Abstract
Colorectal cancer is one of the main causes of cancer incident cases and cancer deaths worldwide. Undetected colon polyps, be them benign or malignant, lead to late diagnosis of colorectal cancer. Computer aided devices have helped to decrease the polyp miss rate. The application of deep learning algorithms and techniques has escalated during this last decade. Many scientific studies are published to detect, localize, and classify colon polyps. We present here a brief review of the latest published studies. We compare the accuracy of these studies with our results obtained from training and testing three independent datasets using a convolutional neural network and autoencoder model. A train, validate and test split was performed for each dataset, 75%, 15%, and 15%, respectively. An accuracy of 0.937 was achieved for CVC-ColonDB, 0.951 for CVC-ClinicDB, and 0.967 for ETIS-LaribPolypDB. Our results suggest slight improvements compared to the algorithms used to date.
1. Introduction
Medical imaging has gained immense importance in healthcare throughout history. It has been used in diagnosing diseases, planning treatments, and assessing results. Furthermore, medical imaging is currently used in preventing illness, usually through screening programs. Aggregating it with demographic and other healthcare data can bring novel insights and help scientists discover breakthrough treatments [1].
A lot of research has been done in automating the delivery of medical imaging results. These results still rely on professional radiologists being present when finalizing them. However, automation can help radiologists be more efficient in their job and deliver results quicker.
A review of deep learning (DL) applications in medical imaging [2] shows that AI algorithms will have a significant impact in the healthcare field. The application areas span from digital pathology and microscopy to brain, eye, chest, breast, cardiac, abdomen, etc. These algorithms are for all types of imaging machines used nowadays: computed tomography (CT), ultrasound, MRI, X-ray, microscope, cervigram, photographs, endoscopy/ colonoscopy, tomosynthesis (TS), mammography, etc. Most of these applications deal with classification, segmentation, or detection problems and convolutional neural networks (CNNs), auto-encoders (AE) or stacked auto-encoders (SAE), recurrent neural networks (RNNs), deep belief networks, and restricted Boltzmann machines (RBM) are the most used architectures for these settings. The architecture of some of the most used algorithms is depicted in Figure 1.
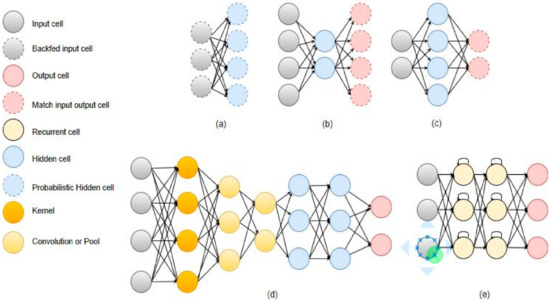
Figure 1.
Graph representation of some of the commonly used architectures in medical imaging. (a) AE, (b) RBM, (c) RNN, (d) CNN, (e) MS-CNN.
In this paper, we focus on colorectal cancer (CRC) and how deep learning algorithms can help detect colon polyps. The World Health Organization, through the International Agency for Research on Cancer, has recognized colorectal cancer as responsible for around 881 thousand deaths, or 9.2% of the total cancer deaths [3]. The main concern is that the incidence rates have been rising, more than 1.85 million cases [3]. This increase could be prevented by conducting effective screening test [4]. However, a 2020 European study on colorectal cancer shows that total cancer mortality rates are predicted to decline, and these numbers for colorectal cancer are 4.2% in men and 8.3% in women [5]. These declines are expected in all age groups [6]. Another study done in the USA shows declining numbers in the USA as well [7]. The implementation of screening programs is an essential factor in the declining numbers various countries have seen. Colonoscopy is the preferred technique among the used screening tests to diagnose CRC. It is also used as a prevention procedure for CRC. CRC starts as growth in the lining of the colon or rectum. These growths are called polyps. Polyps are benign neoplasms; some types can transform into CRC over the years. Within the latter are adenomatous polyps and serrated polyps. Not all polyps develop into CRC. The adenomatous colon polyps (adenomas) and polyps larger than 1 cm have a higher risk of malignancy. Sometimes polyps are flat or hide between the folds of the colon, which makes their detection difficult.
One of the procedures to screen for colon polyps is the colonoscopy, which examines the large bowel and the distal part of the small bowel with a camera. The advantages of this procedure include visualization of the polyps and their removal before they grow bigger and, for biopsy purposes, if the medical personnel suspect a cancerous polyp. According to [7], colonoscopy is very well established as a procedure to prevent the development of CRC playing a significant role in rapid declines in incidence cases during the 2000s but not so much during the recent years. Another study on the impact of CRC screening mortality found that using colonoscopy indicates a more than 50% decline for CRC mortality [6]. Although colonoscopy has shown meaningful improvements, the colon polyp miss rate continues the same. A 2017 retrospective study done with 659 patients indicates that among these patients, the colon polyp miss rate was 17% (372 out of 2158 polyps), and 39% of patients (255 out of 659 patients) had at least one missed polyp [8]. As mentioned before, an undetected polyp, be it benign or malignant, may lead to a late CRC diagnosis, which is associated with a less than 10% survival rate for metastatic CRC. Many elements contribute to missed polyps during a colonoscopy. Two of them are the quality of bowel preparation and the experience of the colonoscopists [9]. While the first problem cannot be fixed by technology, the second one can, and computer-aided tools can assist colonoscopists in detecting polyps and reducing polyp miss rates.
The aims of this study are to give an overview of the recent deep learning algorithms used in colorectal images and videos and introduce a new model for colon polyp detection in images. The rest of the paper is organized as follows. Section 2 presents the recent techniques explicitly used in colon polyp detection, classification and localization in colonoscopy images and videos. Section 3 describes the databases we used to train, validate, and test our proposed model. Section 4 presents the results. We close the paper with Section 5, discussions and conclusions, where we also present the limitations and future work.
The key contributions of this paper are: (i) presenting the state-of-the-art in deep learning techniques to detect, classify, and localize colon polyps; and (ii) introducing the convolutional neural network with autoencoders (CNN-AE) algorithm for detection of polyps with no previous image pre-processing.
2. Background
Researchers have been applying deep learning techniques and algorithms in various healthcare applications. Considerable progress is seen in detecting colon polyps [10,11]. Having a public database of colon polyp images played a big role. Examples of such contributions include using a pre-trained deep convolutional neural network to detect colon polyps [10], dividing images into small patches or in sub-images to increase the database′s size, and then classifying different regions of the same image [12]. Other works include exploring deep learning to automatically classify polyps using various configurations, such as training the CNN model from scratch or modifying different CNN architectures pre-trained in other databases and testing them in an 8-HD-endoscopic image database [13]. Authors in [14] take advantage of transfer learning, a technique where a model is trained on a task and later re-purposed and used for another task similar to the previous one. [14] uses CNN as a feature descriptor and to generate features for the classification of colon polyps. Another CNN was developed to detect hyperplastic and adenomatous polyps and classify them by modifying different low-level CNN layer features learned from non-medical datasets [15].
The authors in [16] use a deep CNN model as a transfer learning scheme. Besides image augmentation strategies for training deep networks, they propose two post-learning methods, automatic false-positive learning and offline learning. Shin & Balasingham (2017) [17] compare a handcraft feature method with a CNN method to classify colorectal images. For the handcraft feature approach, they use the shape and color features together with a support vector machine (SVM) for classification. On the other hand, the CNN approach uses three convolutional layers with pooling to do the same. They compare the strategies by testing them in three public polyp databases. Results show the CNN-based deep learning framework leads better classification performance by achieving an accuracy, sensitivity, specificity, and precision of over 90%. Authors in Korbar et al. [18] build an automatic image analysis method that classifies different types of colorectal polyps on whole-slide images with an accuracy of about 93%. Mahmood & Durr (2018) [19] use a deep CNN together with a conditional random field (CRF) called (CNN-CRF), a framework for estimating the depth of a monocular endoscopy. Estimated depth is used to reconstruct the topography of the surface of the colon from a single image. They train the framework on over 200,000 synthetic images of an anatomically realistic colon, which they generated by developing an endoscope camera model. The validation is done using endoscopy images from a porcine colon, transferred to a synthetic-like domain via adversarial training. The relative error of the CNN-CRF approach is 0.152 for synthetic endoscopy images and 0.242 for real endoscopy images. They show that the depth map can be used to reconstruct the mucosa topography.
Three 2020 studies focus more on polyp classification by approaching the problem in different ways. Carneiro et al. [20] studies the roles of confidence and classification uncertainty in deep learning models and proposes and tests a new Bayesian deep learning method to improve classification accuracy and model interpretability on a privately owned polyp image dataset. Gao et al. [21] use DL methods to establish colorectal lesion detection, positioning, and classification based on white light endoscopic images. The CNN model is used to detect whether the image contains lesions (CRC, colorectal adenoma, and other types of polyps), and the instance segmentation model is used to locate and classify the lesions on the images. They compare some of the most used CNN models to do so, such as ResNet50, AlexNet, VGG19, ResNet18, and GoogleNet. Song et al. [22] developed a computer-aided diagnostic system (CAD) for predicting colorectal polyp histology using deep-learning technology with near-focus narrow-band imaging (NBI) pictures of the privately-owned colorectal polyps image dataset. The performance of the CAD is validated with two test datasets. Polyps were classified into three histological groups. The CAD accuracy (81.3–82.4%) shows to be higher than that of trainee colonoscopists (63.8–71.8%) but comparable with that of expert colonoscopists (82.4–87.3%).
There are other works that are focused on colon polyp detection on colonoscopy videos besides images. Such work includes [23], where authors explore the idea of applying a deep CNN model to a large set of images taken from 20 videos approximately 5 h long (~500,000 frames). In [24], authors develop a three-dimensional (3D) CNN model and train it on 155 short videos. In [25] deep learning method called Y-Net is proposed that consists of two encoder networks with a decoder network that relies on efficient use of pre-trained and un-trained models with novel sum-skip-concatenation operations. The encoders are trained with a learning rate specific to encoders and the same for the decoder. Yu et al. (2017) [26] proposes an offline and online framework by leveraging the 3D fully convolutional network (3D-FCN). Their 3D-FCN framework is able to learn more representative spatial-temporal features from colonoscopy videos by showing more powerful discrimination capability. Their proposed online learning scheme deals with limited training data by harnessing the specific information of an input video in the learning process. They integrate offline learning to the online one to reduce the number of false positives, which brings detection performance improvements. Another work [27] includes using a deep CNN model based on inception network architecture trained in colonoscopy videos. They use only unaltered NBI video frames to train and validate the model. A test dataset of 125 videos of consecutively encountered diminutive polyps was used to test the model. However, the confidence mechanism of the model did not generate sufficient confidence to predict the detection of 19 polyps in the test set, which represented 15% of the polyps. In a more recent study, Poon et al. (2020) [11], the authors design an Artificial Intelligent Endoscopist (AI-doscopist) to localize polyps during colonoscopy with the purpose of evaluating the agreement between endoscopists and AI-doscopist for set localization. Another recent study that deals with colorectal videos is Wang et al. [28], which is the first double-blind, randomized controlled trial to assess the effectiveness of automatic polyp detection using the computer-aided detection (CADe) system during colonoscopy. To the best of our knowledge, this is also the only clinical trial that deals with the use of artificial intelligence (AI) in colorectal image/video detection, localization and/ or classification.
There are studies that train and test models in both images and videos. One of them is Yamada et al. [29], where they develop an AI system that detects early signs of colorectal cancer during colonoscopy by decomposing tensor metrics in the trained model. Their AI system consists of a Faster R-CNN and the VGG16 model. Table 1 summarizes the articles included in this minireview, together with some characteristics of these studies.

Table 1.
Summary of the reviewed work.
Our model is a combination of CNN and autoencoders. This model was trained on three different colon polyp databases, CVC-ColonDB [30], CVC-ClinicDB [31], and ETIS-LaribPolypDB [32]. All these datasets are open source and can be used for research purposes to develop techniques to detect colon and rectal polyps making them in a way the standard datasets in the field.
3. Materials and Methods
3.1. Databases
In this study, we utilize 3 colorectal polyp image datasets, namely CVC-ColonDB, CVC-ClinicDB, and ETIS-LaribPolypDB. The first colorectal polyp image dataset to be made available for researchers is CVC-ColonDB, and it contains 380 images. All the images are part of 15 colonoscopy videos, and each sequence has various numbers of polyp pictures. The same group that published CVC-ColonDB later made available the CVC-ClinicDB dataset, which has 612 images taken from 29 sequences. The third dataset is ETIS-LaribPolypDB which has 196 images, Table 2. Each dataset consists of 2 main folders, the raw original images, and the masked images, the ground truths, of the corresponding one in the original image. Figure 2 shows images of polyps taken during several colonoscopies. As seen from the figure, polyps come in various shapes and sizes, and some of them are not significantly distinguishable from the mucosa of the colon.

Table 2.
Databases used to train and test the CNN-AE model.
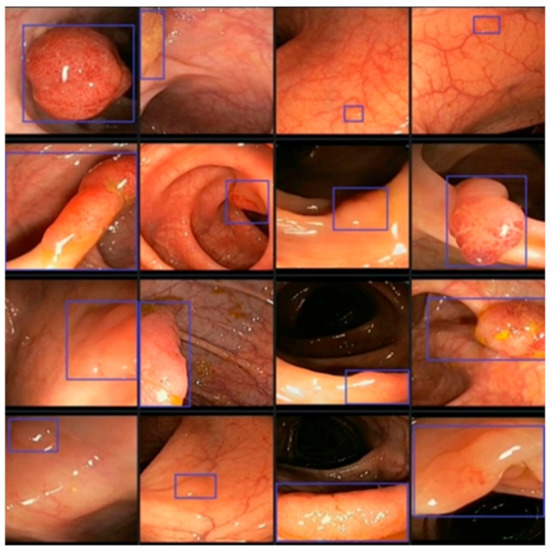
Figure 2.
Different shapes and textures of colon polyps taken from colonoscopy videos.
3.2. The Proposed Model
There are some deep learning libraries that can be used to build a neural network model. One of them is TensorFlow [33], an open-source library created by Google and community contributors, currently on its 2.0 version. We used this library to train and test our convolutional encoder-decoder model. The model uses the same architecture as the SegNet architecture [34], an algorithm programmed using Caffe, another deep learning library created by Berkeley AI Research and community contributors. The training and testing were performed on a computer with NVIDIA Titan X GPU.
Figure 3 shows the architecture of the CNN-Autoencoder model. The model has two parts, the encoder and the decoder. The structure of the encoder is similar to some image classification neural networks such as the convolutional layer, which includes the batch normalization, the rectified linear unit (the ReLu) activation function, and the pooling layer. The decoder part has the inversed layers used in the encoder, such as deconvolution layers and de-max_pool layers.
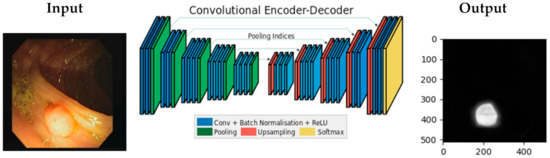
Figure 3.
Convolutional encoder-decoder architecture.
The encoder part has 13 convolutional layers and 5 max_pooling layers, where the first 3 layers of the model have these characteristics: the first convolution layer is with stride 2, followed by the second convolution layer with stride 1, and a non-overlapping 2 × 2 window max_pooling layer with stride 2. As mentioned above, each decoder layer contains the corresponding layer of the encoder, which means the decoder network has 13 layers. The output of the last decoder is fed to the Softmax classifier, which produces for each pixel the probabilities if it is a polyp or a normal colon tissue. The network input-output dimensions are equal:
- use the same layer for the non-shrinking convolution layer.
- use transposed deconvolution for the shrinking convolution layer adjusted with the same parameters.
- use the nearest neighbor upsampling for the max_pooling layer.
Open source medical image datasets lack the number of images in them, often only a couple of hundred images. However, for deep learning algorithms to work, a large amount of data is needed. In the case of image databases, researchers have used image augmentation techniques to increase the number of training images. In our case, we used an image augmentation library in Python called Imgaug Library [35]. Figure 4 shows the results after applying some image augmentations that we used in our model, which include:

Figure 4.
One image of colon polyp after applying different image augmentations.
- Crop—parameter: px = (0, 16) which crops images from each side by 0 to 16 pixels chosen randomly.
- Fliplr—parameter: 0.5 which flips horizontally 50% of all images.
- Flipud—parameter: 0.5 which flips vertically 50% of all images.
- GaussianBlur—parameter: (0, 3.0), blurs each image with varying strength using gaussian blur (sigma between 0 and 3.0).
- Dropout—parameter: (0.02, 0.1), drop randomly 2 to 10% of all pixels (i.e., set them to black).
- AdditiveGaussianNoise—parameter: scale = 0.01*255, adds white noise pixel by pixel to images.
- Affine—parameter: translate_px = {“x”: (-network.IMAGE_HEIGHT // 3, network.IMAGE_WIDTH // 3)}, applies translate/move of images (affine transformation).
By using image augmentation, we not only increased the number of images to train the model but also increased the robustness and reduced overfitting of our model. Another technique to deal with the overfitting problem was the Dropout technique with a rate of 0.2. Each dataset was divided into train set, validation set. Majority of the data in each dataset was used for training, 75%, 15% was used to validate and the other 15% to test the model.
4. Results
We trained the model on the selected databases using only the training sets and then we validated and tested with the validate and test sets. As each database has a different number of images, the time to train the model varied. The same batch size of 100 was used for all datasets. The accuracy and the total training time for each database are depicted in Table 3. The best accuracy was achieved on ETIS-LaribPolypDB’s last batch with a score of 0.967.

Table 3.
The accuracy and the total training time for each dataset.
Apart from the accuracy results from each batch and the final test accuracy, we obtained the images that the algorithm predicted. The test input, test targets, and test predictions were set to gray scale before all the results were drawn. Figure 5 depicts one example from each dataset. The three columns represent the three datasets (left to right: ETIS-LaribPolypDB, CVC-ClinicDB, and CVC-ColonDB) and the three rows, from top to bottom, the test image, the test ground truth (target), and the result of the segment obtained from our model.
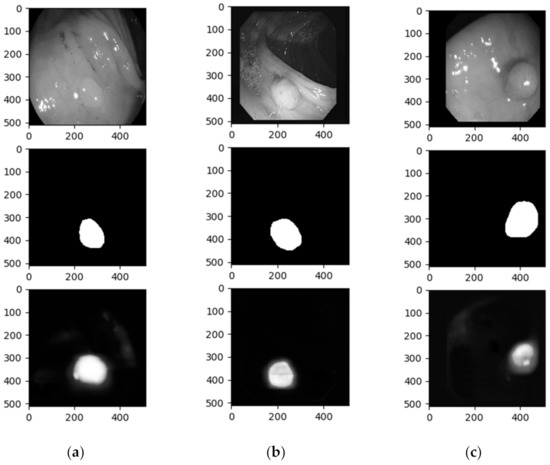
Figure 5.
Images showing the results after training the convolutional encoder -decoder model on (a) ETIS-LaribPolypDB, (b) CVC-ClinicDB, and (c) CVC-ColonDB database.
As we presented in Figure 2, polyps have various shapes and characteristics, ranging from big and recognizable polyps to barely distinguishable circular shapes. In Figure 5, we can see that the polyp in the first column is not easily detectable by the human eye, while the polyp in the last is recognizable. This wide variation induces errors in polyp recognition.
5. Discussion and Conclusions
In the background section we presented many techniques and algorithms used these recent years. A quick glance at summary Tab 1 depicts how diverse these techniques are, but also how diverse the metrics to evaluate them are. Accuracy was one of the most used metrics followed by the other metrics such as precision, recall etc. Although the main topic is the same, colorectal polyps, comparing results is difficult. The first reason is the one we explained above, different metrics. The others are related to the objectives, for what purpose are these algorithms used (classification, segmentation, detection, or classification), and the databases these algorithms are trained.
Among the cited papers we find two other similar studies to ours, meaning they are focused on detection problem and use the same metric and database/s. By using the CNN-Autoencoder model, we obtained the highest accuracy of 96.7, which is slightly better than the current state-of-the-art models that calculated the accuracy, Table 4.

Table 4.
Accuracy comparison for the proposed model and previously published studies on colon polyp detection.
The main challenges with colonoscopy images seem to fall on the shape and texture of the polyps [18,22,26], and the quality of the images [21,22,26,27]. The quality of the images depends on the colonoscopy device itself [21,26] or on the expertise of the endoscopist [18,22,27]. Furthermore, in the case of polyp classification, class imbalance poses another problem [20]. Considering these challenges, we checked the image results and verified that indeed some of the segments the model predicted are not as expected. The unexpected bad masks are shown in Figure 6, and again this shows the implications that shape, and texture of the polyp has, but also the conditions the colonoscopy image was taken. The lighting used during the examination plays a negative role when it comes to colon polyp detection as the models misrecognize the normal tissue as a polyp. This phenomenon happens because the inner surface of the colon is smooth, and the light attached to the colonoscopy used by the endoscopists to exam the colon reflects, confusing the models to consider healthy colon tissues as polyps. We have to mention that the patient needs to prepare well and follow the doctor’s instructions as per the normal colonoscopy session.
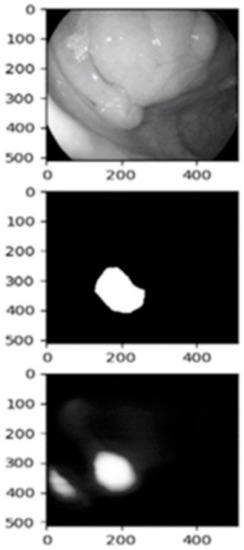
Figure 6.
False detection of a polyp due to lighting conditions.
Technology has helped progress the medical field enormously, especially when it comes to medical imaging. Colorectal cancer has been one of the diseases which has gained attention, and many researchers have worked towards detecting and preventing such disease. CAD systems have shown that the polyp miss rate has gone down. However, research shows deep learning has shown even more progress aiding colonoscopists/ endoscopists to perform better.
In this work, we presented the current state of the art of deep learning techniques in colon polyp detection, classification, segmentation and localization. We contributed by applying a novel algorithm CNN-AE for detection of polyps, which appears promising considering that no image preprocessing was performed prior to training the model. Our model shows better results than the current state of the art, although not very significant. We believe better results may be achieved if we increase the number of images in the dataset. Moreover, having a diverse range of polyp images may improve the algorithms performance. We tested the same model on other medical image databases, namely iris and pressure ulcer datasets, and the results obtained were better than with the colon polyp images. In future work we want to address these issues by making changes to the model and we will add other image augmentations currently not implemented in the Imgaug library. Besides the technical aspect, we want to address the lack of polyp image datasets. We are in the process of creating a bigger and more diverse dataset of colon polyp images. We will test the model as soon as we prepare the dataset which will be made available to researchers for academic purposes as well.
Author Contributions
Conceptualization, O.B., B.G.-Z. and L.B.; methodology, O.B., D.S.-S., B.G.-Z. and L.B.; software, O.B. and D.S.-S.; validation, D.S.-S. and O.B.; formal analysis, O.B. and D.S.-S.; investigation, O.B. and D.S.-S.; resources, B.G.-Z.; writing—original draft preparation, O.B.; writing—review and editing, O.B., B.G.-Z., D.S.-S. and L.B.; visualization, O.B.; supervision, B.G.-Z.; project administration, B.G.-Z. and O.B.; funding acquisition, B.G.-Z. All authors have read and agreed to the published version of the manuscript.
Funding
O.B. received funding from the European Union′s Horizon 2020 CATCH ITN project under the Marie Sklodowska-Curie grant agreement no. 722012.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- World Health Organization and International Agency for Research on Cancer. Cancer Today. 2020. Available online: https://bit.ly/37jXYER (accessed on 17 November 2020).
- Lieberman, D. Quality and colonoscopy: A new imperative. Gastrointest. Endosc. 2005, 61, 392–394. [Google Scholar] [CrossRef]
- Carioli, G.; Bertuccio, P.; Boffetta, P.; Levi, F.; La Vecchia, C.; Negri, E.; Malvezzi, M. European cancer mortality predictions for the year 2020 with a focus on prostate cancer. Ann. Oncol. 2020, 31, 650–658. [Google Scholar] [CrossRef]
- Zauber, A.G. The Impact of Screening on Colorectal Cancer Mortality and Incidence: Has It Really Made a Difference? Dig. Dis. Sci. 2015, 60, 681–691. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Lee, J.; Park, S.W.; Kim, Y.S.; Lee, K.J.; Sung, H.; Song, P.H.; Yoon, W.J.; Moon, J.S. Risk factors of missed colorectal lesions after colonoscopy. Medicine 2017, 96, e7468. [Google Scholar] [CrossRef]
- Bonnington, S.N.; Rutter, M.D. Surveillance of colonic polyps: Are we getting it right? World J. Gastroenterol. 2016, 22, 1925–1934. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed]
- Poon, C.C.; Jiang, Y.; Zhang, R.; Lo, W.W.; Cheung, M.S.; Yu, R.; Zheng, Y.; Wong, J.C.; Liu, Q.; Wong, S.H.; et al. AI-doscopist: A real-time deep-learning-based algorithm for localising polyps in colonoscopy videos with edge computing devices. NPJ Digit. Med. 2020, 3, 73. [Google Scholar]
- Ribeiro, E.; Uhl, A.; Häfner, M. Colonic polyp classification with convolutional neural networks. In Proceedings of the IEEE 29th International Symposium on Computer-Based Medical Systems (CBMS), Dublin, Ireland, 20–24 June 2016. [Google Scholar]
- Ribeiro, E.; Häfner, M.; Wimmer, G.; Tamaki, T.; Tischendorf, J.J.; Yoshida, S.; Tanaka, S.; Uhl, A. Exploring texture transfer learning for colonic polyp classification via convolutional neural networks. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017. [Google Scholar]
- Ribeiro, E.; Uhl, A.; Wimmer, G.; Häfner, M. Exploring Deep Learning and Transfer Learning for Colonic Polyp Classification. Comput. Math. Methods Med. 2016, 2016, 6584725. [Google Scholar] [CrossRef]
- Zhang, R.; Zheng, Y.; Mak, T.W.C.; Yu, R.; Wong, S.H.; Lau, J.Y.; Poon, C.C. Automatic Detection and Classification of Colorectal Polyps by Transferring Low-Level CNN Features from Nonmedical Domain. IEEE J. Biomed. Health Inform. 2017, 21, 41–47. [Google Scholar] [CrossRef]
- Shin, Y.; Qadir, H.A.; Aabakken, L.; Bergsland, J.; Balasingham, I. Automatic Colon Polyp Detection Using Region Based Deep CNN and Post Learning Approaches. IEEE Access 2018, 6, 40950–40962. [Google Scholar] [CrossRef]
- Shin, Y.; Balasingham, I. Comparison of hand-craft feature based SVM and CNN based deep learning framework for automatic polyp classification. In Proceedings of the 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju Island, Korea, 11–15 July 2017. [Google Scholar]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, C.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Deep Learning for Classification of Colorectal Polyps on Whole-slide Images. J. Pathol. Inform. 2017, 8, 30. [Google Scholar] [PubMed]
- Mahmood, F.; Durr, N.J. Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy. Med. Image Anal. 2018, 48, 230–243. [Google Scholar] [CrossRef]
- Carneiro, G.; Pu, L.Z.C.T.; Singh, R.; Burt, A. Deep learning uncertainty and confidence calibration for the five-class polyp classification from colonoscopy. Med. Image Anal. 2020, 62, 101653. [Google Scholar] [CrossRef]
- Gao, J.; Guo, Y.; Sun, Y.; Qu, G. Application of Deep Learning for Early Screening of Colorectal Precancerous Lesions under White Light Endoscopy. Comput. Math. Methods Med. 2020, 2020, 8374317. [Google Scholar] [CrossRef]
- Song, E.M.; Park, B.; Ha, C.A.; Hwang, S.W.; Park, S.H.; Yang, D.H.; Ye, B.D.; Myung, S.J.; Yang, S.K.; Kim, N.; et al. Endoscopic diagnosis and treatment planning for colorectal polyps using a deep-learning model. Sci. Rep. 2020, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Urban, G.; Tripathi, P.; Alkayali, T.; Mittal, M.; Jalali, F.; Karnes, W.; Baldi, P. Deep Learning Localizes and Identifies Polyps in Real Time With 96% Accuracy in Screening Colonoscopy. Gastroenterology 2018, 155, 1069–1078.e8. [Google Scholar] [CrossRef] [PubMed]
- Misawa, M.; Kudo, S.E.; Mori, Y.; Cho, T.; Kataoka, S.; Yamauchi, A.; Ogawa, Y.; Maeda, Y.; Takeda, K.; Ichimasa, K.; et al. Artificial Intelligence-Assisted Polyp Detection for Colonoscopy: Initial Experience. Gastroenterology 2018, 154, 2027–2029.e3. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.K.; Yildirim, S.; Farup, I.; Pederse, M.; Hovde, O. Y-Net: A deep Convolutional Neural Network for Polyp Detection. 2018. Available online: http://arxiv.org/abs/1806.01907 (accessed on 10 June 2020).
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Integrating Online and Offline Three-Dimensional Deep Learning for Automated Polyp Detection in Colonoscopy Videos. IEEE J. Biomed. Health Inform. 2017, 21, 65–75. [Google Scholar] [CrossRef]
- Byrne, M.F.; Chapados, N.; Soudan, F.; Oertel, C.; Pérez, M.L.; Kelly, R.; Iqbal, N.; Chandelier, F.; Rex, D.K. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019, 68, 94. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, X.; Berzin, T.M.; Brown, J.R.G.; Liu, P.; Zhou, C.; Lei, L.; Li, L.; Guo, Z.; Lei, S.; et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): A double-blind randomised study. Lancet Gastroenterol. Hepatol. 2020, 5, 343–351. [Google Scholar] [CrossRef]
- Yamada, M.; Saito, Y.; Imaoka, H.; Saiko, M.; Yamada, S.; Kondo, H.; Takamaru, H.; Sakamoto, T.; Sese, J.; Kuchiba, A.; et al. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bernal, J.; Sánchez, J.; Vilariño, F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognit. 2012, 45, 3166–3182. [Google Scholar] [CrossRef]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2–4 November 2016; pp. 265–283. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Jung, A. imgaug 0.2.5 [Internet]. 2017. Available online: http://imgaug.readthedocs.io/en/latest (accessed on 10 June 2020).
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).