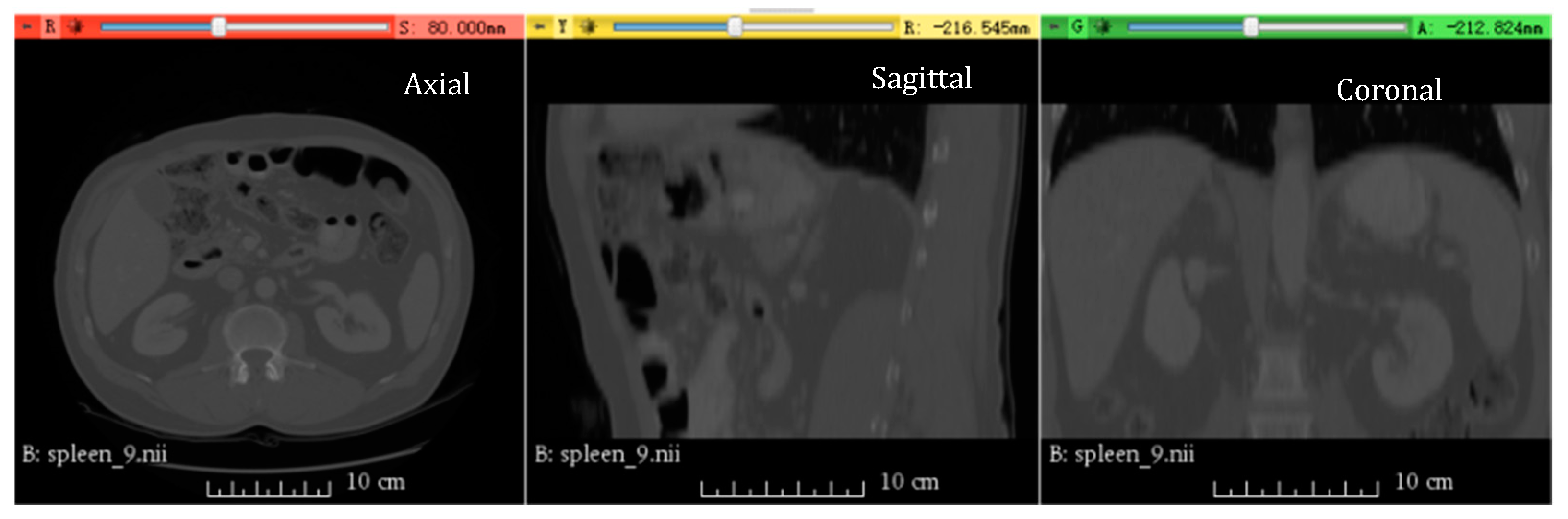
3.4.2. Medical Image Segmentation Using Spleen Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bai, W.; Oktay, O.; Sinclair, M.; Suzuki, H.; Rajchl, M.; Tarroni, G.; Rueckert, D. Semi-supervised learning for network-based cardiac MR image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Min, S.; Chen, X.; Xie, H.; Zha, Z.J.; Zhang, Y. A mutually attentive co-training framework for semi-supervised recognition. IEEE Trans. Multimed. 2020, 23, 899–910. [Google Scholar] [CrossRef]
- Feng, Z.; Nie, D.; Wang, L.; Shen, D. Semi-supervised learning for pelvic MR image segmentation based on multi-task residual fully convolutional networks. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018. [Google Scholar]
- Gu, L.; Zheng, Y.; Bise, R.; Sato, I.; Imanishi, N.; Aiso, S. Semi-supervised learning for biomedical image segmentation via forest oriented super pixels (voxels). In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Tseng, K.K.; Zhang, R.; Chen, C.M.; Hassan, M.M. DNetUnet: A semi-supervised CNN of medical image segmentation for super-computing AI service. J. Supercomput. 2021, 77, 3594–3615. [Google Scholar] [CrossRef]
- Blum, A.; Mitchell, T. Combining labeled and unlabeled data with co-training. In Proceedings of the Eleventh Annual Conference on Computational Learning Theory, Madison, WI, USA, 24–26 July 1998. [Google Scholar]
- Xia, Y.; Yang, D.; Yu, Z.; Liu, F.; Cai, J.; Yu, L.; Roth, H. Uncertainty-aware multi-view co-training for semi-supervised medical image segmentation and domain adaptation. Méd. Image Anal. 2020, 65, 101766. [Google Scholar] [CrossRef]
- Peng, J.; Estrada, G.; Pedersoli, M.; Desrosiers, C. Deep co-training for semi-supervised image segmentation. Pattern Recognit. 2020, 107, 107269. [Google Scholar] [CrossRef]
- Tseng, C.M.; Huang, T.W.; Liu, T.J. Data labeling with novel decision module of tri-training. In Proceedings of the 2020 2nd International Conference on Computer Communication and the Internet (ICCCI), Nagoya, Japan, 26–29 June 2020; pp. 82–87. [Google Scholar]
- Li, Z.; Lin, L.; Zhang, C.; Ma, H.; Zhao, W. Automatic image annotation based on co-training. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019. [Google Scholar]
- Wang, P.; Peng, J.; Pedersoli, M.; Zhou, Y.; Zhang, C.; Desrosiers, C. Self-paced and self-consistent co-training for semi-supervised image segmentation. Med. Image Anal. 2021, 73, 102146. [Google Scholar] [CrossRef]
- Ning, Z.; Zhong, S.; Feng, Q.; Chen, W.; Zhang, Y. SMU-Net: Saliency-Guided Morphology-Aware U-Net for Breast Lesion Segmentation in Ultrasound Image. IEEE Trans. Méd. Imaging 2021, 41, 476–490. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Shen, W.; Park, S.; Fishman, E.K.; Yuille, A.L. Abdominal multi-organ segmentation with organ-attention networks and statistical fusion. Méd. Image Anal. 2019, 55, 88–102. [Google Scholar] [CrossRef]
- Abdelgayed, T.S.; Morsi, W.G.; Sidhu, T.S. Fault detection and classification based on co-training of semisupervised machine learning. IEEE Trans. Ind. Electron. 2017, 65, 1595–1605. [Google Scholar] [CrossRef]
- Du, L.; Wang, Y.; Xie, W. A semi-supervised method for SAR target discrimination based on co-training. In Proceedings of the IGARSS 2019-2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019. [Google Scholar]
- Zhou, B.; Wang, Y.; Liu, W.; Liu, B. Identification of working condition from sucker-rod pumping wells based on multi-view co-training and hessian regularization of SVM. In Proceedings of the 2018 14th IEEE International Conference on Signal Processing (ICSP), Beijing, China, 12–16 August 2018. [Google Scholar]
- Duan, X.; Thomsen, N.B.; Tan, Z.H.; Lindberg, B.; Jensen, S.H. Weighted score based fast converging CO-training with application to audio-visual person identification. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence (ICTAI), Boston, MA, USA, 6–8 November 2017. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Dirvanauskas, D.; Maskeliūnas, R.; Raudonis, V.; Damaeviius, R.; Scherer, R. Hemigen: Human embryo image generator based on generative adversarial networks. Sensors 2019, 19, 3578. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Chen, J.; Fredericksen, M.; Hughes, D.P.; Chen, D.Z. Deep adversarial networks for biomedical image segmentation utilizing unannotated images. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Nie, D.; Trullo, R.; Lian, J.; Wang, L.; Petitjean, C.; Ruan, S.; Shen, D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans. Biomed. Eng. 2018, 65, 2720–2730. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Yao, J.; Jiang, T. Retinal Vessel Segmentation Network Based on Patch-GAN. In Intelligent Life System Modelling, Image Processing and Analysis; Springer: Singapore, 2021. [Google Scholar]
- Lahiri, A.; Ayush, K.; Kumar Biswas, P.; Mitra, P. Generative adversarial learning for reducing manual annotation in semantic segmentation on large scale miscroscopy images: Automated vessel segmentation in retinal fundus image as test case. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Yang, D.; Xu, D.; Zhou, S.K.; Georgescu, B.; Chen, M.; Grbic, S.; Comaniciu, D. Automatic liver segmentation using an adversarial image-to-image network. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Jin, D.; Xu, Z.; Tang, Y.; Harrison, A.P.; Mollura, D.J. CT-realistic lung nodule simulation from 3D conditional generative adversarial networks for robust lung segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Abirami, N.; Vincent, D.R.; Kadry, S. P2P-COVID-GAN: Classification and segmentation of COVID-19 lung infections from CT images using GAN. Int. J. Data Warehous. Min. 2021, 17, 101–118. [Google Scholar] [CrossRef]
- Xun, S.; Li, D.; Zhu, H.; Chen, M.; Wang, J.; Li, J.; Huang, P. Generative adversarial networks in medical image segmentation: A review. Comput. Biol. Med. 2022, 140, 105063. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2242–2251. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intelli. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zhou, S.; Nie, D.; Adeli, E.; Yin, J.; Lian, J.; Shen, D. High-resolution encoder–decoder networks for low-contrast medical image segmentation. IEEE Trans. Image Process. 2019, 29, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Méd. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Weng, Y.; Zhou, T.; Li, Y.; Qiu, X. Nas-unet: Neural architecture search for medical image segmentation. IEEE Access 2019, 7, 44247–44257. [Google Scholar] [CrossRef]
- Han, L.; Huang, Y.; Dou, H.; Wang, S.; Ahamad, S.; Luo, H.; Zhang, J. Semi-supervised segmentation of lesion from breast ultrasound images with attentional generative adversarial network. Comput. Methods Programs Biomed. 2020, 189, 105275. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, Y.; Ping, B.; Li, D.; Du, J.; Qin, Y.; Xiang, J. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: A pilot study. J. Cancer 2019, 10, 4876. [Google Scholar] [CrossRef] [PubMed]
- Theckedath, D.; Sedamkar, R.R. Detecting affect states using VGG16, ResNet50 and SE-ResNet50 networks. SN Comput. Sci. 2020, 1, 79. [Google Scholar] [CrossRef]
- Zhang, K.; Guo, Y.; Wang, X.; Yuan, J.; Ding, Q. Multiple feature reweight densenet for image classification. IEEE Access 2019, 7, 9872–9880. [Google Scholar] [CrossRef]
- Liu, X.; Song, L.; Liu, S.; Zhang, Y. A review of deep-learning-based medical image segmentation methods. Sustainability 2021, 13, 1224. [Google Scholar] [CrossRef]
- Pravitasari, A.A.; Iriawan, N.; Almuhayar, M. UNet-VGG16 with transfer learning for MRI-based brain tumor segmentation. TELKOMNIKA (Telecommun. Comput. Electron. Control) 2020, 18, 1310. [Google Scholar] [CrossRef]
- Soomro, M.H.; Coppotelli, M.; Conforto, S.; Schmid, M.; Giunta, G.; Del Secco, L.; Laghi, A. Automated segmentation of colorectal tumor in 3D MRI using 3D multiscale densely connected convolutional neural network. J. Healthc. Eng. 2019, 2019, 1075434. [Google Scholar] [CrossRef] [PubMed]


| Learning Model | Dice Value | Number of Layers |
|---|---|---|
| VGG16 | 0.741 | 16 |
| ResNet50 | 0.726 | 50 |
| Dense121 | 0.690 | 121 |
| Experiments | Dataset 1 | Training Dataset Setting (Labeled Slices/Unlabeled Slices) | Dataset 2 | Training Dataset Setting (Labeled Slices/Unlabeled Slices) |
|---|---|---|---|---|
| Semi-supervised learning using co-training | Hippocampus | 100/100 100/1000 100/2000 100/2900 | Spleen | 100/100 100/400 100/800 |
| Semi-supervised learning using GAN (semi-GAN) | ||||
| Semi-supervised learning using Diff-CoGAN |
| Initialization | Dice | IoU | HD | ASD |
|---|---|---|---|---|
| seg1only | 0.460 (0.045) | 0.072 (0.003) | 28.209 (111.717) | 10.639 (21.102) |
| seg2only | 0.731 (0.018) | 0.285 (0.016) | 10.079 (21.403) | 3.459 (1.715) |
| Data Setting | Dice | IOU | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Co-Training | Semi-GAN | Diff-CoGAN | Co-Training | Semi-GAN | Diff-CoGAN | |||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100/100 | 0.774 (0.015) | 0.785 (0.017) | 0.774 (0.015) | 0.783 (0.018) | 0.780 (0.014) | 0.805 (0.012) | 0.543 (0.021) | 0.551 (0.022) | 0.543 (0.022) | 0.547 (0.023) | 0.545 (0.021) | 0.578 (0.019) |
| 100/1000 | 0.793 (0.014) | 0.796 (0.019) | 0.783 (0.015) | 0.802 (0.012) | 0.797 (0.013) | 0.808 (0.011) | 0.576 (0.020) | 0.582 (0.024) | 0.534 (0.022) | 0.581 (0.019) | 0.586 (0.019) | 0.589 (0.018) |
| 100/2000 | 0.801 (0.01) | 0.811 (0.011) | 0.686 (0.028) | 0.802 (0.013) | 0.804 (0.012) | 0.812 (0.011) | 0.593 (0.018) | 0.600 (0.017) | 0.424 (0.027) | 0.581 (0.019) | 0.593 (0.018) | 0.601 (0.017) |
| 100/2900 | 0.804 (0.011) | 0.814 (0.011) | 0.790 (0.014) | 0.806 (0.011) | 0.805 (0.012) | 0.814 (0.011) | 0.595 (0.018) | 0.606 (0.017) | 0.576 (0.020) | 0.590 (0.018) | 0.597 (0.018) | 0.605 (0.017) |
| Data Setting | HD | ASD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Co-Training | Semi-GAN | Diff-CoGAN | Co-Training | Semi-GAN | Diff-CoGAN | |||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100/100 | 8.276 (12.405) | 7.384 (8.190) | 7.921 (9.190) | 7.265 (8.200) | 8.081 (14.777) | 7.264 (7.756) | 3.148 (1.425) | 2.724 (1.154) | 3.087 (1.269) | 2.606 (1.081) | 3.037 (1.628) | 2.624 (0.906) |
| 00/1000 | 6.813 (9.286) | 6.550 (7.673) | 7.316 (13.288) | 6.807 (7.847) | 6.537 (7.426) | 6.533 (6.917) | 2.596 (1.626) | 2.468 (1.398) | 2.473 (1.490) | 2.571 (1.017) | 2.420 (1.076) | 2.458 (1.032) |
| 100/2000 | 6.117 (8.939) | 5.990 (7.354) | 8.852 (20.004) | 7.034 (8.010) | 5.966 (7.420) | 5.963 (7.182) | 2.054 (0.626) | 2.050 (0.837) | 2.902 (1.936) | 2.728 (1.122) | 2.044 (0.549) | 2.043 (0.885) |
| 100/2900 | 5.902 (7.203) | 5.777 (6.683) | 6.163 (8.888) | 6.038 (7.518) | 5.832 (6.506) | 5.765 (6.656) | 1.952 (0.815) | 1.866 (0.340) | 2.110 (1.002) | 2.030 (0.841) | 1.917 (0.506) | 1.863 (0.420) |
| Data Setting | HD | ASD | ||||||
|---|---|---|---|---|---|---|---|---|
| Diff-CoGAN (without) | Diff-CoGAN (with) | Diff-CoGAN (without) | Diff-CoGAN (with) | |||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100/100 | 8.433 (12.695) | 7.306 (8.646) | 8.081 (14.777) | 7.264 (7.756) | 3.300 (1.583) | 2.660 (1.136) | 3.037 (1.628) | 2.624 (0.906) |
| 100/1000 | 7.251 (8.028) | 7.108 (6.887) | 6.537 (7.426) | 6.533 (6.917) | 2.894 (1.255) | 2.849 (0.962) | 2.420 (1.076) | 2.458 (1.032) |
| 100/2000 | 6.678 (10.805) | 6.261 (6.084) | 5.966 (7.420) | 5.963 (7.182) | 2.233 (0.845) | 2.282 (0.533) | 2.044 (0.549) | 2.043 (0.885) |
| 100/2200 | 8.256 (15.609) | 5.932 (6.790) | 6.045 (8.258) | 5.880 (6.366) | 2.420 (1.184) | 2.009 (0.485) | 2.031 (0.554) | 1.967 (0.413) |
| 100/2400 | 5.928 (7.087) | 5.851 (6.326) | 5.905 (6.381) | 5.851 (6.993) | 1.970 (0.646) | 1.945 (0.423) | 1.979 (0.492) | 1.929 (0.471) |
| 100/2900 | 5.833 (6.151) | 5.795 (6.918) | 5.832 (6.506) | 5.765 (6.656) | 1.942 (0.435) | 1.877 (0.511) | 1.917 (0.506) | 1.863 (0.420) |
| Initialization | Dice | IoU | HD | ASD |
|---|---|---|---|---|
| seg1only | 0.161 (0.041) | 0.019 (0.001) | 83.886 (136.112) | 40.423 (68.744) |
| seg2only | 0.886 (0.007) | 0.410 (0.018) | 28.307 (188.846) | 7.665 (10.764) |
| Data Setting | Dice | IoU | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Co-Training | Semi-GAN | Diff-CoGAN | Co-Training | Semi-GAN | Diff-CoGAN | |||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100/100 | 0.874 (0.019) | 0.943 (0.002) | 0.893 (0.010) | 0.944 (0.002) | 0.900 (0.014) | 0.944 (0.002) | 0.714 (0.029) | 0.812 (0.008) | 0.731 (0.021) | 0.813 (0.008) | 0.747 (0.023) | 0.815 (0.007) |
| 100/400 | 0.904 (0.010) | 0.945 (0.002) | 0.896 (0.011) | 0.947 (0.002) | 0.914 (0.010) | 0.946 (0.002) | 0.760 (0.016) | 0.820 (0.006) | 0.744 (0.020) | 0.822 (0.007) | 0.771 (0.017) | 0.828 (0.006) |
| 100/900 | 0.933 (0.003) | 0.947 (0.002) | 0.899 (0.013) | 0.947 (0.002) | 0.930 (0.003) | 0.948 (0.002) | 0.792 (0.013) | 0.829 (0.006) | 0.757 (0.020) | 0.828 (0.006) | 0.800 (0.016) | 0.832 (0.005) |
| Data Setting | HD | ASD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Co-Training | Semi-GAN | Diff-CoGAN | Co-Training | Semi-GAN | Diff-CoGAN | |||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100/ 100 | 21.244 (221.23) | 15.078 (127.9) | 18.502 (185.37) | 14.080 (111.72) | 17.587 (155.476) | 13.351 (94.51) | 6.226 (21.12) | 3.851 (5.170) | 5.605 (17.15) | 3.602 (3.919) | 5.079 (15.048) | 3.400 (2.285) |
| 100/ 400 | 12.336 (140.521) | 9.053 (31.39) | 15.661 (154.10) | 12.187 (64.307) | 12.099 (152.556) | 8.740 (31.70) | 3.633 (10.94) | 2.866 (1.554) | 4.551 (9.385) | 3.660 (3.401) | 3.570 (11.839) | 2.729 (1.189) |
| 100/ 900 | 7.344 (86.641) | 7.370 (76.36) | 15.151 (268.41) | 9.575 (102.35) | 7.253 (58.916) | 6.369 (32.36) | 2.300 (6.600) | 2.043 (3.590) | 3.040 (9.370) | 2.485 (5.063) | 2.307 (3.177) | 1.849 (0.630) |
| Data Setting | HD | ASD | ||||||
|---|---|---|---|---|---|---|---|---|
| Diff-CoGAN (without) | Diff-CoGAN (with) | Diff-CoGAN (without) | Diff-CoGAN (with) | |||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100–100 | 15.065 (94.445) | 16.065 (127.375) | 17.587 (155.476) | 13.351 (94.517) | 5.185 (13.472) | 4.035 (5.021) | 5.079 (15.048) | 3.400 (2.285) |
| 100–400 | 16.238 (218.456) | 9.224 (21.845) | 12.099 (152.556) | 8.740 (31.700) | 4.408 (17.928) | 2.963 (1.128) | 3.570 (11.839) | 2.729 (1.189) |
| 100–900 | 11.667 (180.558) | 7.839 (91.117) | 7.253 (58.916) | 6.369 (32.365) | 2.697 (6.920) | 2.162 (5.497) | 2.307 (3.177) | 1.849 (0.630) |
| Data Setting | Dice | IoU | HD | ASD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Co-Training | SemiGAN | Co-Training | SemiGAN | Co-Training | SemiGAN | Co-Training | SemiGAN | |||||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100–100 | <0.05 | <0.01 | <0.05 | <0.01 | >0.05 | <0.01 | >0.05 | <0.01 | >0.05 | >0.05 | >0.05 | >0.05 | <0.05 | <0.05 | >0.05 | >0.05 |
| 100–1000 | <0.05 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | >0.05 | <0.01 | <0.01 | <0.01 | >0.05 | >0.05 | <0.01 |
| 100–2000 | <0.05 | <0.05 | <0.01 | <0.01 | >0.05 | <0.01 | >0.05 | <0.01 | <0.05 | >0.05 | <0.01 | <0.01 | >0.05 | >0.05 | <0.01 | <0.01 |
| 100–2900 | >0.05 | >0.05 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | >0.05 | >0.05 | <0.01 | <0.01 | >0.05 | >0.05 | <0.01 | <0.01 |
| Data Setting | Dice | IoU | HD | ASD | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Co-Training | SemiGAN | Co-Training | SemiGAN | Co-Training | SemiGAN | Co-Training | SemiGAN | |||||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | |
| 100–100 | <0.01 | >0.05 | >0.05 | >0.05 | <0.01 | >0.05 | <0.05 | >0.05 | <0.05 | <0.05 | >0.05 | >0.05 | <0.01 | >0.05 | <0.05 | >0.05 |
| 100–400 | <0.05 | >0.05 | <0.01 | >0.05 | <0.01 | <0.01 | <0.01 | <0.01 | >0.05 | <0.05 | <0.01 | <0.01 | <0.05 | <0.05 | <0.01 | <0.01 |
| 100–900 | <0.05 | >0.05 | >0.05 | <0.05 | <0.01 | >0.05 | >0.05 | <0.05 | <0.05 | <0.05 | <0.01 | <0.01 | >0.05 | <0.01 | <0.01 | <0.05 |
| Data Setting | Diff-CoGAN (without) Using Hippocampus Dataset | Data Setting | Diff-CoGAN (without) Using Spleen Dataset | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HD | ASD | HD | ASD | ||||||
| G1 | G2 | G1 | G2 | G1 | G2 | G1 | G2 | ||
| 100–100 | <0.05 | >0.05 | <0.05 | >0.05 | 100–100 | <0.05 | <0.01 | >0.05 | <0.01 |
| 100–1000 | <0.05 | <0.05 | <0.05 | <0.05 | 100–400 | <0.05 | >0.05 | <0.05 | <0.01 |
| 100–2000 | <0.05 | <0.05 | <0.05 | <0.05 | 100–900 | <0.05 | <0.05 | <0.05 | <0.05 |
| 100–2900 | <0.01 | >0.05 | <0.01 | >0.05 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Jamil, N.; Hamzah, R. An Improved Co-Training and Generative Adversarial Network (Diff-CoGAN) for Semi-Supervised Medical Image Segmentation. Information 2023, 14, 190. https://doi.org/10.3390/info14030190
Li G, Jamil N, Hamzah R. An Improved Co-Training and Generative Adversarial Network (Diff-CoGAN) for Semi-Supervised Medical Image Segmentation. Information. 2023; 14(3):190. https://doi.org/10.3390/info14030190
Chicago/Turabian StyleLi, Guoqin, Nursuriati Jamil, and Raseeda Hamzah. 2023. "An Improved Co-Training and Generative Adversarial Network (Diff-CoGAN) for Semi-Supervised Medical Image Segmentation" Information 14, no. 3: 190. https://doi.org/10.3390/info14030190
APA StyleLi, G., Jamil, N., & Hamzah, R. (2023). An Improved Co-Training and Generative Adversarial Network (Diff-CoGAN) for Semi-Supervised Medical Image Segmentation. Information, 14(3), 190. https://doi.org/10.3390/info14030190






