Dual-Channel Heterogeneous Graph Network for Author Name Disambiguation
Abstract
:1. Introduction
- Heterogeneous graphs incorporate more comprehensive information into node clustering, which allows our method to capture more comprehensive information.
- The semantic information extracted by fastText is merged with the relational information extracted from the heterogeneous graph, and the nodes are clustered using DBSCAN.
- Extensive experiments on real-world datasets prove the effectiveness of our method.
2. Related Work
2.1. Heterogeneous Graph Network
2.2. Author Name Disambiguation
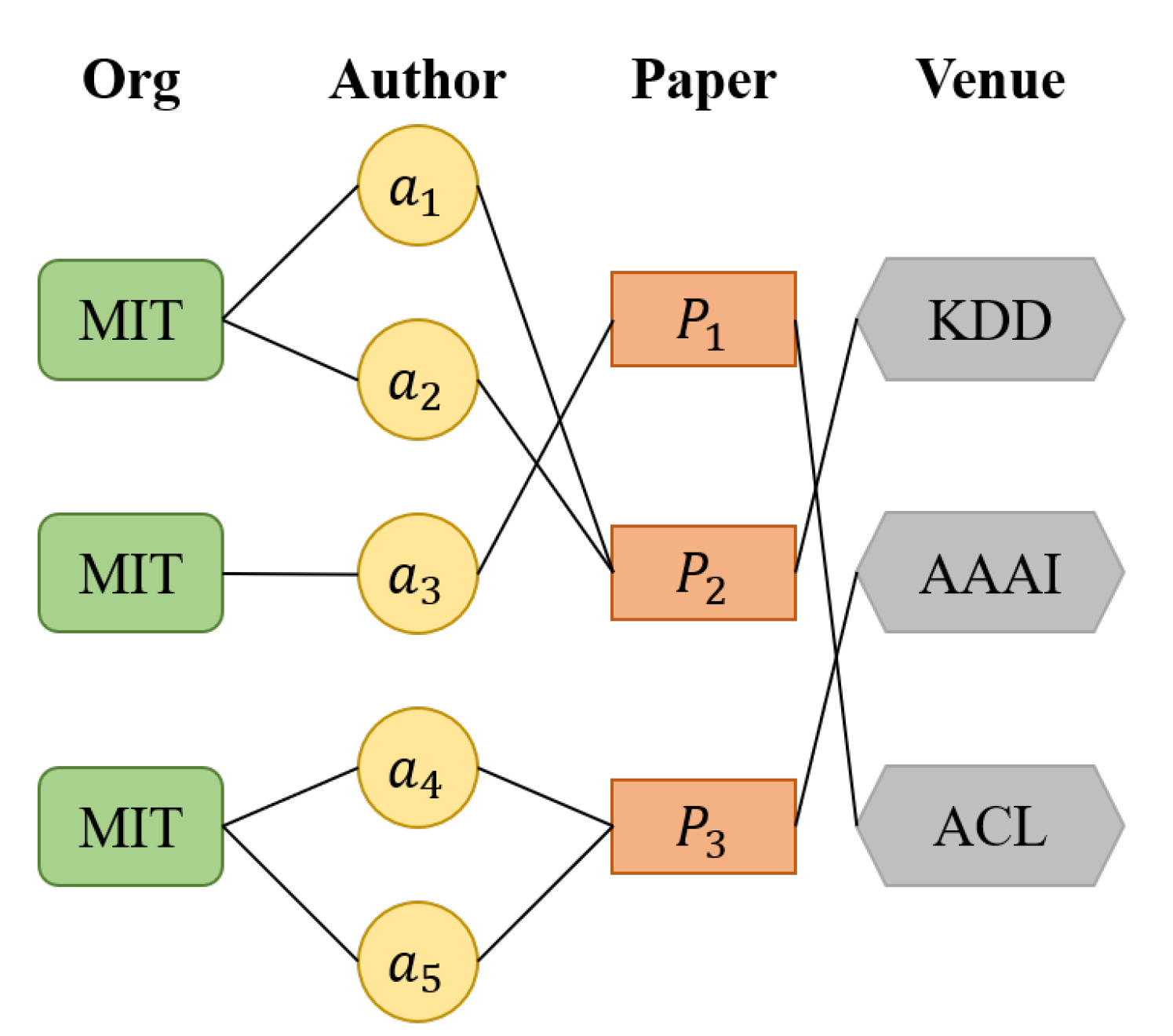
3. Problem Formulation
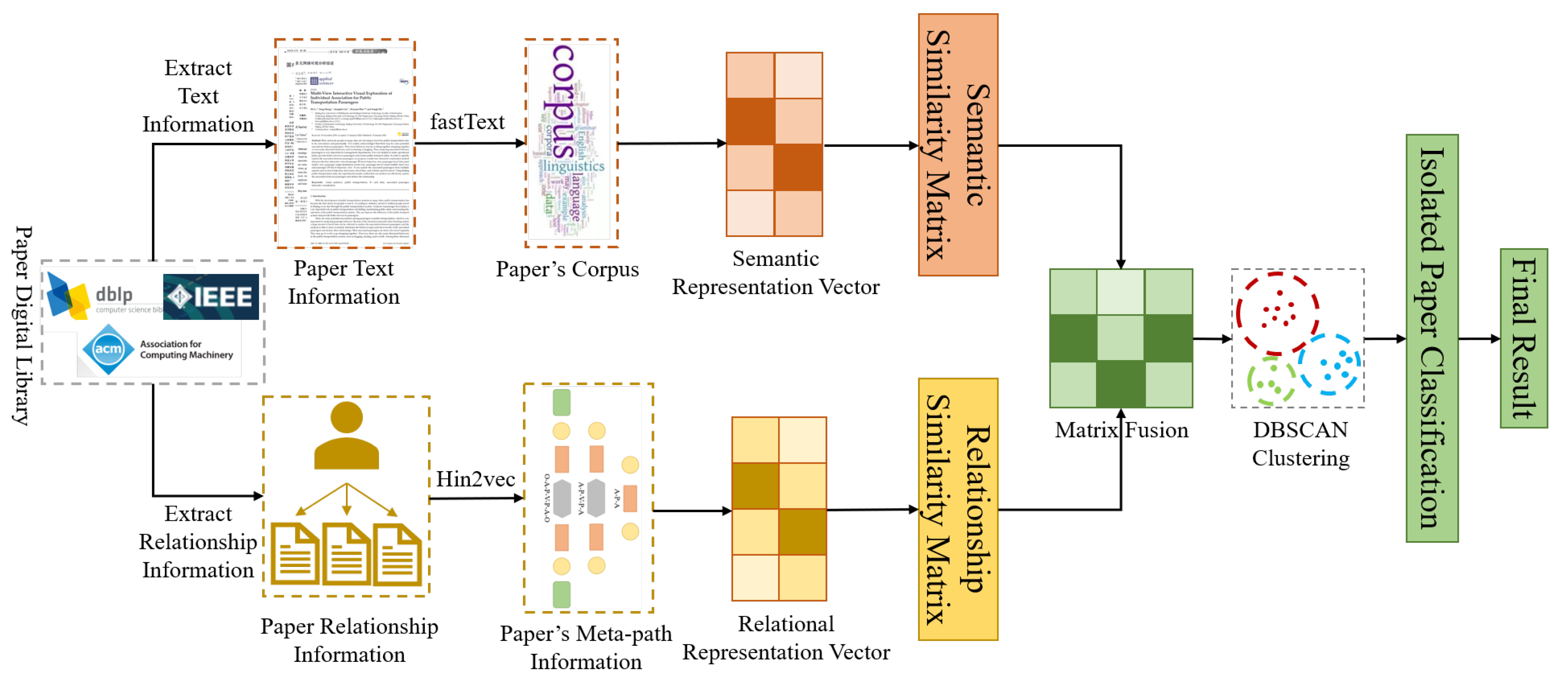
4. DHGN: The Proposed Method for Author Name Disambiguation
4.1. Use FastText to Construct Semantic Representation Vector
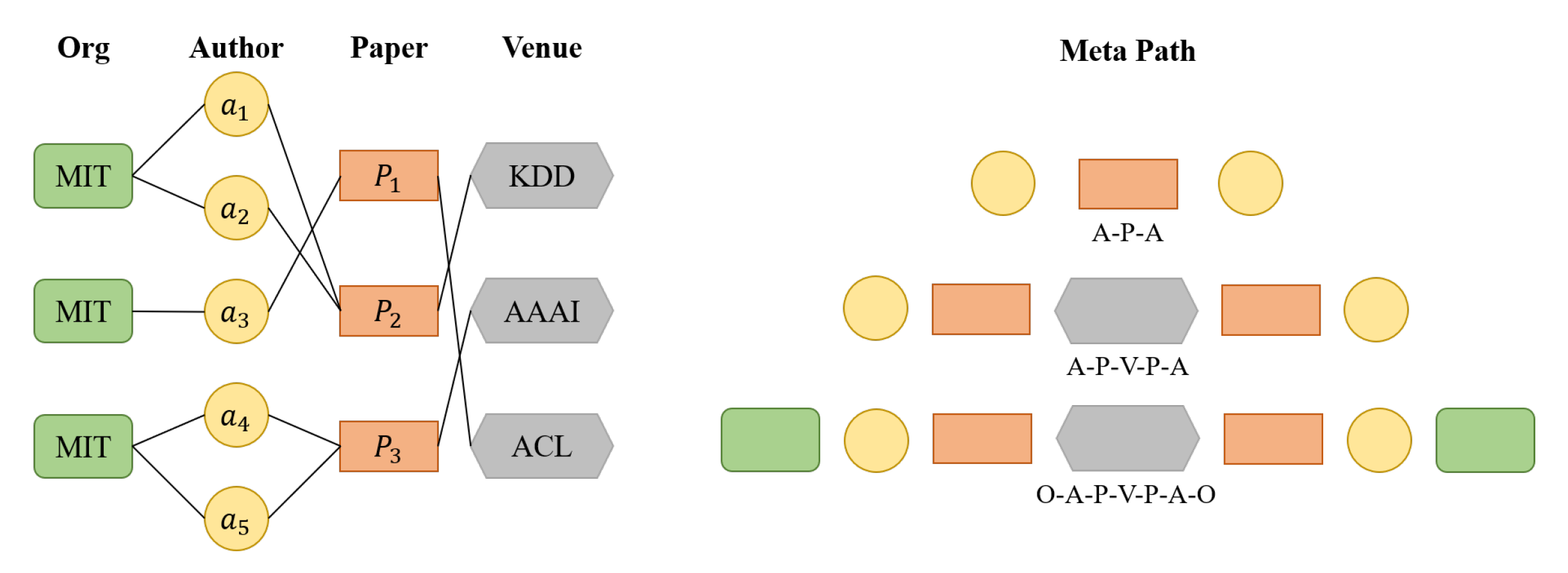
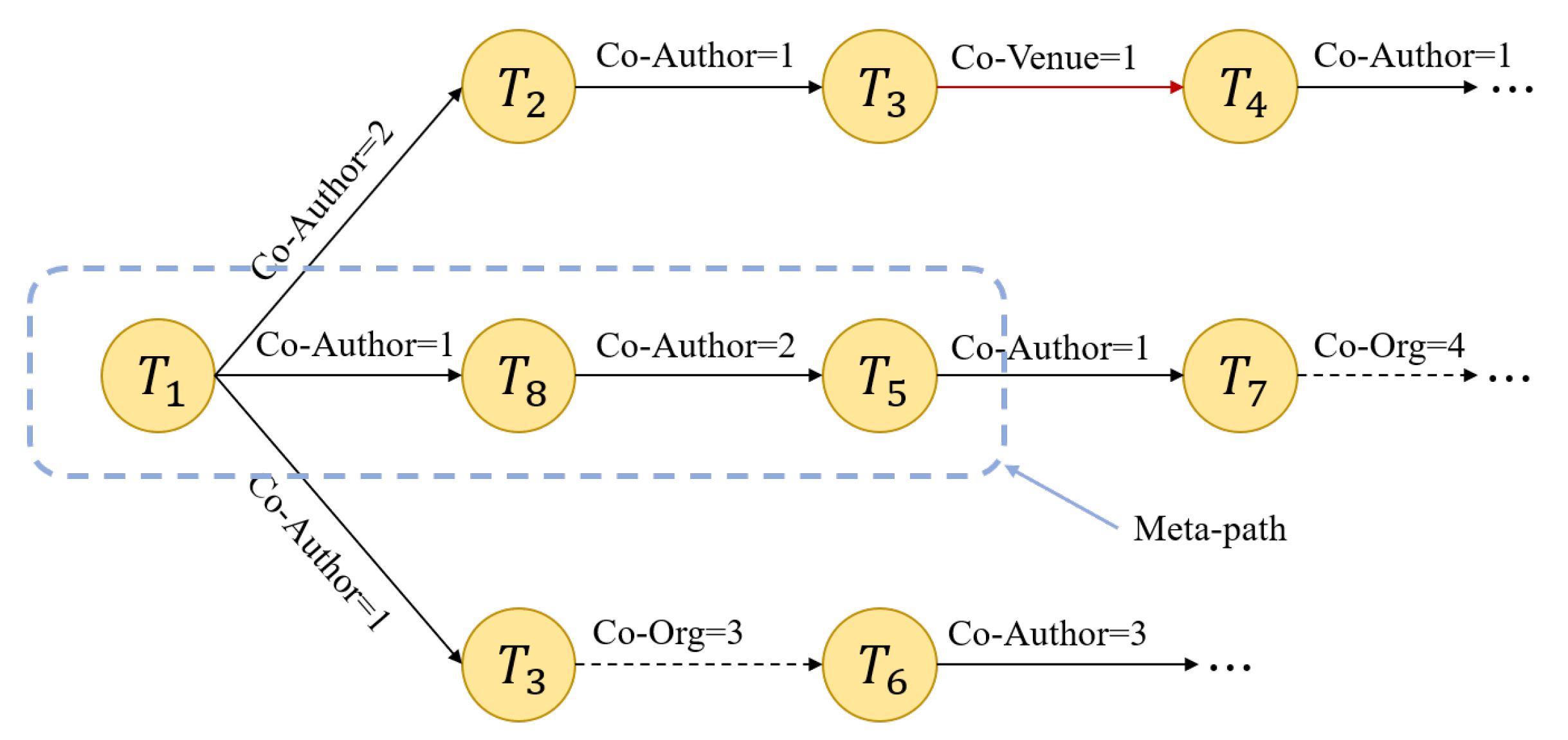
4.2. Use Heterogeneous Graph to Construct Relational Representation Vector
4.3. Use DBSCAN for Node Clustering
5. Experimental Results and Analysis
5.1. Datasets
- (1)
- The author’s name. The name of the same author is usually written in different ways in different papers. For example, the author’s name is “Huang JianCheng”, which can be written in many ways, such as “Huang_JianCheng”, “Huang_Jian_Cheng”, and “JianCheng Huang”. It is necessary to unify the name into one written form during data preprocessing.
- (2)
- The author’s institution, name of the journal, name of the conference, abstract, and title. This information usually contains a large number of special symbols. Special symbols need to be removed during preprocessing, and the institution names need to be converted to the lowercase English form.
- (3)
- Keywords. Observations indicate that keywords generally do not contain special symbols. Keywords need to be converted to lowercase.
- (4)
- Escape characters. Some papers contain escape characters. For example, “\u03b2” means “” in the paper. We found that removing such escape characters has little impact on our research results, so we deleted them. We also found that if special symbols are removed in step (2), escape characters cannot be effectively removed. Therefore, it is necessary to remove escape characters before performing step (2).
5.2. Baselines
5.3. Results
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Levin, M.; Krawczyk, S.; Bethard, S.; Jurafsky, D. Citation-based bootstrapping for large-scale author disambiguation. J. Am. Soc. Inf. Sci. Technol. 2012, 63, 1030–1047. [Google Scholar] [CrossRef] [Green Version]
- Bo, D.; Wang, X.; Shi, C.; Zhu, M.; Lu, E.; Cui, P. Structural deep clustering network. In Proceedings of the Web Conference 2020, Taipei, Taiwan, 20–24 April 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 1400–1410. [Google Scholar]
- Wang, R.; Mou, S.; Wang, X.; Xiao, W.; Ju, Q.; Shi, C.; Xie, X. Graph Structure Estimation Neural Networks. In Proceedings of the Web Conference 2021, Ljubljana, Slovenia, 19–23 April 2021; Association for Computing Machinery: New York, NY, USA, 2021; pp. 342–353. [Google Scholar]
- Hu, B.; Zhang, Z.; Shi, C.; Zhou, J.; Li, X.; Qi, Y. Cash-out user detection based on attributed heterogeneous information network with a hierarchical attention mechanism. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January 27–1 February 2019; AAAI Press: Palo Alto, CA, USA, 2019; Volume 33, pp. 946–953. [Google Scholar]
- Sun, Y.; Han, J. Mining heterogeneous information networks: Principles and methodologies. Synth. Lect. Data Min. Knowl. Discov. 2012, 3, 1–159. [Google Scholar] [CrossRef]
- Nandanwar, S.; Moroney, A.; Murty, M.N. Fusing diversity in recommendations in heterogeneous information networks. In Proceedings of the Eleventh ACM International Conference on Web Search and Data Mining, Los Angeles, CA, USA, 5–9 February 2018; pp. 414–422. [Google Scholar]
- Shi, C.; Hu, B.; Zhao, W.X.; Philip, S.Y. Heterogeneous information network embedding for recommendation. IEEE Trans. Knowl. Data Eng. 2018, 31, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Anchiêta, R.T.; Sousa, R.F.d.; Pardo, T.A. Modeling the Paraphrase Detection Task over a Heterogeneous Graph Network with Data Augmentation. Information 2020, 11, 422. [Google Scholar] [CrossRef]
- Han, H.; Zha, H.; Giles, C.L. A model-based k-means algorithm for name disambiguation. In Proceedings of the 2nd International Semantic Web Conference (ISWC-03) Workshop on Semantic Web Technologies for Searching and Retrieving Scientific Data, Sanibel Island, FL, USA, 20–23 October 2003. [Google Scholar]
- Han, H.; Giles, L.; Zha, H.; Li, C.; Tsioutsiouliklis, K. Two supervised learning approaches for name disambiguation in author citations. In Proceedings of the 2004 Joint ACM/IEEE Conference on Digital Libraries, Tucson, AZ, USA, 11 June 2004; pp. 296–305. [Google Scholar]
- Kang, I.S.; Na, S.H.; Lee, S.; Jung, H.; Kim, P.; Sung, W.K.; Lee, J.H. On co-authorship for author disambiguation. Inf. Process. Manag. 2009, 45, 84–97. [Google Scholar] [CrossRef]
- Shin, D.; Kim, T.; Jung, H.; Choi, J. Automatic method for author name disambiguation using social networks. In Proceedings of the 2010 24th IEEE International Conference on Advanced Information Networking and Applications, Perth, Australia, 20–23 April 2010; pp. 1263–1270. [Google Scholar]
- Schulz, C.; Mazloumian, A.; Petersen, A.M.; Penner, O.; Helbing, D. Exploiting citation networks for large-scale author name disambiguation. EPJ Data Sci. 2014, 3, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Shen, D.; Kou, Y.; Nie, T. Author name disambiguation for citations on the deep web. In International Conference on Web-Age Information Management; Springer: Berlin/Heidelberg, Germany, 2010; pp. 198–209. [Google Scholar]
- Sun, Y.; Han, J. Meta-path-based search and mining in heterogeneous information networks. Tsinghua Sci. Technol. 2013, 18, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Cai, H.; Zheng, V.W.; Chang, K.C.C. A comprehensive survey of graph embedding: Problems, techniques, and applications. IEEE Trans. Knowl. Data Eng. 2018, 30, 1616–1637. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Zhang, Z.; Luo, P.; Yu, P.S.; Yue, Y.; Wu, B. Semantic path based personalized recommendation on weighted heterogeneous information networks. In Proceedings of the 24th ACM International on Conference on Information and Knowledge Management, Melbourne, Australia, 18–23 October 2015; pp. 453–462. [Google Scholar]
- Joulin, A.; Grave, E.; Bojanowski, P.; Mikolov, T. Bag of tricks for efficient text classification. arXiv 2016, arXiv:1607.01759. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Hinton, G.E. Learning distributed representations of concepts. In Proceedings of the Eighth Annual Conference of the Cognitive Science Society, Amherst, MA, USA, 15–17 August 1986; Volume 1, p. 12. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; Curran Associates Inc.: Long Beach, CA, USA, 2017; pp. 5998–6008. [Google Scholar]
- Shi, C.; Li, Y.; Zhang, J.; Sun, Y.; Philip, S.Y. A survey of heterogeneous information network analysis. IEEE Trans. Knowl. Data Eng. 2016, 29, 17–37. [Google Scholar] [CrossRef]
- Dong, Y.; Chawla, N.V.; Swami, A. metapath2vec: Scalable representation learning for heterogeneous networks. In Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and DATA Mining, Halifax, NS, Canada, 13–17 August 2017; pp. 135–144. [Google Scholar]
- Fu, T.y.; Lee, W.C.; Lei, Z. Hin2vec: Explore meta-paths in heterogeneous information networks for representation learning. In Proceedings of the 2017 ACM on Conference on Information and Knowledge Management, 6–10 November 2017; pp. 1797–1806. [Google Scholar]
- Gori, M.; Monfardini, G.; Scarselli, F. A new model for learning in graph domains. In Proceedings of the 2005 IEEE International Joint Conference on Neural Networks, Montreal, QC, Canada, 31 July–4 August 2005; Volume 2, pp. 729–734. [Google Scholar]
- Agesen, O. The cartesian product algorithm. In European Conference on Object-Oriented Programming; Springer: Berlin/Heidelberg, Germany, 1995; pp. 2–26. [Google Scholar]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. Density-based spatial clustering of applications with noise. In Proceedings of the International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; Volume 240, p. 6. [Google Scholar]
- Yin, J.; Wang, J. A model-based approach for text clustering with outlier detection. In Proceedings of the 2016 IEEE 32nd International Conference on Data Engineering (ICDE), Helsinki, Finland, 16–20 May 2016; pp. 625–636. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Cao, S.; Lu, W.; Xu, Q. Grarep: Learning graph representations with global structural information. In Proceedings of the 24th ACM International on Conference on Information and Knowledge Management, Melbourne, Australia, 18–23 October 2015; pp. 891–900. [Google Scholar]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]

| Parameter | Parameter Description | Parameter Settings |
|---|---|---|
| sentence | training text per line | Academic paper information |
| size | dimension of word vector | 100 |
| windows | context word range | 5 |
| sg | CBOW or Skip-gram | 0 |
| hs | optimal strategy | 1 |
| min_n | minimum number of characters | 3 |
| max_n | maximum number of characters | 6 |
| Frequency | Total Number of Words |
|---|---|
| less than or equal to 10 times | 316,802 |
| 10–100 times | 49,972 |
| more than 100 times | 19,738 |
| Parameter | Parameter Description | Parameter Settings |
|---|---|---|
| walk | times of random Walks | 5 |
| walk_length | random walk length | 20 |
| embed_size | dimension of word vector | 100 |
| n_epoch | number of training sessions | 5 |
| batch_size | number of training sample | 20 |
| Parameter | Parameter Description | Parameter Settings |
|---|---|---|
| eps | cluster neighborhood threshold | 0.2 |
| min_simples | minimum number of samples in cluster | 4 |
| metric | distance measurement | precomputes |
| Field | Data Type | Meaning | Example |
|---|---|---|---|
| Name | String | Name to be disambiguated | Li_guo |
| Name ID | String | Author ID | sCKCrny5 |
| ID | String | Paper ID | UG32p2zs |
| Field | Data Type | Meaning | Example |
|---|---|---|---|
| ID | String | Paper ID | P9a1gcvg |
| Title | String | Paper Title | Rapid determination of central nervous drugs in plasma by solid-phase extraction and GC-FID and GC-MS |
| Venue | String | Journal/conference | Chinese Pharmaceutical Journal |
| Author.name | String | Author’s name | Li Guo |
| Author.org | String | Author’s organization | Institute of Pharmacology and Toxicology |
| Keywords | String | Paper’s keywords | Cholecystokinin-4; Enzymatic synthesis; Peptide; |
| Abstract | String | Paper’s abstract | The enzymatic synthesis of CCK-8 tripeptide derivative Phac-Met-Asp(OMe)-Phe-NH |
| Year | Int | Paper’s publication year | 2019 |
| GSDPMM | LightGBM | TF-IDF | GraRep | metapath2vec | Deepwalk | Ours | |
|---|---|---|---|---|---|---|---|
| Precision | 0.5987 | 0.5896 | 0.3721 | 0.6190 | 0.6179 | 0.6024 | 0.6834 |
| Recall | 0.5921 | 0.8818 | 0.8755 | 0.6010 | 0.6172 | 0.6142 | 0.6872 |
| F1 | 0.5181 | 0.5796 | 0.4094 | 0.6103 | 0.6135 | 0.6029 | 0.6242 |
| Name | Paper’s Number | Author’s Number | Precision | Recall | F1 |
|---|---|---|---|---|---|
| Kenji_kaneko | 148 | 5 | 0.7258 | 0.7701 | 0.7473 |
| guohua_chen | 828 | 24 | 0.8057 | 0.6524 | 0.7210 |
| hai_jin | 56 | 5 | 1.0000 | 0.6051 | 0.7540 |
| guoliang_li | 744 | 36 | 0.7149 | 0.8467 | 0.7752 |
| jiang_he | 416 | 10 | 1.0000 | 0.9855 | 0.9927 |
| jianping_wu | 219 | 21 | 0.9980 | 0.7229 | 0.8384 |
| peng_shi | 815 | 11 | 0.7288 | 0.8545 | 0.7867 |
| xiaoyang_zhang | 447 | 30 | 0.9755 | 0.9624 | 0.9689 |
| mei_han | 388 | 19 | 0.9786 | 0.9108 | 0.9435 |
| d_zhang | 223 | 16 | 0.8905 | 0.8971 | 0.8938 |
| akira_ono | 195 | 9 | 0.7508 | 0.9504 | 0.8389 |
| bin_gao | 398 | 16 | 0.9603 | 0.8812 | 0.9190 |
| chao_yuan | 555 | 63 | 0.7587 | 0.9367 | 0.8383 |
| hong_yan_wang | 84 | 18 | 0.7589 | 0.8718 | 0.8115 |
| c_c_lin | 105 | 9 | 0.8249 | 0.8205 | 0.8227 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Zhang, P.; Cui, Y.; Du, R.; Zhang, Y. Dual-Channel Heterogeneous Graph Network for Author Name Disambiguation. Information 2021, 12, 383. https://doi.org/10.3390/info12090383
Zheng X, Zhang P, Cui Y, Du R, Zhang Y. Dual-Channel Heterogeneous Graph Network for Author Name Disambiguation. Information. 2021; 12(9):383. https://doi.org/10.3390/info12090383
Chicago/Turabian StyleZheng, Xin, Pengyu Zhang, Yanjie Cui, Rong Du, and Yong Zhang. 2021. "Dual-Channel Heterogeneous Graph Network for Author Name Disambiguation" Information 12, no. 9: 383. https://doi.org/10.3390/info12090383
APA StyleZheng, X., Zhang, P., Cui, Y., Du, R., & Zhang, Y. (2021). Dual-Channel Heterogeneous Graph Network for Author Name Disambiguation. Information, 12(9), 383. https://doi.org/10.3390/info12090383







