Abstract
In this work, we propose a CAD (computer-aided diagnosis) system using advanced deep-learning models and computer vision techniques that can improve diagnostic accuracy and reduce transmission risks using the YOLOv7 (You Only Look Once, version 7) object detection architecture. The proposed system is capable of accurate object detection, which provides a bounding box denoting the area in the X-rays that shows some possibility of TB (tuberculosis). The system makes use of CNNs (Convolutional Neural Networks) and YOLO models for the detection of the consolidation of cavitary patterns of the lesions and their detection, respectively. For this study, we experimented on the TBX11K dataset, which is a publicly available dataset. In our experiment, we employed class weights and data augmentation techniques to address the data imbalance present in the dataset. This technique shows a promising improvement in the model’s performance and thus better generalization. In addition, it also shows that the developed model achieved promising results with a mAP (mean average precision) of 0.587, addressing class imbalance and yielding a robust performance for both obsolete pulmonary TB and active TB detection. Thus, our CAD system, rooted in state-of-the-art deep-learning and computer vision methodologies, not only advances diagnostic accuracy but also contributes to the mitigation of TB transmission risks. The substantial improvement in the model’s performance and the ability to handle class imbalance underscore the potential of our approach for real-world TB detection applications.
1. Introduction
An important worldwide health issue is the highly contagious bacterial illness tuberculosis caused by Mycobacterium tuberculosis. This airborne bacterial disease, which primarily affects the lungs but can also damage other organs, is spread through respiratory droplets expelled by infected people. According to the report [1], TB is a major global health threat, with approximately 8–10 million new TB patients and 2–3 million deaths from the disease each year. About 10.6 million people were diagnosed with TB in 2021, a 4.5% increase over the 10.1 million cases that were reported in 2020. The report [1] shows the number of new cases of TB per 100,000 individuals annually had increased by 3.6% between 2020 and 2021. A decreasing tendency of about 2% per year that had been present for the previous two decades was reversed by this rise [1]. If left untreated, TB has a high mortality rate. Furthermore, the prevalence and mortality rate are found to be higher in countries with lower human development indexes [2].
TB can be active or obsolete. The term “active TB” describes the condition in which the bacteria are present in the body actively, causing clinical symptoms and the potential for transmission to others. On the other hand, “obsolete TB” refers to a condition in which the bacteria are still present but do not result in an active illness, causing a latent infection [3]. However, early diagnosis and treatment with antibiotics greatly improve survival chances and help control the spread of active infection [4,5,6].
The most common TB detection approaches include TST (Tuberculin Skin Test) and IGRAs (Interferon-Gamma Release Assays). The ancient TST has limitations due to technical errors and BCG (Bacillus Calmette–Guérin) vaccination influence. Identifying TST conversion is important for LTBI (latent TB infection) preventive therapy. IGRAs offer higher specificity but have logistical constraints and are more expensive [7]. This might also be a challenge for developing countries where most of the population cannot afford such diagnostic tests.
However, a chest X-ray is the most common and less expensive screening method for TB. Early TB screening through X-ray has significant implications for the early detection, treatment, and prevention of TB [8,9,10,11]. However, radiologists’ examination of X-rays is often prone to errors, with experienced radiologists from top hospitals only achieving an accuracy of 68.7% [9,10]. This is due to the difficulty distinguishing TB areas from other areas in X-rays, which the human eye is not sensitive enough to detect.
The medical field’s evaluation of chest X-rays can significantly benefit from the integration of an efficient CAD system paired with precise computer vision models for disease detection. Detecting TB early is crucial for implementing effective measures to mitigate risks and prevent the spread of infection. Additionally, such a system can expedite the diagnostic process, reducing the risk of infection extension during prolonged testing procedures. Typically, doctors focus on specific areas in chest X-ray images, searching for consolidation or cavitary lesions in the lungs as indicators of infection [3]. Utilizing computer vision technology alongside advanced neural network (NN) architectures enhances the ability to annotate, locate, and analyze chest X-ray images, predicting the presence of similar patterns [12].
AI (Artificial Intelligence) aims to make machines mimic and perform like humans in everyday tasks by utilizing large amounts of data. This enables machines to surpass human capabilities in tasks such as image analysis and video processing [13,14,15]. Particularly, CNN is a deep-learning algorithm designed to take an image as input and acquire knowledge about the image from different perspectives, leveraging the unique properties of a CNN [16]. This helps the CNN-based AI systems to simulate human-like understanding and excel in tasks such as image analysis, surpassing human capabilities in these areas. This technology, along with CAD software, can help medical professionals make effective decisions. CADs are useful decision-support tools that aid physicians and radiologists in increasing their precision, effectiveness, and patient outcomes by providing additional insights and reducing diagnostic errors [17].
In this study, we present a computer vision model based on YOLOv7, a CNN-based single-shot object detector, accompanied by an effective CAD system trained on the TBX11K dataset. Throughout our study, we converted Pascal VOC annotations to the YOLO format using scripts for each annotation and the associated images, ensuring compatibility with YOLO algorithms. Previously, the authors in [12] applied object detection and segmentation models such as Faster RCNN (Faster Region-Based Convolutional Neural Network), SSD (Single-Shot Detector), RetinaNet, and FCOS (Fully Convolutional One-Stage) detectors on the imbalanced TBX11K dataset. However, to address this imbalance, we introduced class weights and image augmentations, resulting in the synthesis of a balanced dataset. Following this, YOLOv7, an approach not previously tested for this specific object detection problem, was employed with hyperparameter evolution. This implementation results in more generalized outcomes and a higher mAP than the other formally recorded approaches. Additionally, the improved efficiency and detection speed of the YOLOv7 model made it well suited for the seamless integration into our cloud-based CAD system, developed on the foundation of this object detection model. Despite the efforts, the approach and the system can be further improved. Also, the medical data are always hard to obtain; the model might perform better in a high abundance of data since neural networks are very data-hungry [13].
2. Related Work
Most of the recent approaches in the field cover the classification aspect from chest X-rays and some infection location techniques without employing the actual annotated dataset for training and validation. For instance, in the proposed model from [18], ImageNet fine-tuned NFNets (Normalization-Free Networks) are employed for chest X-ray classification using the Score-Cam algorithm to highlight diagnostic regions. The proposed method achieves a high accuracy, AUC, sensitivity, and specificity for both binary and multiclass classification, with results reaching 96.91% accuracy, 99.38% AUC, 91.81% sensitivity, and 98.42% specificity on the multiclass dataset.
Likewise, the study from [19] proposes a TB detection system employing advanced deep-learning models. Segmentation networks are utilized to extract the region of interest from CXR images, leading to an improved classification performance. Among the tested CNN models, the proposed model achieves the highest accuracy of 99.1% on three publicly available CXR datasets, demonstrating the superiority of segmented lung CXR images over raw ones.
The authors in [12] conducted a comparison of object detection and classification methods for TB detection using annotated chest X-ray images. The goal was to distinguish between TB and non-TB X-rays, with a focus on predicting the boundary box of TB in the X-ray images. They evaluated several models, including SSD with VGGNet-16 backbone, RetinaNet with ResNet-50 backbone and FPN, and Faster R-CNN with ResNet-50 backbone and FPN.
In the case of classification models, the results showed that using pre-trained backbones significantly improved the performance of the models. SSD with pre-trained VGGNet-16 achieved an accuracy of 84.7% and an AUC of 93.0%. RetinaNet with pre-trained ResNet-50 achieved an accuracy of 87.4% and an AUC of 91.8%. Faster R-CNN with pre-trained ResNet-50 achieved an accuracy of 89.7% and an AUC of 93.6%.
However, without pre-training, the performance of the models decreased. SSD without pre-trained weights achieved an accuracy of 88.2%, RetinaNet without pre-trained weights achieved an accuracy of 79.0%, and Faster R-CNN without pre-trained weights achieved an accuracy of 81.3%.
The evaluation also focused on the models’ ability to detect active TB and latent TB cases. Faster R-CNN with ResNet-50 backbone and FPN demonstrated the highest accuracy of 53.3% and sensitivity of 21.9% for active TB detection. For latent TB detection, RetinaNet with ResNet-50 backbone and FPN achieved the highest accuracy of 37.8% and sensitivity of 12.7%.
The study also evaluates the performance of object detection models to make the boundary box prediction of the infected regions of active and latent TB. Here, many approaches have been considered with separate backgrounds and with the option of using pre-trained weights. Then, the respective AP scores are compared with each other, and the models are compared. It is illustrated in Table 1.

Table 1.
TB area detection results with different architecture on TBX11K dataset [12].
Table 1 compares different deep-learning architectures for TB detection on the TBX11K dataset, emphasizing the impact of backbones, pre-trained models, and data configurations. Faster R-CNN with ResNet-50 and FPN consistently excels in detecting active TB and latent TB, showcasing its clinical potential.
Overall, the study emphasized the importance of using appropriate backbone networks and pre-trained weights for improved performance in TB detection from chest X-ray images. The authors acknowledged the challenge of data imbalance when detecting latent TB cases. Further research and optimization are necessary to improve the detection rates for latent TB, as it poses additional challenges due to its subtler manifestations in X-ray images compared to active TB. Similarly, the method did not consider one of the state-of-the-art computer vision algorithms: YOLO.
YOLO has been found to be promising for the automation of the detection of TB. A basic CNN’s maximum speed cap is exceeded by YOLO, which achieves an excellent balance between speed and accuracy. One of the swiftest all-purpose item detectors is YOLO. Furthermore, YOLO is the best model for applications that require quick, accurate object recognition because it generalizes objects better than other models. Because of these outstanding and valuable benefits, it merits being strongly suggested and receiving more exposure over time [20].
YOLO in CAD is its ability to detect multiple objects in a single image. This is particularly useful in medical imaging, where multiple abnormalities or lesions may be present in a single image. By detecting and localizing multiple objects in a single pass, YOLO can help improve the efficiency and accuracy of CAD systems [21]. Additionally, YOLO can be trained on datasets of annotated medical images, allowing it to learn and recognize patterns that are characteristic of various diseases and conditions. This can help improve the accuracy of CAD systems, as they can be trained to detect subtle changes in images that may be difficult for human experts to detect. Overall, YOLO has the potential to significantly improve the accuracy and efficiency of CAD systems, leading to earlier and more accurate diagnoses of various medical conditions With the introduction of YOLO and its architectural successors/descendants, YOLOs are widely used in many applications, primarily because of their quicker conclusions and respectable accuracy, due to which YOLO is a perfect fit for boundary box detection in medical images along with its quick inference in the CAD software [22].
The YOLO architecture in Figure 1 is a pioneering object detection model that combines speed and accuracy. It consists of 24 convolutional layers followed by 2 fully connected layers. Notably, the YOLO architecture incorporates alternating 1 × 1 convolutional layers, which effectively reduce the feature space from the preceding layers. This reduction aids in capturing and consolidating essential information while maintaining computational efficiency. To leverage the power of transfer learning, the YOLO model adopts a pre-trained approach. The convolutional layers are initially pre-trained on the ImageNet classification task using a resolution of 224 × 224 input images. This pre-training step enables the network to learn discriminative features and high-level representations from a large-scale dataset. Subsequently, the resolution is doubled for object detection, enhancing the spatial resolution of the final predictions [23].
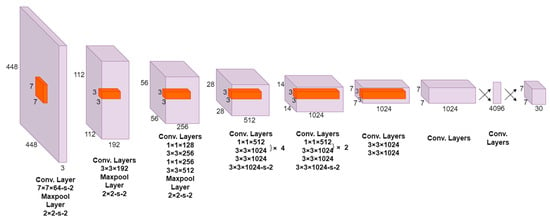
Figure 1.
Architecture of the YOLO object detection model.
The output of the YOLO network is a tensor with dimensions of 7 × 7 × 30. This tensor represents a grid that divides the input image into spatial cells, with each cell responsible for predicting multiple bounding boxes and corresponding class probabilities. The 7 × 7 grid allows for efficient detection across the image while maintaining a balance between localization accuracy and computational complexity. The 30-dimensional tensor encompasses various predictions, including bounding box coordinates, class probabilities, and other parameters needed for object detection tasks [23].
Similarly, the above architecture evolved into many iterations and YOLOV7 (the latest at the time) was considered during the experiments. The YOLOv7 architecture, known for its efficiency and real-time object detection capabilities, has undergone advancements in the form of extended efficient layer aggregation networks (ELANs) and model scaling. The ELAN introduces variations to the VoVNet architecture, considering factors such as memory access cost, input/output channel ratio, number of branches, and element-wise operations. Additionally, the ELAN analyzes the gradient path to enable the learning of diverse features and achieve faster and more accurate inference. Building upon the ELAN, the extended-ELAN (E-ELAN) further enhances the network’s learning ability by employing expand, shuffle, and merge cardinality operations within the computational blocks. This allows for continuous learning without disrupting the original gradient path [24].
When it comes to model scaling in concatenation-based architectures like YOLO, a compound scaling approach is necessary due to the interdependence of scaling factors. Traditional scaling methods used in other architectures may not be directly applicable to concatenation-based models. For instance, when scaling up or down the depth of a computational block in YOLO, it affects the in-degree and out-degree of the subsequent transition layer, which can impact hardware usage and model performance. To address this, a compound model scaling method is proposed for concatenation-based models. This method considers both depth and width scaling, ensuring that changes in one factor are accompanied by corresponding adjustments in the other to maintain the optimal structure and preserve the desired properties of the model [24].
This paper addresses a significant research gap in the domain of TB detection from chest X-ray images, particularly in relation to the utilization of advanced computer vision techniques. While previous studies have underscored the importance of appropriate backbone networks and pre-trained weights for enhanced performance, an underexplored area pertains to the integration of state-of-the-art algorithms like YOLO, whose efficiency and inference speed beat all of the other algorithms. This paper also addresses the challenge of data imbalance, particularly in detecting latent TB cases with subtler manifestations. This study not only emphasizes the value of optimizing TB detection rates but also introduces a novel approach through the comprehensive CAD system. By incorporating CNNs and YOLOv7 architecture, we bridge the gap between accuracy and speed, thereby contributing to the advancement of automated TB detection in CAD systems.
3. Materials and Methods
This section first discusses the architecture of both a CAD and object detection system to detect TB. Section 3.1 presents the dataset. The data preprocessing step is presented in Section 3.2. Section 3.3 and Section 3.4 are about class weights and data augmentation to tackle the data imbalance problem. Section 3.5 discusses more on the experiments. Section 3.6 covers hyperparameter tuning with an approach like hyperparameter evolution. Finally, Section 3.7 is about the deployment of the model in a CAD system.
The architectural design of the system in Figure 2 revolves around distinct views and routes, aligning with specific functionalities of the computer vision model. The server effectively handles image data received via POST requests, utilizing the TB detection computer vision model to compute potential infection region coordinates. These coordinates are then transmitted to the front end, where the image is rendered with infection predictions. To facilitate communication between the front end and the backend application programming interface (API), the Django REST Framework is employed.
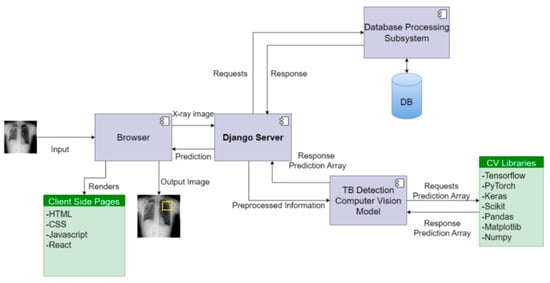
Figure 2.
Block diagram of the CAD system for TB detection.
The system ensures ease of use for doctors by incorporating features such as create, retrieve, update, and delete (CRUD) functionality, thereby enhancing accessibility and supporting informed decision-making. In addition, the platform maintains detailed records of each doctor’s interactions, enabling further analysis of infections.
The backend server relies on the Postgresql database engine, while pivotal libraries like TensorFlow, Scikit, Keras, PyTorch, Pandas, and NumPy are utilized to effectively serve the model. This configuration allows for the seamless integration of CAD software, thereby augmenting the analysis of TB infections in medical imaging.
Figure 3 illustrates the schematics of the deep-learning models for TB classification and detection. First and foremost, the image preprocessing was performed to improve the image quality, reduce noise, and standardize the format of the X-ray images. To determine whether the X-ray depicts a sick patient, a healthy patient, or a patient with TB, a CNN was employed. The model was trained on a labeled dataset that contained X-ray pictures of patients with and without TB after the CNN determined that the patient had been diagnosed with TB. Assuming the infection classification task was effective, the next phase involved locating and identifying items in an X-ray by employing the YOLO object detection technique. In this instance, it was used to identify particular TB subtypes in the X-ray pictures, such as pulmonary TB and obsolete TB. An extensive and varied dataset of X-ray pictures with associated labels for which X-rays were tagged and were necessary for an accurate diagnosis. Deep-learning-based medical image analysis has demonstrated considerable promise for supporting medical practitioners with early diagnosis and therapy planning. However, it is essential to validate the models and ensure they are integrated into the medical workflow responsibly to avoid any potential harm caused by misdiagnosis.
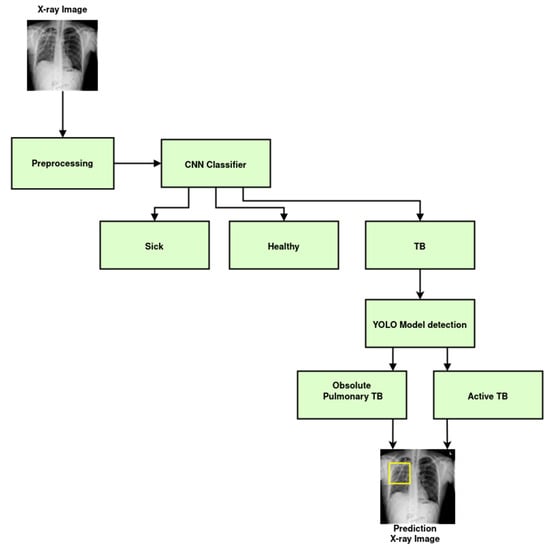
Figure 3.
ML schematics diagram of the TB classification and detection system.
3.1. Dataset
The TBX11K dataset, a publicly available and labeled dataset, was utilized to train and evaluate the performance of our model. According to the authors in [12], there are a total of 11,200 chest X-ray images; the dataset encompasses various categories, including 5000 healthy cases, 5000 cases with diseases other than TB, and 1200 cases exhibiting manifestations of TB. Notably, each X-ray image corresponds to a distinct individual. Within the subset of TB X-rays, there are 924 cases of active TB, 212 cases of latent TB, 54 cases exhibiting both active and latent TB simultaneously, and 10 cases where the TB type cannot be definitively determined under current medical conditions. All images possess a standardized size of 512 × 512 pixels.
The writers from [13] meticulously curated the dataset to guarantee its quality and dependability. Bounding box annotations for TB locations in the X-ray pictures were carried out by seasoned radiologists from prominent hospitals. The annotation process required two parts. The first involved labeling the bounding boxes and assigning TB types (active or latent) for each box by a radiologist with 5–10 years of experience in TB diagnosis. The correctness and consistency of the bounding box labels and TB-type designations were subsequently confirmed by cross-checking the annotations with another radiologist with more than 10 years of expertise in diagnosing TB. To maintain consistency across the dataset, efforts were also made to align the classified TB kinds with the image-level labels.
The TBX11K dataset represents a significant contribution as the first publicly available dataset for TB detection, encompassing not only classification but also bounding box annotations. With its carefully curated diverse cases, including different stages and variations of TB, as well as other common chest pathologies, the dataset serves as a valuable benchmark for the development and evaluation of algorithms and models aimed at TB diagnosis using chest X-ray images.
The dataset split presented in Table 2 illustrates the comprehensive partitioning strategy adopted for the TBX11K dataset. This meticulous allocation aims to ensure a balanced and representative distribution of classes, facilitating the robust training and evaluation of our proposed TB detection system. The dataset encompasses three primary classes: Non-TB, Sick and Non-TB, and TB for the classification task. Within the TB class, further distinctions are made between active TB and latent TB. The training set consists of 6600 samples, with 3000 dedicated to the Non-TB class, 3000 to the Sick and Non-TB class, and 600 to various TB classes. These are the numbers for the unannotated images and are useful for the classification task. However, there are 1200 annotated images emphasizing both active TB and latent TB for the images consisting of the TB infection that can be used by the object detection model.

Table 2.
Proposed dataset split for TBX11K dataset [12].
3.2. Preprocessing
A comprehensive analysis of gray scale X-ray images was conducted to explore the characteristics of chest-related complications. A subset of 799 X-ray images, each measuring 512 × 512 pixels, was extracted from the TBX11K dataset. To facilitate further investigation, the annotation format of the dataset was transformed into the YOLO format, which includes detailed information about the boundary boxes, such as the class label, x and y coordinates of the box center, box height, and box width. The original dataset utilized annotations in the old coco_json and pascal_voc formats, which were incompatible with the latest YOLO frameworks. Therefore, an annotation conversion process was employed to convert the annotations into the YOLO format, associating the boundary box information with the corresponding images and class labels. To facilitate model training and evaluation, the dataset was randomly partitioned into three sets: training, testing, and validation, with a split ratio of 70:20:10, respectively. Subsequently, initial model iteration was conducted, wherein no hyperparameters were tuned. This initial training process served as a baseline to establish a starting point for the further refinement and optimization of the model.
3.3. Class Weights
The TBX-11 dataset had a huge data imbalance between active and obsolete TB cases. This data imbalance caused overfitting and the AP for the obsolete pulmonary TB was found to be very small and for the active TB, it was found to be very high. The mAP of both classes for the Intersection over Union (IoU) greater than or equal to 0.5 shrunk and did not generalize so well. Due to the imbalance in the class, the algorithms tend to become biased towards the majority values present on active TB and did not perform well on the obsolete pulmonary TB. To tackle this problem, appropriate class weights were assigned to the minority (obsolete TB) class during the training of the TB detection model.
The technique called focal loss was used to address this problem, which dynamically adjusted the loss function based on the difficulty of predicting each class [25]. Focal loss assigned higher weights to the obsolete pulmonary TB class, which helped the model to prioritize that minority class and resulted in a slightly better outcome. It is primarily intended to address the issue of class imbalance in object detection tasks, where the proportion of background (negative) instances to foreground (positive) examples is disproportionately high by reshaping the standard cross-entropy loss such that it down-weights the loss assigned to well-classified examples.
The introduction of a new loss function, focal loss, which is a more potent substitute for prior methods for addressing class imbalance, is formulated as:
FL(pt) = −(1 − pt)γ . log(pt)
It is used for computing ‘p’ with the loss computation, resulting in greater numerical stability. The loss function is a cross-entropy loss that is dynamically scaled, with the scaling factor decrementing to zero as confidence in the correct class rises. Intuitively, this scaling factor can quickly concentrate the model on difficult examples while automatically de-weighting the contribution of easy examples during training. Focal loss attempts to solve this issue by down-weighting the loss ascribed to well-classified examples and minimizing the loss contribution from cases that are easily classified [25].
3.4. Image Augmentations
The TBX-11 dataset had a huge data imbalance between active and obsolete TB cases. We tried solving this problem using focal loss, which dynamically adjusted the loss function in favor of the obsolete pulmonary TB class. The approach spiked the mAP@0.5 to 300 percent improvement; however, the model could still not generalize well. Another approach was to perform image augmentation on the minority class to increase the sample size and equate it with the sample size of the majority class [26]. Image augmentation is a technique used to increase the diversity of training data by applying various transformations to the existing data. The dataset had 148 obsolete pulmonary TB cases and 651 active cases, as shown in Figure 4.
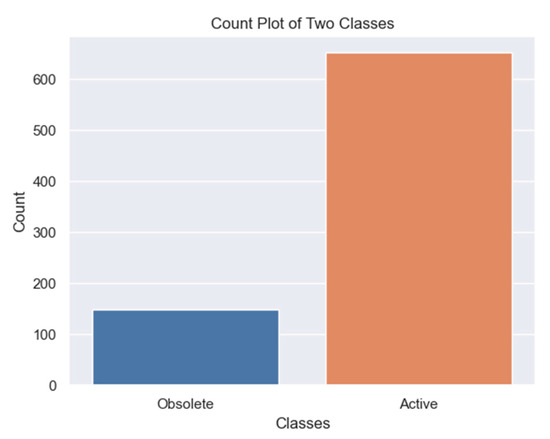
Figure 4.
Data distribution of both obsolete and active classes before the minority class image augmentation.
Four (4) different image augmentation approaches were incorporated in order to equate the sample size of the obsolete class with the active class. The geometric transformations like reflection about the Y-axis, rotation of −5 and 5 degrees, and scaling of 0.8 were incorporated into the experiment because they were the transformations that did not generate noise in the chest X-ray images and did not hamper the model training [26].
When it comes to X-ray images, rotation is a commonly used augmentation technique, as shown in Figure 5 and Figure 6. To apply a clockwise and anticlockwise rotation transformation to the X-ray image by 5 degrees, the Image library was used. The rotation also had to persist in the respective bounding box, which was also performed with the Image library.
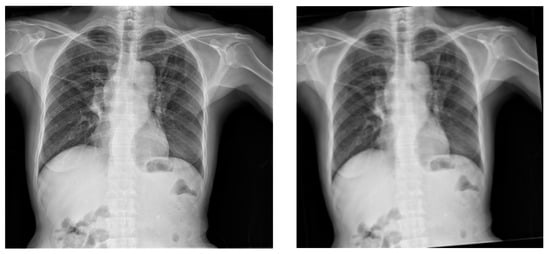
Figure 5.
Original X-ray (left image) and the output of the application of anticlockwise rotation of the X-ray images by 5 degrees (right image).
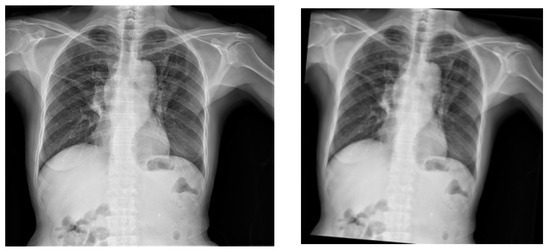
Figure 6.
Original X-ray (left image) and the output of the application of clockwise rotation of the X-ray images by 5 degrees (right image).
Similarly, scaling is also a frequently employed augmentation technique, as illustrated in Figure 7. To implement a scaling transformation with a scaling factor of 0.8, the Image library was utilized. This involved adjusting the size of the X-ray image uniformly to 80% of its original dimensions. Notably, the scaling transformation was extended to maintain consistency within the corresponding bounding box; a process seamlessly executed using the Image library. This ensures that the scaling effect persists coherently across both the X-ray image and its associated bounding box.
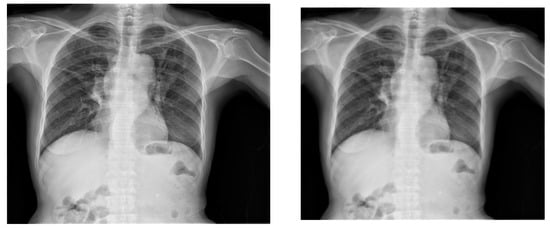
Figure 7.
Original X-ray (left) and 0.8 scaled X-ray (right).
Another data augmentation method to broaden the diversity of the training data is to reflect or horizontally flip the X-ray pictures, as illustrated in Figure 8. By providing differences in the direction or location of components inside the X-ray, flipping an image horizontally can aid in the model’s ability to generalize. This augmentation step is proposed by Elgendi et. al. [26], who had a similar problem set for thorax chest X-ray images. This augmentation technique might hurt the model’s performance in the context of non-symmetric and lateral medical objects, but it might not be the case in the context of small non-lateral objects [27]. The YOLOv7 architecture, with its focus on object detection inside a small region of interest [24], might be less sensitive to spatial distortions, particularly when the lesions are confined within a small boundary box. Most of the annotations of the TBX11k dataset resemble this property. According to the authors in [3], TB results on mostly non-lateral airspace opacities, cavitation, and effusion. Plus, TB usually affects the mid and upper lungs. The lower lungs and regions near the heart are uncommon sites of tuberculosis involvement. That may be one of the reasons why the non-symmetric and lateral properties of lungs including the placement of the heart will not interfere with the results of this particular object detection task. Objects like non-lateral airspace opacities, cavitation, and effusion in the mid and upper lungs are the subject of interest for our algorithm, thus making horizontal flipping a decent augmentation choice.
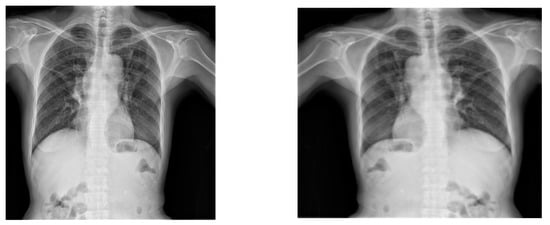
Figure 8.
Original X-ray (left) and horizontal flipping of the X-ray (right).
Hence, the sample size of the minority class grew by four times after the augmentations resulted in 592 obsolete cases’ X-ray images, as illustrated in Figure 9. Thus, data augmentation had a positive impact on improving the performance of the models, particularly for obsolete cases’ X-rays. Data augmentation increased the diversity of the training data. This augmented data helped the model learn from a wider range of examples. Further, it provided a more comprehensive representation of the minority class X-rays, allowing the model to generalize better and capture important patterns.
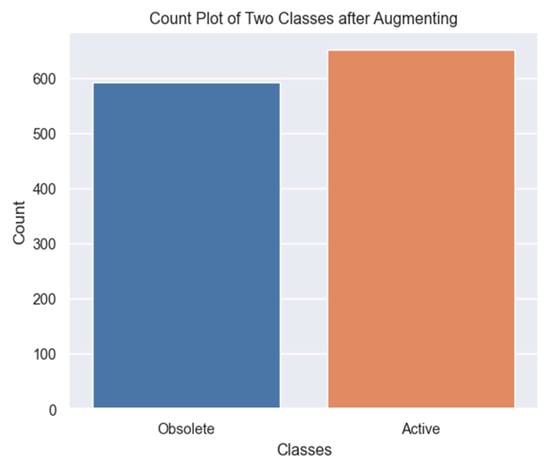
Figure 9.
Final distribution of the image numbers for both obsolete and active classes after minority class data augmentation.
3.5. Experiments
YoloV7 incorporates E-ELANs (extended efficient layer aggregation networks) as its underlying architecture. The E-ELAN retains the original architecture’s gradient transmission path without modification. Instead, it leverages group convolution to augment the cardinality of additional features and combines the features from different groups through a shuffle and merges the cardinality approach. This operation enhances feature learning across diverse feature maps and improves the utilization of parameters and computations [25]. This architecture can further enhance the trivial YOLO architecture, which works well with medical images, allowing it to learn and recognize patterns that are characteristic of various diseases and conditions [22]. Therefore, this architecture was incorporated into the experiments.
During the experiment, we used the Adam Optimizer [28] with learning rates of 0.01, mini-batch size of 32 images, momentum of 0.937, and weight decay of 0.0005 with total epochs of 600. We used a Linux server with Intel(R) Core(TM) i9-9900X @ 4.40 GHz CPU, 2.5 TB hard disk space with an SSD, and a Cuda-enabled RTX 6000 Quadro GPU alongside Nvidia Titan V GPU. The networks were implemented using the PyTorch and YOLOv7 libraries in Python 3.10.3.
3.6. Hyperparameter Evolution
In deep learning, hyperparameters play a crucial role in the training process, but determining the best values for them can be difficult. Traditional methods like grid searches become impractical due to the large number of hyperparameters, unknown relationships between them, and the time-consuming nature of evaluating each set of hyperparameters. Genetic algorithms are a viable solution for hyperparameter searches [29].
In Figure 10, the mAP@0.5 is represented in the Y-axis and the values of the hyperparameters are illustrated in the X-axis. The approach, as shown in Figure 10, seeks to maximize the fitness, which is a metric used to evaluate the hyperparameter selection of the model. In the YOLO library, we defined a default fitness function as a weighted combination of metrics where mAp@0.5 and mAP@0.5:0.95 were given the highest priority. Then, the evolution is performed about a base scenario, which we seek to improve upon. Here, the main genetic operators are crossover and mutation. In this work, the mutation is used to create new offspring based on a combination of the best parents from all previous generations and give out the best possible offspring (hyperparameters) to maximize the fitness. This step was carried out in the final best-performing PYtorch model from the experiments.
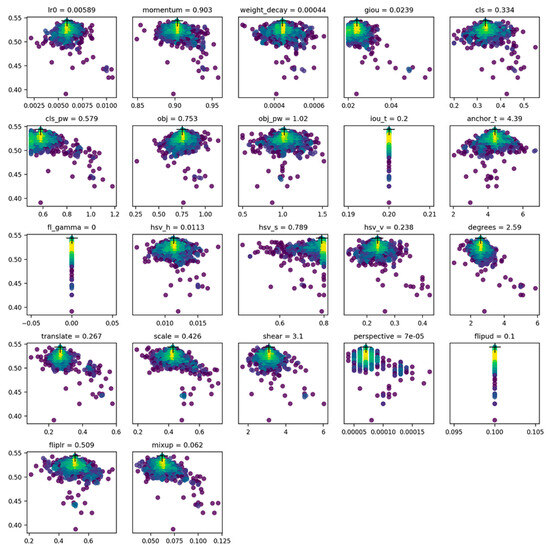
Figure 10.
Parameter selection after applying genetic evolution algorithm for hyperparameter tuning, adapted from [30].
3.7. Deployment in CAD System
The proposed system functions as an assistive technology for medical professionals, offering efficient annotation of potential TB infections in two classes: obsolete pulmonary TB and active TB. The primary focus is on early detection to mitigate future risks. To access the platform, doctors undergo a verification process overseen by the site’s administrator, requiring the submission of relevant credentials. Once verified, doctors gain access to the chest X-ray infection detection assistance platform, which includes a computer vision model deployed in a Django server.
The system’s architecture is structured around individual views and routes that align with specific functionalities of the computer vision model. The server handles image data received via POST requests, utilizing the TB detection computer vision model to compute potential infection region coordinates. These coordinates are then sent to the frontend, where the image is rendered with infection predictions using the Django REST Framework to communicate with the backend API. Each doctor’s account retains records of their interactions with the system, facilitating further analysis of infections.
The platform ensures ease of use for doctors through features like create, retrieve, update, and delete (CRUD) functionality, enhancing accessibility and supporting informed decision-making. The system relies on the Postgresql database engine on the server side, while key libraries like TensorFlow, Scikit, Keras, PyTorch, Pandas, and NumPy are employed to serve the model effectively from the backend server. This setup enables the seamless integration of CAD software, enhancing the analysis of TB infections in medical imaging.
4. Results and Discussions
This section evaluates the model, focusing on precision, recall, AP, and mAP. In Section 4.1, we discuss these metrics’ role in assessing object detection algorithms for chest X-ray images. After this, Section 4.2 details the results of experiments addressing class imbalance, image weights, minority class augmentation, and hyperparameter evolution. Visualization and analysis of the model predictions are covered in Section 4.3, offering insights into the true positives, false positives, and false negatives of the random samples from the professional’s perspective. The learning curve analysis, addressing overfitting concerns, is presented in Section 4.4. This structured approach provides a concise yet thorough exploration of our TB detection model’s development and performance.
4.1. Performance Metrics
Precision, recall, average precision (AP), and mean average precision (mAP) are key performance metrics utilized in this study to evaluate the accuracy and reliability of object detection algorithms for TB detection using chest X-ray images.
Precision in the context of object detection for active TB and obsolete pulmonary TB refers to the proportion of correctly detected objects belonging to a specific class (either active TB or obsolete pulmonary TB) out of all the objects predicted as that class. Precision indicates the algorithm’s ability to accurately identify and classify objects of interest, thereby minimizing false positives.
To calculate the precision for a specific class, we use the following formula:
Recall in the context of object detection for active TB and obsolete pulmonary TB refers to the proportion of correctly detected objects belonging to a specific class (either active TB or obsolete pulmonary TB) out of all the ground truth objects of that class. Recall indicates the algorithm’s ability to capture and identify a high percentage of objects of interest, minimizing false negatives.
To calculate the recall for a specific class, we use the following formula:
Intersection over Union (IoU) is a widely used performance metric in object detection, including for the detection of active TB and obsolete pulmonary TB. IoU measures the overlap between the predicted bounding box and the ground truth bounding box for an object of interest. It quantifies the spatial agreement between the predicted and ground truth regions and is particularly useful in evaluating the accuracy of object localization. It is used to deduce true positives, false positives, and false negatives to calculate precision and recall.
To calculate IoU, we use the following formula:
A higher IoU score indicates a better alignment between the predicted and ground truth regions, implying a more accurate localization of the TB-related abnormalities.
AP and mAP are key performance metrics used in object detection, including the detection of active TB and obsolete pulmonary TB. AP evaluates the detection algorithm’s precision–recall trade-off by considering precision at different recall levels. It is calculated by averaging precision values at different recall levels. mAP evaluates the algorithm’s precision–recall trade-off across multiple classes by averaging AP values for each class. A higher mAP score indicates a better overall performance.
4.2. Results
The precision–recall curves are studied for various experiments of the model. The major aspect of the results encompasses the study of the model with the class imbalance problem, introducing the class weights, minority class augmentation, and the hyperparameter evolution. The effects of each approach are documented, and a final model is trained based on the results of the previous experiments.
Figure 11 displays the results obtained from training the YOLO model in its original form, without applying any augmentation techniques, assigning class weights, or performing hyperparameter tuning. The figure indicates that the AP for the active TB class is 0.489, while for the obsolete TB class, it is only 0.009. At an IoU threshold of 0.5, the mAP for all classes is 0.249. Figure 12 illustrates the bias observed in the model, where the majority class has a significant influence on the minority class due to the limited number of training samples. This is further depicted in the histogram shown in Figure 4 and Figure 9. Consequently, the model’s ability to generalize effectively is hindered by this imbalance.
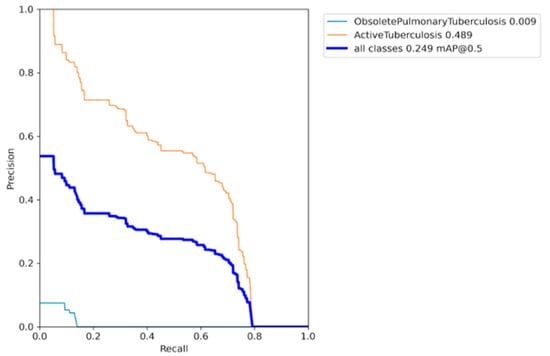
Figure 11.
Precision–recall curve of the base model with class imbalance.
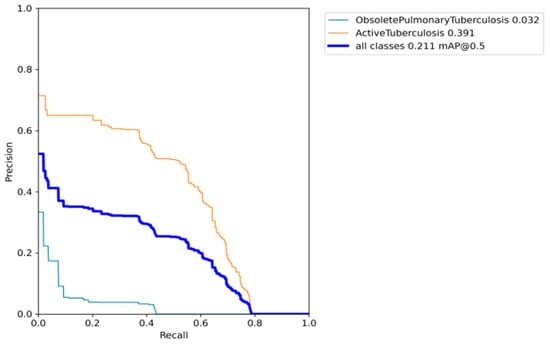
Figure 12.
Precision–recall curve when the base model is introduced with image weights.
To address the issue of dataset imbalance, the focal loss technique was implemented, which dynamically adjusted the loss function to assign optimal weights to the minority class [25]. Figure 11 visually demonstrates the significant improvement in the AP of the obsolete pulmonary TB class, surpassing a remarkable 300 per cent improvement. These results were achieved during the training of the original YOLO model, without the utilization of any augmentation or hyperparameter evolution. However, it should be noted that the effectiveness of the focal loss technique became more apparent due to the assignment of class weights. Despite these enhancements, the model still exhibited limitations in generalizing across all classes, yielding a mAP of 0.211. It is important to mention that the AP of the majority class suffered degradation as a result of the lower class weights applied.
In order to address the unsatisfactory effectiveness of assigning class weights to the model, we experimented with augmenting only the minority class. As a result, the sample size of the minority class increased fourfold and approached parity with the majority class, as depicted in Figure 12. This successfully resolved the issue of imbalanced datasets. Remarkable improvements were observed in the AP scores of the minority class, as illustrated in Figure 13. These outcomes were obtained while training the YOLO model without assigning class weights or performing hyperparameter evolution. This enabled us to visualize the effectiveness of this augmentation method on our model. The model demonstrated improved generalization; however, there was potential for further maximizing the AP and mAP scores.
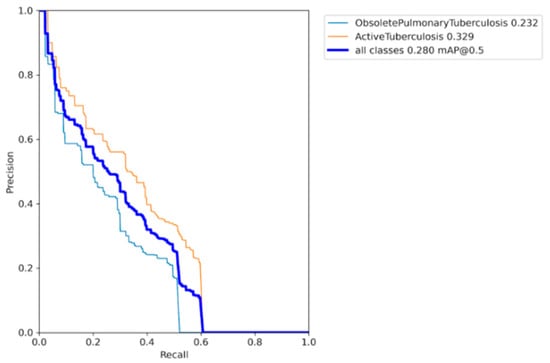
Figure 13.
Precision–recall curve for the model with minority class image augmentation.
Based on the conclusions drawn from the aforementioned experiments, we implemented all the methods that demonstrated improvements in the precision, recall, AP, and mAP of the model. The weights of the model corresponding to the epoch that yielded the best results on the test set were saved as a PyTorch model. Subsequently, this specific model served as the starting point for hyperparameter evolution. A genetic algorithm was identified as a suitable approach for searching the fittest hyperparameters that would maximize our results [29]. Precision, recall, AP, and mAP were defined as fitness parameters during the evolution process. The evolution was conducted over 1000 generations, with each generation consisting of 30 epochs dedicated to searching for the optimal hyperparameters. Ultimately, in generation number 733, the fittest hyperparameters were identified, significantly optimizing our results. Table 3 provides a list of the fittest hyperparameters obtained from this evolutionary process.

Table 3.
List of hyperparameters and corresponding values selected for the final TB detection model.
After conducting our comprehensive experiments with image weights assignment, minority class augmentation, and evolving the hyperparameter, we achieved promising results for our TB detection model. The AP for the obsolete pulmonary TB class reached 0.499, while the AP for the active TB class achieved a value of 0.675. As a result, the mAP at an IoU threshold of 0.5 amounted to 0.587, as illustrated in Figure 14.
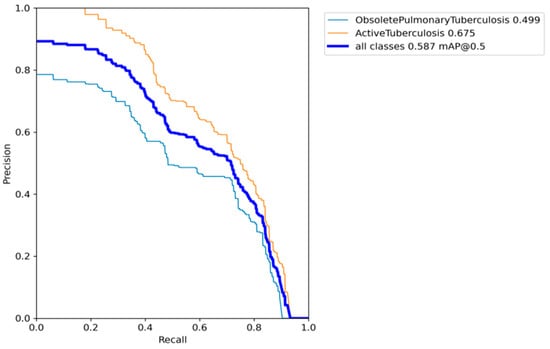
Figure 14.
Precision–recall curve for the model with image weights assignment, minority class augmentation, and evolving the hyperparameter.
All of the outcomes are summarized in Table 4, which provides an overview of the successful experiments conducted during the development of our TB detection model. Notably, our approach effectively addressed the class imbalance issue discussed in [12], mitigating the bias towards the minority class and yielding a robust TB detection model with a commendable mAP score. Table 4 serves as a concise summary of our TB detection computer vision model architecture.

Table 4.
TB detection models’ evaluation with respect to mAP of IOU threshold at 0.5 for various training approaches.
Table 5 compares the features of the final TB object detection model using YOLO architecture for both obsolete and active cases. The models have an input size of 512 × 512, 37.2 million parameters, 415 layers, and a size of 71.3 MB, demonstrating their capacity for accurate TB detection with efficient deployment potential.

Table 5.
Feature of the final TB detection model.
4.3. Visualization and Analysis
A random subset comprising 16 chest X-ray images was extracted from the dataset for analysis. Employing our computer vision model, various regions of imaging anomalies were detected within these images. To establish a benchmark, the predictions of the model were juxtaposed with interpretations provided by a radiologist, regarded as the reference standard. The radiologist identified a total of 25 abnormalities, consisting of 24 cases of airspace opacity, indicating a robust indication of TB infection, and 1 instance of cavitation. In terms of the model’s performance, there were 23 true positives, 2 false positives, and 2 false negatives. Specifically, the AI accurately identified 22 instances of airspace opacity and correctly flagged 1 case of cavitation. However, the model failed to detect two instances of airspace opacity and erroneously labeled two healthy lung regions as abnormalities. Figure 15 provides visual representations of the true positive, false positive, and false negative cases. Here, the orange box indicates an active TB infection, the blue box indicates an obsolete pulmonary TB infection, the red box indicates a false negative, and the red box outside the prediction box represents a false positive.
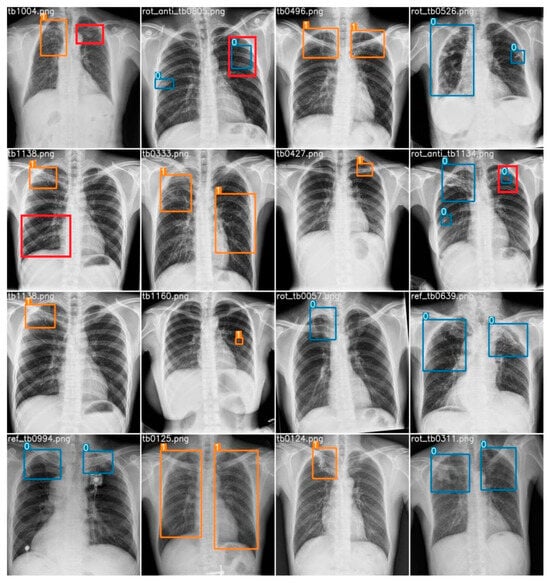
Figure 15.
Prediction of all 16 random test samples with indication of false positives (red box outside prediction) and false negatives (red box). Here, blue box stands for obsolete cases and orange box stands for active cases.
4.4. Learning Curve Analysis
This study explores the model’s performance on the challenging TBX11k chest X-ray dataset, showcasing the learning curves for the infection’s classification loss and the boundary box prediction loss for both the training and the validation set.
The YOLOv7 model underwent training on the TBX11k dataset, known for its challenging chest X-ray samples with limited data. The evaluation encompassed both training and validation sets, revealing a well-controlled classification loss but a slight overfitting in box prediction towards the end of training. Despite this, Figure 16 shows the satisfactory performance of the model, achieving a classification loss below 0.1 and a box prediction loss below 0.06 on the validation set, showcasing its proficiency in identifying and localizing lesions in chest X-rays with limited data.
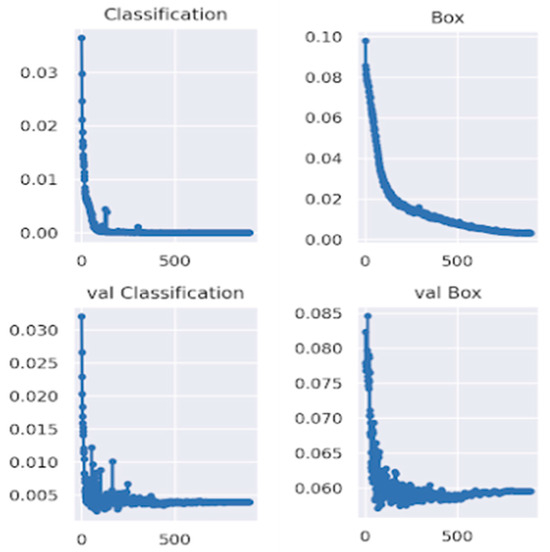
Figure 16.
Learning curves of box classification loss and box prediction loss for both training and validation sets.
To address overfitting, the incorporation of additional annotated data is proposed, aiming to augment the sample size for enhanced generalization. It is pertinent to note that during training, optimal weights were saved, and checkpoint weights were retained for practical deployment in a CAD system. This strategic approach ensures the model’s adaptability and effectiveness in real-world clinical scenarios, further emphasizing its potential for improved performance with increased dataset diversity.
5. Conclusions and Future Work
In this research study, we have presented a comprehensive CAD system for TB detection using advanced deep-learning models and computer vision techniques. TB continues to be a global health concern due to its contagious nature and severe implications for public health. Chest X-rays are widely used for screening, but the accuracy of visual inspection by human experts is limited and time-consuming. Our proposed CAD system aims to overcome these limitations and enhance the diagnostic accuracy for TB detection, thereby facilitating early intervention and reducing transmission risks.
The utilization of CNNs and YOLOv7 architecture enables the accurate detection and classification of TB patterns in chest X-ray images. By leveraging the TBX11K dataset, we demonstrated the effectiveness of the proposed system in distinguishing between active TB and obsolete pulmonary TB cases. Mitigation of data imbalance in the dataset through class weights and data augmentation techniques, including focal loss and image augmentation, resulted in an improved model performance and robust generalization, as illustrated in Figure 14.
After incorporating genetic algorithms to optimize the hyperparameters in the YOLO model, our research findings indicate that the proposed CAD system achieved a promising mAP of 0.587, as illustrated in Figure 14. Our model outperforms previous formally recorded models, which are recorded in Table 1. Figure 16 also illustrated that the classification loss generalized well in the validation set with respectable numbers, concluding there was no overfitting of our model for the boundary box classification task. Also, the professional’s evaluation of our model in the randomly chosen test set depicts a decent result and practicality of our model, as shown in Figure 15. The developed CAD system holds great promise for the medical community, providing a more efficient and accurate approach to TB detection in chest X-rays. With seamless integration through a Django server and leveraging key deep-learning libraries like TensorFlow and PyTorch, our system can aid healthcare professionals in the early identification and diagnosis of TB, potentially leading to improved patient outcomes and reduced disease transmission through effective pattern recognition.
However, it is important to acknowledge that further research and development are necessary to refine and enhance the CAD system for TB detection. One significant challenge is the scarcity of medical data, which poses difficulties for neural network models. By obtaining a more extensive and diverse dataset for training and validation purposes, the performance of the TB detection model can be improved [31,32,33,34]. It will also help the model to mitigate the slight overfitting of the boundary box prediction loss, as illustrated in Figure 16.
Incorporating multi-modal data sources, such as clinical information and patient history, can also contribute to enhancing the accuracy and reliability of the CAD system [35]. By considering a broader range of information and integrating it into the analysis, a more comprehensive understanding of TB patterns and characteristics can be achieved, which might be the interest of future research.
There is also a need for further evaluation of the model’s clinical applicability through a larger prospective study, assessing its impact on clinical outcomes. While our current study did not encompass such a study due to limitations in data and resources available at the time, our approach utilizes existing resources to enhance TB detection technology, positioning it as a relevant tool for pattern recognition.
Importantly, we stress that our model should not replace the expertise of radiologists but serve as an assistive technology. The mAP of 0.587 on the validation set instills confidence in the model’s pattern recognition capabilities, making it a promising assistive tool for radiologists. The claim is further supported by the analysis of our model conducted by a professional radiologist, as presented in Figure 15. The example result of the model’s predictions on random samples on test set images suggests a fair applicability, strengthening confidence in its potential clinical utility in the context of the pattern recognition of TB infection.
Author Contributions
Conceptualization, A.T., R.B. and J.C.F.; methodology, A.T., A.M., R.B. and J.C.F.; software, A.P., A.B., R.B. and J.C.F.; validation, R.B. and J.C.F.; formal analysis, R.B. and J.C.F.; investigation, R.B. and J.C.F.; resources, R.B.; data curation, A.B., R.B. and J.C.F.; writing—original draft preparation, A.B.; writing—review and editing, A.T., R.B. and J.C.F.; visualization, S.W., R.B. and J.C.F.; supervision, R.B.; project administration, A.P., R.B. and J.C.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received funds from Erasmus+, NEEM project under GA 101083048.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data will be available as per request for research.
Acknowledgments
This paper was carried out in the Scopus of Erasmus+ European Project NEEM. The computing resource was provided by Kathmandu University High Performance Computing (KUHPC) lab.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022 (accessed on 1 August 2023).
- Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3278873/ (accessed on 1 August 2023).
- Bhalla, A.S.; Goyal, A.; Guleria, R.; Gupta, A.K. Chest tuberculosis: Radiological review and imaging recommendations. Indian J. Radiol. Imaging 2015, 25, 213–225. [Google Scholar] [CrossRef]
- Chauhan, A.; Chauhan, D.; Rout, C. Role of gist and PHOG features in computer-aided diagnosis of tuberculosis without segmentation. PLoS ONE 2014, 9, e112980. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kim, H.E.; Jeong, J.; Kim, H.J. A novel approach for tuberculosis screening based on deep convolutional neural networks. In Medical Imaging 2016: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2016; Volume 9785, pp. 750–757. [Google Scholar]
- Jaeger, S.; Karargyris, A.; Candemir, S.; Folio, L.; Siegelman, J.; Callaghan, F.; Xue, Z.; Palaniappan, K.; Singh, R.K.; Antani, S.; et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 2013, 33, 233–245. [Google Scholar] [CrossRef]
- Zellweger, J.P.; Sotgiu, G.; Corradi, M.; Durando, P. The diagnosis of latent tuberculosis infection (LTBI): Currently available tests, future developments, and perspectives to eliminate tuberculosis (TB). La Med. Del Lav. 2020, 111, 170–183. [Google Scholar] [CrossRef]
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 2013, 33, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Konstantinos, A. Testing for tuberculosis. Aust. Prescr. 2010, 33, 12–18. [Google Scholar] [CrossRef]
- Available online: https://apps.who.int/iris/handle/10665/252424 (accessed on 1 August 2023).
- Van Cleeff, M.R.A.; Kivihya-Ndugga, L.E.; Meme, H.; Odhiambo, J.A.; Klatser, P.R. The role and performance of chest X-ray for the diagnosis of tuberculosis: A cost-effectiveness analysis in Nairobi, Kenya. BMC Infect. Dis. 2005, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, Y.H.; Ban, Y.; Wang, H.; Cheng, M.M. Rethinking computer-aided tuberculosis diagnosis. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 2646–2655. [Google Scholar]
- Bansal, M.A.; Sharma, D.R.; Kathuria, D.M. A systematic review on data scarcity problem in deep learning: Solution and applications. ACM Comput. Surv. 2022, 54, 1–29. [Google Scholar] [CrossRef]
- Alshehri, F.; Muhammad, G. A comprehensive survey of the Internet of Things (IoT) and AI-based smart healthcare. IEEE Access 2021, 9, 3660–3678. [Google Scholar] [CrossRef]
- Muhammad, G.; Alshehri, F.; Karray, F.; El Saddik, A.; Alsulaiman, M.; Falk, T.H. A comprehensive survey on multimodal medical signals fusion for smart healthcare systems. Inf. Fusion 2021, 76, 355–375. [Google Scholar] [CrossRef]
- Muhammad, G.; Alhamid, M.F.; Long, X. Computing and processing on the edge: Smart pathology detection for connected healthcare. IEEE Netw. 2019, 33, 44–49. [Google Scholar] [CrossRef]
- Lieberman, R.; Kwong, H.; Liu, B.; Huang, H.K. Computer-assisted detection (CAD) methodology for early detection of response to pharmaceutical therapy in tuberculosis patients. In Medical Imaging 2009: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2009; Volume 7260, pp. 847–854. [Google Scholar]
- Acharya, V.; Dhiman, G.; Prakasha, K.; Bahadur, P.; Choraria, A.; Sushobhitha, M.; Sowjanya, J.; Prabhu, S.; Kautish, S.; Viriyasitavat, W.; et al. AI-assisted tuberculosis detection and classification from chest X-rays using a deep learning normalization-free network model. Comput. Intell. Neurosci. 2022, 2022, 2399428. [Google Scholar] [CrossRef] [PubMed]
- Nafisah, S.I.; Muhammad, G. Tuberculosis detection in chest radiograph using convolutional neural network architecture and explainable artificial intelligence. Neural Comput. Appl. 2022, 19, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Du, J. Understanding of object detection based on CNN family and YOLO. In Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2018; Volume 1004, p. 012029. [Google Scholar]
- Luo, Y.; Zhang, Y.; Sun, X.; Dai, H.; Chen, X. Intelligent solutions in chest abnormality detection based on YOLOv5 and ResNet50. J. Healthc. Eng. 2021, 2021, 2267635. [Google Scholar] [CrossRef]
- Diwan, T.; Anirudh, G.; Tembhurne, J.V. Object detection using YOLO: Challenges, architectural successors, datasets and applications. Multimed. Tools Appl. 2022, 82, 9243–9275. [Google Scholar] [CrossRef] [PubMed]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. [Google Scholar]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023; pp. 7464–7475. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Nishio, M.; Noguchi, S.; Matsuo, H.; Murakami, T. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: Combination of data augmentation methods. Sci. Rep. 2020, 10, 17532. [Google Scholar] [CrossRef]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2023, 152, 106391. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 1–74. [Google Scholar] [CrossRef]
- Available online: https://docs.ultralytics.com/yolov5/tutorials/hyperparameter_evolution/ (accessed on 1 August 2023).
- Yang, R.; Yu, Y. Artificial convolutional neural network in object detection and semantic segmentation for medical imaging analysis. Front. Oncol. 2021, 11, 638182. [Google Scholar] [CrossRef]
- Li, W.; Kazemifar, S.; Bai, T.; Nguyen, D.; Weng, Y.; Li, Y.; Xia, J.; Xiong, J.; Xie, Y.; Owrangi, A.; et al. Synthesizing CT images from MR images with deep learning: Model generalization for different datasets through transfer learning. Biomed. Phys. Eng. Express 2021, 7, 025020. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.C.; Ye, J.C. Generalization capability of deep learning. In Geometry of Deep Learning: A Signal Processing Perspective; Springer: Berlin/Heidelberg, Germany, 2022; pp. 243–266. [Google Scholar]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.; Mahmood, F. Synthetic data in machine learning for medicine and healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Acharya, Y.; Vink, I.; Ebisi, M.; Arja, S. A descriptive analysis of patient history based on its relevance. Int. J. Med. Sci. Educ. 2018, 35, 144–148. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).