Medical QA Oriented Multi-Task Learning Model for Question Intent Classification and Named Entity Recognition
Abstract
1. Introduction
- A multi-task learning model based on ALBERT-BILSTM is proposed for intent classification and named entity recognition of Chinese online medical questions.
- The experimental results demonstrate that the proposed method in this paper outperforms the benchmark methods and improves the model generalization ability compared to the single-task model.
2. Related Work
2.1. Medical Named Entity Recognition
2.2. Intent Classification
2.3. Multi-Task Learning
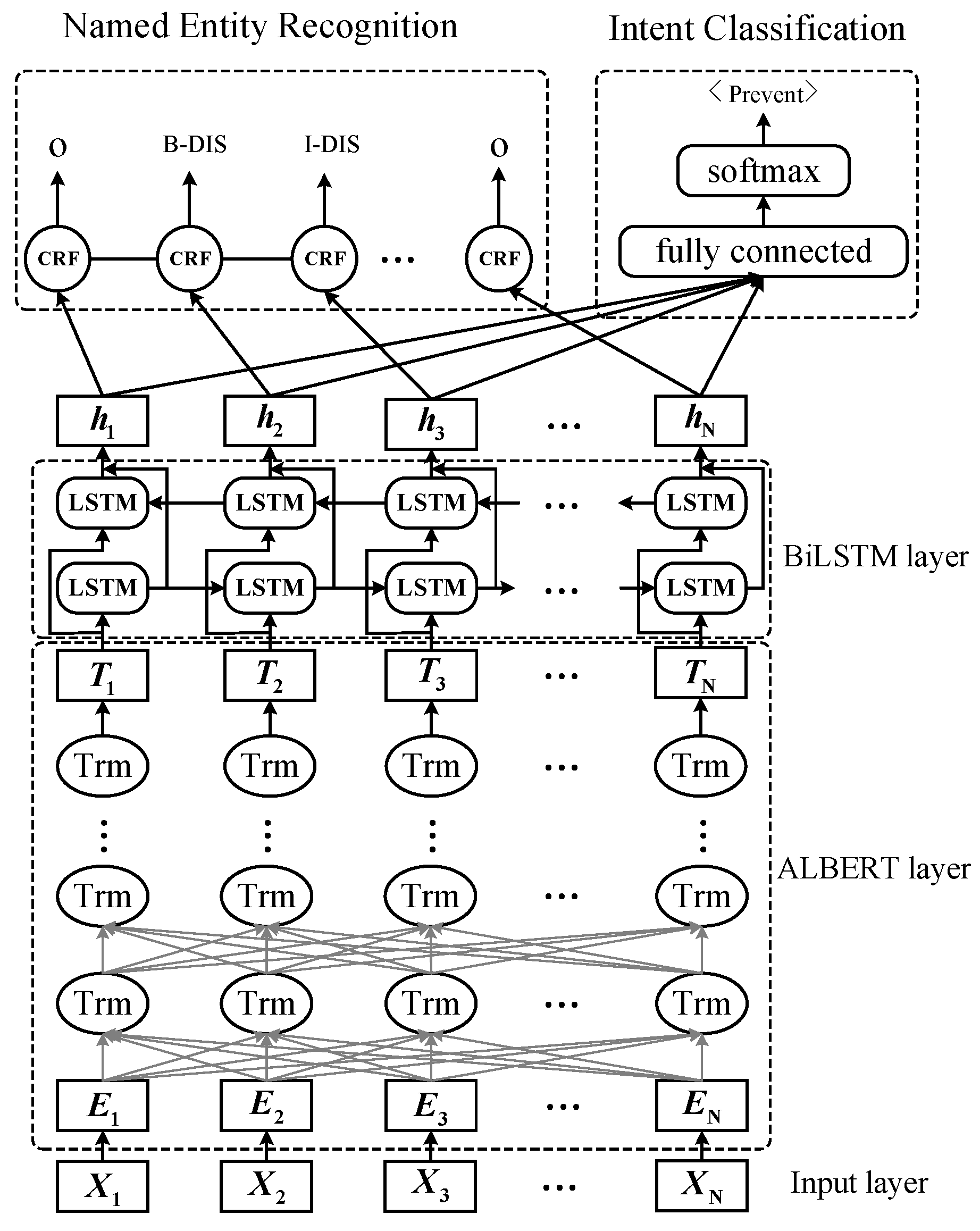
3. Methodology and Model
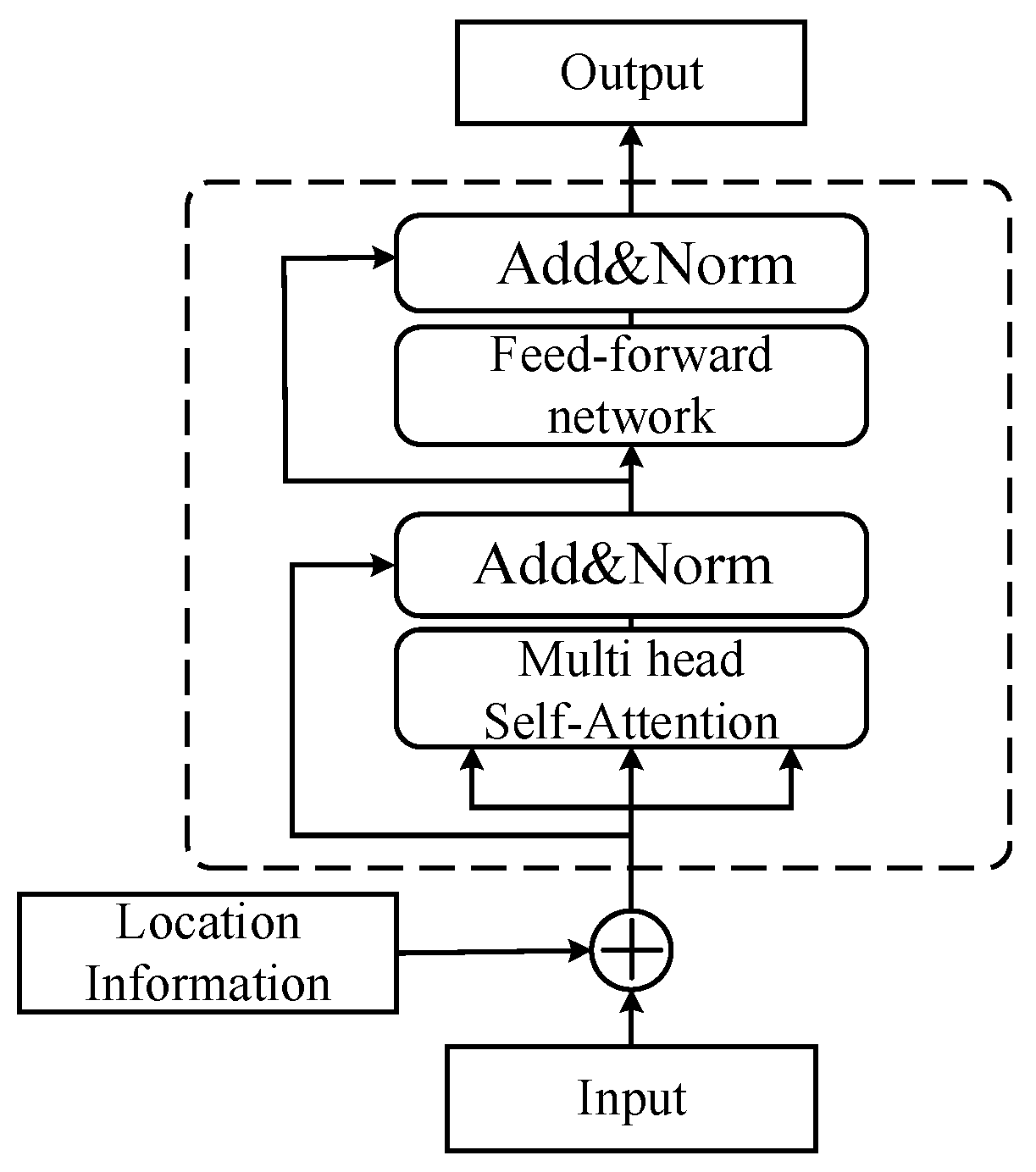
3.1. ALBERT Pre-Trained Language Model
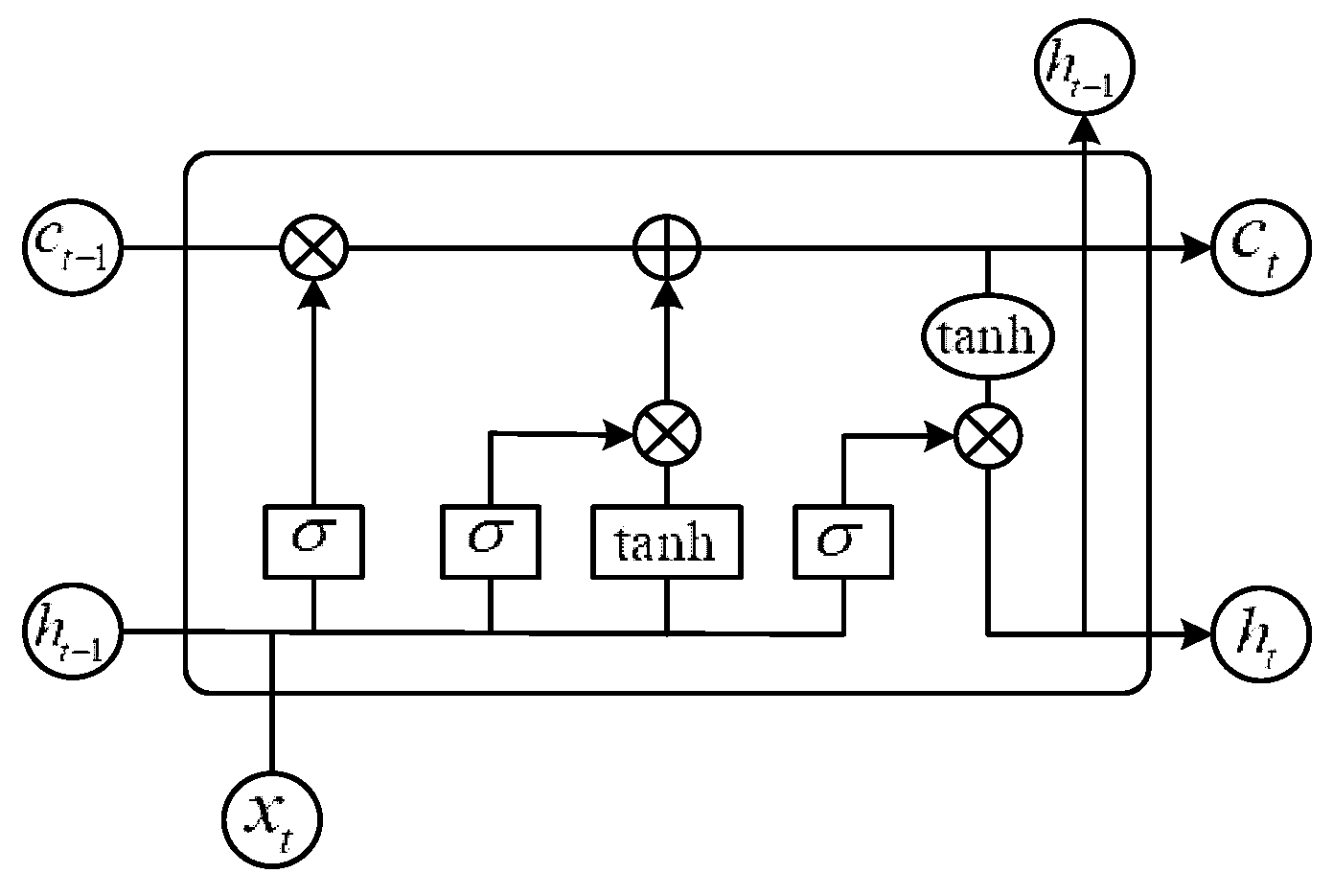
3.2. BILSTM Module
3.3. Decoding Unit
3.4. Multi-Task Learning Step
| Algorithm 1 Multi-task learning training process |
| Input: two task datasets DC and DN |
| Batch size K for each task |
| Maximum number of iterations T, learning rates α and β |
| Random initialization parameter θ0 |
| for t = 1 ⋯ T do |
| /*Prepare the data for both tasks*/ |
| Randomly divide DC and DN into small batch sets |
| BC = {JC,1, …, JC,n} |
| BN = {JN,1, …, JN,m} |
| end |
| Merge all small batch samples B′ = BC ∪ BN |
| Random sorting B′ |
| for each J B′ do |
| Calculate the loss L(θ) on the small batch sample |
| /* calculate only the loss of J on the corresponding task */ |
| Update the parameters: |
| end |
| end |
4. Experiments and Results Analysis
4.1. Dataset
4.2. Evaluation Metrics
4.3. Experimental Environment and Parameter Settings
4.4. Benchmark
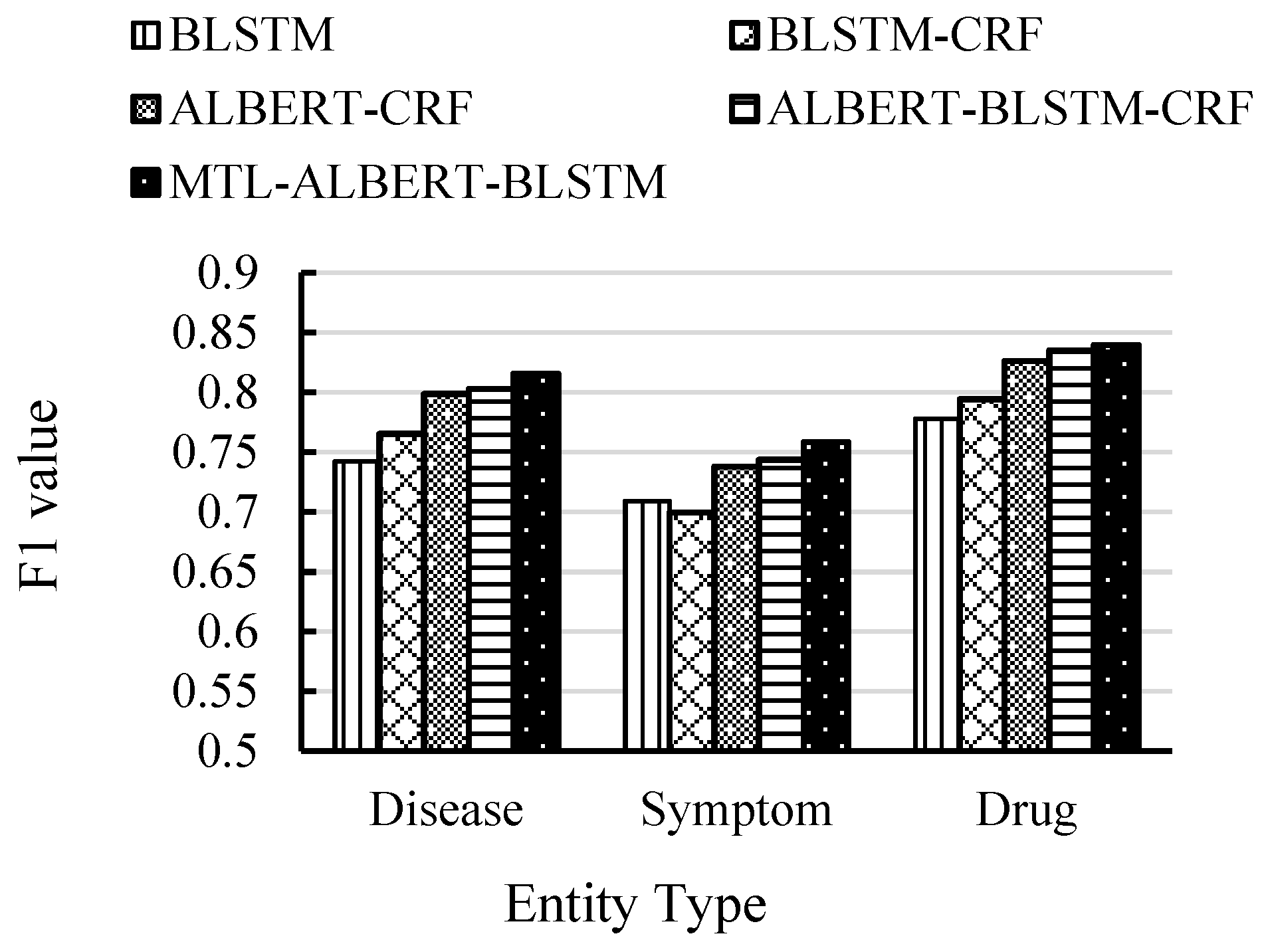
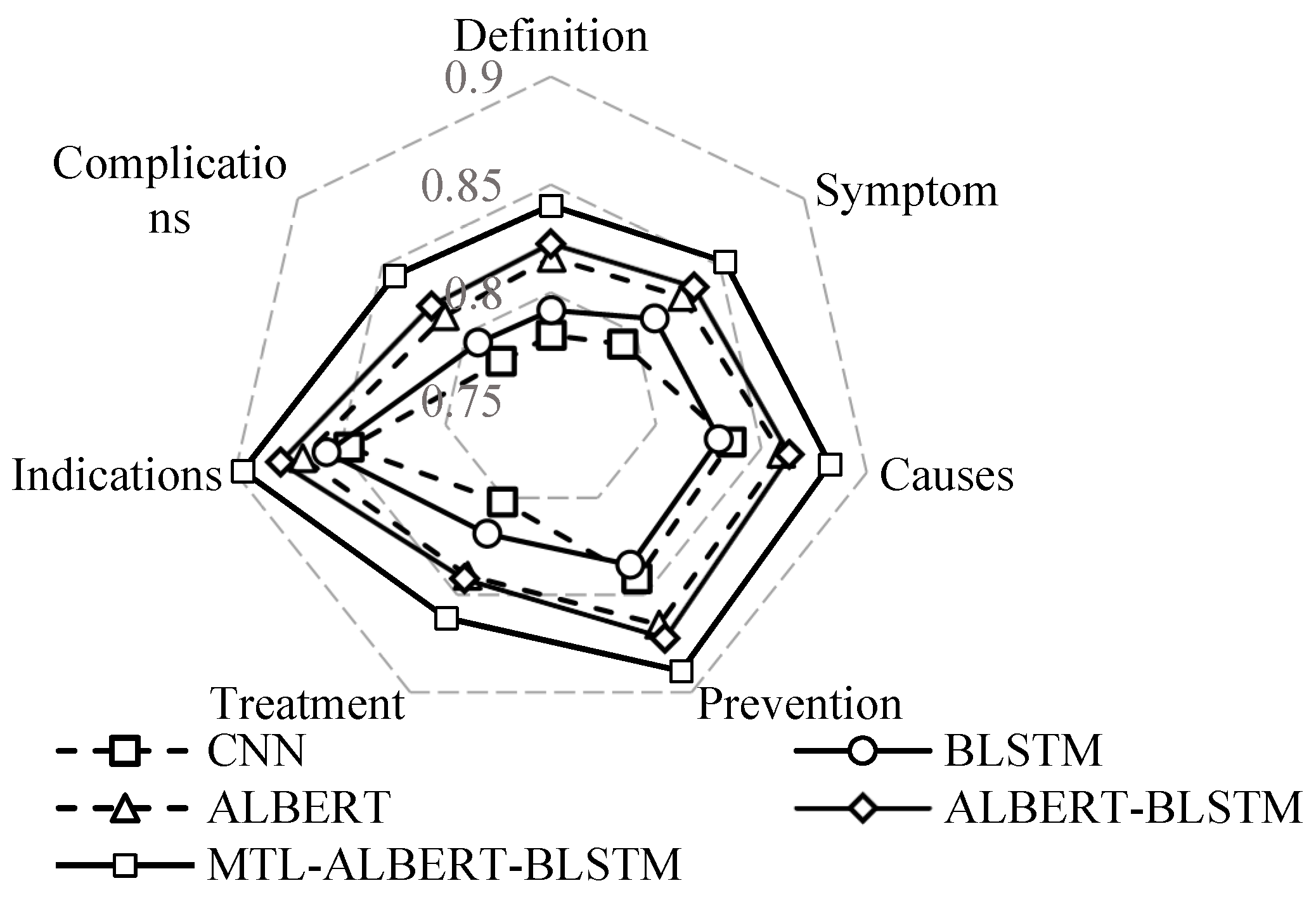
4.5. Experimental Results and Analysis
5. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gerner, M.; Nenadic, G.; Bergman, C.M. LINNAEUS: A species name identification system for biomedical literature. BMC Bioinform. 2010, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.I.; Tsunoda, T.; Tamura, A.; Takagi, T. Toward information extraction: Identifying protein names from biological papers. Pac. Symp. Biocomput. 1998, 707, 707–718. [Google Scholar]
- He, L.; Yang, Z.; Lin, H.; Li, Y. Drug name recognition in biomedical texts: A machine-learning-based method. Drug Discov. Today 2014, 19, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, C.; Li, T.; Wu, H.; Zhao, X.; Ye, K.; Liao, J. Named entity recognition from Chinese adverse drug event reports with lexical feature based BiLSTM-CRF and tri-training. J. Biomed. Inform. 2019, 96, 103252. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xu, W.; Yu, K. Bidirectional LSTM-CRF Models for Sequence Tag-grog. arXiv 2015, arXiv:1508.01991. [Google Scholar]
- Yang, P.; Yang, Z.; Luo, L.; Lin, H.; Wang, J. An attention-based approach for chemical compound and drug named entity recognition. J. Comput. Res. Dev. 2018, 55, 1548–1556. [Google Scholar]
- Li, L.; Guo, Y. Biomedical named entity recognition with CNN-BILSTM-CRF. J. Chin. Inf. Process. 2018, 32, 116–122. [Google Scholar]
- Su, Y.; Liu, J.; Huang, Y. Entity Recognition Research in Online Medical Texts. Acta Sci. Nat. Univ. Pekin. 2016, 52, 1–9. [Google Scholar]
- Qin, Q.; Zhao, S.; Liu, C. A BERT-BiGRU-CRF Model for Entity Recognition of Chinese Electronic Medical Records. Complexity 2021, 2021, 6631837. [Google Scholar] [CrossRef]
- Ji, B.; Li, S.; Yu, J.; Ma, J.; Tang, J.; Wu, Q.; Tan, Y.; Liu, H.; Ji, Y. Research on Chinese medical named entity recognition based on collaborative cooperation of multiple neural network models. J. Biomed. Inform. 2020, 104, 103395. [Google Scholar] [CrossRef]
- Minaee, S.; Kalchbrenner, N.; Cambria, E.; Nikzad, N.; Chenaghlu, M.; Gao, J. Deep learning--based text classification: A comprehensive review. ACM Comput. Surv. (CSUR) 2021, 54, 1–40. [Google Scholar] [CrossRef]
- Ravuri, S.; Stolcke, A. A comparative study of recurrent neural network models for lexical domain classification. In Proceedings of the 41st IEEE International Conference on Acoustics, Speech, and Signal Processing, Shanghai, China, 20–25 March 2016; pp. 6075–6079. [Google Scholar]
- Zhang, S.; Grave, E.; Sklar, E.; Elhadad, N. Longitudinal analysis of discussion topics in an online breast cancer community using convolutional neural networks. J. Biomed. Inform. 2017, 69, 1–9. [Google Scholar] [CrossRef]
- Yao, L.; Mao, C.; Luo, Y. Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. BMC Med. Inform. Decis. Mak. 2019, 19, 31–39. [Google Scholar] [CrossRef]
- Jang, B.; Kim, M.; Harerimana, G.; Kang, S.U.; Kim, J.W. Bi-LSTM model to increase accuracy in text classification: Combining Word2vec CNN and attention mechanism. Appl. Sci. 2020, 10, 5841. [Google Scholar] [CrossRef]
- Zhang, Q.; Yuan, Q.; Lv, P.; Zhang, M.; Lv, L. Research on Medical Text Classification Based on Improved Capsule Network. Electronics 2022, 11, 2229. [Google Scholar] [CrossRef]
- Zaib, M.; Sheng, Q.Z.; Emma Zhang, W. A short survey of pre-trained language models for conversational ai-a new age in nlp. In Proceedings of the Australasian Computer Science Week Multiconference, Melbourne, VIC, Australia, 4–6 February 2020; pp. 1–4. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; Association for Computational Linguistics: Stroudsburg, PA, USA, 2019; pp. 4171–4186. [Google Scholar]
- Song, Z.; Xie, Y.; Huang, W.; Wang, H. Classification of traditional chinese medicine cases based on character-level bert and deep learning. In Proceedings of the 2019 IEEE 8th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 24–26 May 2019; pp. 1383–1387. [Google Scholar]
- Yao, L.; Jin, Z.; Mao, C.; Zhang, Y.; Luo, Y. Traditional Chinese medicine clinical records classification with BERT and domain specific corpora. J. Am. Med. Inform. Assoc. 2019, 26, 1632–1636. [Google Scholar] [CrossRef]
- Lan, Z.; Chen, M.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. Albert: A lite bert for self-supervised learning of language representations. arXiv 2019, arXiv:1909.11942. [Google Scholar]
- Zhang, Z.; Jin, L. Clinical short text classification method based on ALBERT and GAT. In Proceedings of the 2022 7th International Conference on Intelligent Computing and Signal Processing (ICSP), Xi’an, China, 15–17 April 2022; pp. 401–404. [Google Scholar]
- Yang, Q.; Shang, L. Multi-task learning with bidirectional language models for text classification. In Proceedings of the International Joint Conference on Neural Network (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar]
- Ruder, S. An overview of multi-task learning in deep neural networks. arXiv 2017, arXiv:1706.05098. [Google Scholar]
- Liu, P.; Qiu, X.; Huang, X. Recurrent neural network for text classification with multi-task learning. arXiv 2016, arXiv:1605.05101. [Google Scholar]
- Wu, Q.; Peng, D. MTL-BERT: A Multi-task Learning Model Utilizing Bert for Chinese Text. J. Chin. Comput. Syst. 2021, 42, 291–296. [Google Scholar]
- Chowdhury, S.; Dong, X.; Qian, L.; Li, X.; Guan, Y.; Yang, J.; Yu, Q. A multitask bi-directional RNN model for named entity recognition on Chinese electronic medical records. BMC Bioinform. 2018, 19, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, T.; Zhao, S.; Wang, F. A neural multi-task learning framework to jointly model medical named entity recognition and normalization. In Proceedings of the AAAI Conference on Artificial Intelligence, Budapest, Hungary, 27 January–1 February 2019; Volume 33, pp. 817–824. [Google Scholar] [CrossRef]
- Peng, Y.; Chen, Q.; Lu, Z. An Empirical Study of Multi-Task Learning on BERT for Biomedical Text Mining. BioNLP 2020, 2020, 205. [Google Scholar]
- Yang, Z.; Dai, Z.; Yang, Y.; Carbonell, J.; Salakhutdinov, R.R.; Le, Q.V. Xlnet: Generalized autoregressive pretraining for language understanding. In Proceedings of the 33rd International Conference on Neural Information Processing Systems (NeurIPS), Vancouver, BC, Canada, 8–14 December 2019; pp. 5753–5763. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural machine translation by jointly learning to align and translate. arXiv 2014, arXiv:1409.0473. [Google Scholar]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer normalization. arXiv 2016, arXiv:1607.06450. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short memory. Neural Comput. 2014, 9, 1735–1780. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, X.; Wang, H.; Cheng, J.; Li, P.; Ding, Z. Chinese medical question answer matching using end-to-end character-level multi-scale CNNs. Appl. Sci. 2017, 7, 767. [Google Scholar] [CrossRef]
- Chen, N.; Su, X.; Liu, T.; Hao, Q.; Wei, M. A benchmark dataset and case study for Chinese medical question intent classification. BMC Med. Inform. Decis. Mak. 2020, 20, 125. [Google Scholar] [CrossRef]
| Category | Quantity |
|---|---|
| Definition | 963 |
| Symptom | 1215 |
| Causes | 2177 |
| Prevention | 1205 |
| Treatment | 2506 |
| Indications | 1572 |
| Complications | 879 |
| Method | P | R | F1 |
|---|---|---|---|
| BILSTM | 0.7515 | 0.7432 | 0.7473 |
| BILSTM-CRF | 0.7578 | 0.7641 | 0.7606 |
| ALBERT-CRF | 0.7869 | 0.7954 | 0.7911 |
| ALBERT-BILSTM-CRF | 0.7926 | 0.8014 | 0.7970 |
| MTL-ALBERT-BILSTM | 0.8103 | 0.8036 | 0.8069 |
| Method | P | R | F1 |
|---|---|---|---|
| CNN | 0.8315 | 0.7921 | 0.8113 |
| BILSTM | 0.8377 | 0.8086 | 0.8229 |
| ALBERT | 0.8582 | 0.8391 | 0.8485 |
| ALBERT-BILSTM | 0.8634 | 0.8473 | 0.8553 |
| MTL-ALBERT-BILSTM | 0.8842 | 0.8654 | 0.8747 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tohti, T.; Abdurxit, M.; Hamdulla, A. Medical QA Oriented Multi-Task Learning Model for Question Intent Classification and Named Entity Recognition. Information 2022, 13, 581. https://doi.org/10.3390/info13120581
Tohti T, Abdurxit M, Hamdulla A. Medical QA Oriented Multi-Task Learning Model for Question Intent Classification and Named Entity Recognition. Information. 2022; 13(12):581. https://doi.org/10.3390/info13120581
Chicago/Turabian StyleTohti, Turdi, Mamatjan Abdurxit, and Askar Hamdulla. 2022. "Medical QA Oriented Multi-Task Learning Model for Question Intent Classification and Named Entity Recognition" Information 13, no. 12: 581. https://doi.org/10.3390/info13120581
APA StyleTohti, T., Abdurxit, M., & Hamdulla, A. (2022). Medical QA Oriented Multi-Task Learning Model for Question Intent Classification and Named Entity Recognition. Information, 13(12), 581. https://doi.org/10.3390/info13120581






