Abstract
Ontology matching is a rapidly emerging topic crucial for semantic web effort, data integration, and interoperability. Semantic heterogeneity is one of the most challenging aspects of ontology matching. Consequently, background knowledge (BK) resources are utilized to bridge the semantic gap between the ontologies. Generic BK approaches use a single matcher to discover correspondences between entities from different ontologies. However, the Ontology Alignment Evaluation Initiative (OAEI) results show that not all matchers identify the same correct mappings. Moreover, none of the matchers can obtain good results across all matching tasks. This study proposes a novel BK multimatcher approach for improving ontology matching by effectively generating and combining mappings from biomedical ontologies. Aggregation strategies to create more effective mappings are discussed. Then, a matcher path confidence measure that helps select the most promising paths using the final mapping selection algorithm is proposed. The proposed model performance is tested using the Anatomy and Large Biomed tracks offered by the OAEI 2020. Results show that higher recall levels have been obtained. Moreover, the F-measure values achieved with our model are comparable with those obtained by the state of the art matchers.
1. Introduction
The evolution of semantic web technologies and the growth of big data volumes maintained by various database models have resulted in many disparate and independent data sources [1]. However, data growth will pose many issues if we cannot keep pace with these improvements. To succeed, it is crucial to determine how traditional information systems can be transferred into more integrated systems. In this context, ontologies play an essential role in addressing semantic heterogeneity to achieve semantic interoperability among the various web applications and services [2]. Semantic web languages have a sharp learning curve, and a shift in viewpoint is necessary, particularly in individuals with qualifications in software engineering, object focused programming, or relational databases.
During the early 1990s, researchers in the field of computer science began investigating ontologies. The claim was that ontologies could facilitate information sharing by users and software agents regarding particular topics. The given definition of ontology was a conceptual representation of an entity, its characteristics and correlations within a domain [3]. Over the past 10 years, ontologies have gained increasing attention in many different fields, including academia, industry, biomedicine, finance, engineering, law, and governmental agencies [4]. Furthermore, ontologies have gained significant importance as a component of biomedical research investigations because they supply the formalism, objectivity, and common terminology required to report research findings that can enable direct exchange and reuse by scientists and computers [5]. However, integrating and sharing data are still challenging because ontologies are semantically heterogeneous.
Ontology matching has grown in popularity, particularly in the biomedical, biological, and geographical domains [6,7]. From an abstract perspective, ontology matching aims to identify how ontologies relate to one another. The matching process can be completed by detecting any two given entities’ interrelated or comparable elements. Precisely, the two entities must be tallied to yield the appropriate set of correspondences [3]. It is challenging to match biomedical ontologies because of their huge size, vocabulary complexity, and rising semantic richness, including new forms of interactions between classes making the task computationally challenging [6]. Several studies have presented alternative approaches to address the ontology matching problem. They differ principally in terms of the type of information that each ontology encodes and how that knowledge is applied in the context of detecting equivalences across features or structures in ontologies [8,9,10,11,12]. Furthermore, additional factors, such as matching settings (e.g., weights and cut thresholds) and external BK resources, influence the matching process. However, BK sources must include lexical or structural knowledge that the source and target ontologies do not have, to recognize novel mappings.
1.1. Background Knowledge (BK)
The definition of BK varies in different techniques. Ren and Deng [13] define BK as the critical information required to understand a situation or problem. The BK based matching or indirect matching approach or context based matching is the opposite of direct matching. It detects mappings between ontologies for alignment by taking advantage of external resources [14]. Placing ontologies in the context of other ontologies may improve direct matching, as illustrated in Figure 1 [15]. Recently, attention has been directed toward finding a different solution to automatic methods by employing BK as a mediator to identify the input ontologies’ correspondence [16]. BK resources are linked data, lexical databases, one or several ontologies, a BK repository, and existing mappings.
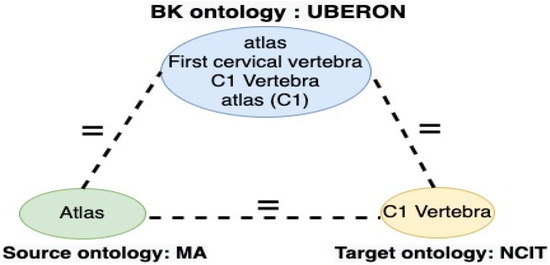
Figure 1.
Matching utilizing a BK Source.
Semantic heterogeneity is a significant problem during ontology matching [17]. The efficiency of direct matching is diminished by heterogeneous ontologies, as reflected in the definition of the same concept with different labels or structuring based on distinct modeling perspectives [14]. Every suggested approach involved the utilization of BK as a complementary solution to current automatic methods. Such aspects have been explored by several studies [18,19,20]. Although lexicon based alignment (e.g., WordNet) has been attempted in several studies [21,22,23], other types of BK have not been extensively employed [7,17]. BK based matching techniques aim to address semantic heterogeneity by exploring an external resource to cover the semantic gap among matched ontologies. However, existing BK based matching systems, such as AML [6] or LogMapBio [24], have built the indirect matching process into their internal design. Therefore, the reuse of such systems is contingent on adjusting their code, which can be difficult.
Generic frameworks, such as Scarlet and GBKOM for BK based ontology matching, are the only standard BK based matchers; however, the former is significantly outdated and lacks functionality [14]. Meanwhile, only a singular matcher is employed by GBKOM to take advantage of external BK sources to bridge the semantic gap between ontologies for alignment. However, greater performance can be obtained by using a multimatcher, as we will show in this work. Different ontology matchers may not always detect the exact correspondences. Accordingly, multiple competing matchers are typically used to reinforce possible matches to attain reliable results. Subsequently, the final alignment outcomes are strengthened by combining the generated mappings into a single one.
1.2. Contributions
This work presents the approach of combining and aggregating several mapping alignments to demonstrate the effectiveness of the multimatcher model for BK based ontology matching. Several matchers are currently available. However, the OAEI results indicate that not all matchers discover the same correct mappings. As a result, none of them is capable of achieving excellent performance in all matching tasks. Our Multimatcher BK based ontology matching strategy estimates that it would be more effective to merge alignments generated by the different matcher. Therefore, it uncovers new mappings between the ontologies that are being matched and enhances the final alignment. Our model uses a path driven inferencing strategy. The pathways between the source and target ontologies are established first. Then, a matcher confidence value for the constructed paths is built using our suggested measure, which the final mapping judgment process will use to help determine if the pathways are effective or not. This proposed model consists of three main components: (1) matcher aggregation strategies, (2) BK path driven inferencing, and (3) merging paths and final mapping selection. The proposed model will enhance direct matching results by providing better recall and F-measure than existing methods. The three primary contributions of this work are as follows:
- An algorithm to improve mapping correspondence quality using different matchers and several aggregation strategies;
- A matcher path confidence measure that indicates the generated path matchers, which will be exploited by final mapping judgment;
- An algorithm to select the final mapping from several paths based on the matcher path confidence measure and false mapping repository to enhance the direct matching performance.
We have used the Anatomy and Large Biomed tracks supplied by the OAEI 2020 to evaluate our model’s performance to illustrate the enhancement gain with the BK matching process in mapping quality, recall, and F-measure. Moreover, the model offers a comprehensive range of linked parameters and allows multiple setups.
1.3. Organization
The remainder of this work is organized in the following manner. Section 2 introduces the required preliminaries on ontology matching. Section 3 reviews the related work. Section 4 proposes a BK multimatcher model. Section 5 explains the experimental and result analysis. Section 6 concludes the study with a discussion and recommendations for future research.
2. Preliminaries
The following fundamental terms are used throughout the study:
Ontology: Ontologies are the tools that allow us to formally describe a domain by its objects and the relationships that exist between them. Ontology is defined in this study as a collection of classes, properties, and instances for a specific topic of interest. The set of classes, properties, and instances that make up the given ontology is often referred to as the entity of the ontology.
Matcher: a matcher is a system used to find mappings between ontologies, such as AML [6], LogMap, and LogMapLt [24].
Ontology matching system: A standard ontology matching system inputs two ontologies representing the source and the target and attempts to identify similar entities [3].
Correspondence: Correspondence is defined as the mapping of an entity between the source and the target ontologies. This task may include additional information regarding the mapping (e.g., relation, score, and matcher).
<e, e′, r, s, m>: Represents a basic correspondence. In this context, e represents an entity from the source ontology, and e′ is an entity from the target ontology. r represents the equivalence between the entities. s represents the degree of confidence reflecting the reliability of a correspondence in the range [0, 1], and m denotes a matcher given by a series of single- or multimatcher.
Alignment: The series of correspondences among the pairs of entities represents the alignment for the specific source and target ontologies. According to this definition, the alignment constitutes the standard results of an ontology alignment system.
Aggregation strategy. A satisfactory output alignment is not always achieved with just one ontology entity matcher. Accordingly, multiple matchers are frequently integrated to generate a singular confidence value representing an aggregated value. The quality of the alignments is highly dependent on the suitable aggregation approach. However, determining an effective combination strategy is a complicated task. A complex procedure is manually carried out by an expert or a generic method (e.g., maximum, minimum, average, and vote) [25].
Biomedical ontology matching: This is concerned with determining an ontology alignment made up of biomedical concept correspondences. In most cases, the matching procedure requires the use of external BK sources.
BK: BK has different definitions in various techniques. BK is defined as the essential information needed to comprehend a scenario or problem in ontology matching. We identify it as a collection of external ontologies that give lexical or semantic information on the domain of the ontologies to align.
Once the final alignments are established, multiple performance scores are generally determined to measure system performance. In this work, a reference alignment encompassing the ground truth of the mappings between specific ontologies is needed. Two measures, typically referred to as recall and precision, are employed to evaluate the alignment. Recall, known as completeness, assesses the proportion of accurate alignments identified to the overall number of available accurate alignments. Meanwhile, precision is known as correctness and assesses the proportion of identified alignments that are indeed accurate. For example, reference alignment, R, and particular alignment, A, are defined as follows:
In most cases, recall and precision are needed for alignment performance comparison. Furthermore, the F-measure can be employed for a trade off between the two measures and is given by:
The collaborative international initiative (OAEI) is designed to assess the increasing number of ontology matching systems. This initiative is primarily geared toward an open and equal comparison of systems and algorithms to ensure that the ideal matching techniques can be determined by everyone [26]. Furthermore, the initiative includes a range of tracks (e.g., anatomy, conference, and large biomedical ontologies), and the outcomes of the evaluated systems are disclosed for further analysis.
3. Related Work
In this section, we will look at relevant research on the four main topics of this work: BK framework architectures, BK based ontology matching, BK ontology selection, and aggregation strategies.
3.1. GBKOM BK Based Ontology Matching
Existing matchers, such as GOMMA [27], LogMap [28], or AML [6], use BK based matching modules closely associated with their internal architectures. GOMMA was the first system to use a mapping composition to implement a BK based method in 2012. LogMap is a large scale ontology matching system capable of dealing with massive ontologies. BK is used in two versions of the LogMap ontology matcher. LogMap-BK uses the UMLS Metathesaurus, while LogMapBio supplies a selection of the biomedical ontology from the NCBO BioPortal as BK. AML is a framework for ontology matching based on an AgreementMaker, one of the most used ontology matching systems. AML is a lightweight system focused on the biomedical sector but applicable to other ontologies. Nevertheless, reusing these modules demands a detailed study and customization of their code, which is not easy.
However, GBKOM is an exception [14]. The GBKOM BK-based ontology matching is a flexible framework. It is openly accessible on GitHub, can be added to any current matcher, and is suitable for undertaking experimental evaluations. The GBKOM instance employs YAM++ as a single matcher with BK from UBERON and DOID, two biomedical ontologies. GBKOM uses the LogMap Repair module to remove the incoherent mapping of generated alignments. In this register, we extend this work using several aggregations of alignments provided by different matchers to increase the matching quality compared with using a single matcher. The study revealed that employing multimatchers and composing mappings for ontologies is highly successful.
3.2. BK Based Ontology Matching
BK can be represented in various ways, including domain ontologies, pre-existing alignments, and web sources [7]. The amount of structured knowledge that is publicly available has dramatically increased. Several large knowledge graphs, including BabelNet, DBpedia, and Wikidata, are accessible [8]. Nonetheless, these knowledge bases are rarely used for automated matching. Much earlier research has employed lexicons to accomplish alignment, such as WordNet as a generic source [21,22,23]. However, the biological domain is an exception: domain specific BK is widely available and frequently utilized [17].
Given that many biomedical ontologies overlap, correspondences to a mediating ontology must be used to enhance the delivery of final correspondences between the ontologies. A straightforward and effective strategy is to compose existing mappings to generate new mappings quickly. Studies by [29,30] derived mappings from existing mappings to third ontologies, referred to as intermediate ontologies. For example, we assume the transitivity of the correspondences. The composition of a particular mapping between schemes S1, S2, and schemes S2 and S3 will lead to a new mapping between S1 and S3.
Chen et al. [31] proposed dynamically composing mappings by picking ontologies from BioPortal. Annane et al. [16] proposed using one or more intermediary ontologies as a composition based strategy to align living science ontologies indirectly. The suggested technique aims to increase alignment efficiency and quality by reusing ontology alignments. This approach matches existing alignments between the BioPortal’s ontologies by integrating source and target entities into the global maps graph using a path based mechanism. The paths connecting the concept to the graph allow new maps to be created. Although various BK sources are accessible in the biological domain, this is not the case in other fields. Therefore, such procedures are not readily applicable.
3.3. BK Ontology Selection
Research on BK selection has also been carried out in the biological domain. Faria et al. [32] suggested a measure known as mapping gain (MG) that is based on the new alignment found in a baseline alignment. MG is used to examine the individual use of BK sources. The source with the most significant MG value is selected. Hartung et al. [33] presented a new measure for ontology matching termed effectiveness, based on how much information is shared between the two ontologies being matched. This metric is based mainly on the overlap in an intermediate ontology in terms of concepts. For example, the higher the overlap, the higher the efficiency.
Tigrine et al. [34] incorporated the problem into an information retrieval paradigm. Ontologies and BKs are compared in terms of content and structure. This technique’s selection procedure is automated and independent of domain. Quinx et al. [35] proposed a similar methodology to find appropriate BK sources using a keyword based vector similarity technique. Chen et al. [31] used a fast selection strategy to determine a suitable collection of mediating ontologies due to the high number of ontologies available in BioPortal. The fast selection methodology finds labels present in the input ontologies and research into ontologies containing synonyms in BioPortal. Such specialized organized resources remain scarce outside the biomedical field. In contrast with the current work, a GKBOM selects a fragment of the BK resource related to the source ontology.
3.4. Aggregation Techniques
A single algorithm cannot easily achieve a quality alignment on its own because of the multiplicity of human made data models. Accordingly, the matching process is approached with a set of matchers or matching algorithms [3,36]. The setting of various matchers is manually performed by experienced ontology matching system users, domain experts and ontology developers [37]. However, setting up and configuring such systems with several matchers, combination methods, and individual parameter settings are difficult, even for specialists. The ontology matching community has already addressed these challenges when combining several similarity measures in the same matcher [6,27,38] and has provided several solutions [39,40,41].
There are many combination methods, some of which are basic and others more advanced, as illustrated in Figure 2. Several commonly used fundamental approaches are mentioned in the literature, including Average, Maximum, Minimum, and Cut threshold. The Average approach calculates the average similarity of all individual matchers who have discovered a specific relation. It indicates that all matchers are given the same weight. This technique aggregates the relationships contained in the different alignments and calculates a final score based on the average confidence of the different alignments. This calculation is carried out despite the sort of relationship between the two elements. The Maximum method finds the maximum similarity value across all possible matchers. On the other hand, the Minimum technique selects the lowest similarity value from any particular matcher. The Cut threshold technique has numerous modifications; in its simplest version, it means that a preset cut threshold selects which relations would be included in a final alignment [3]. Advanced combination methods are described in [37,42,43].
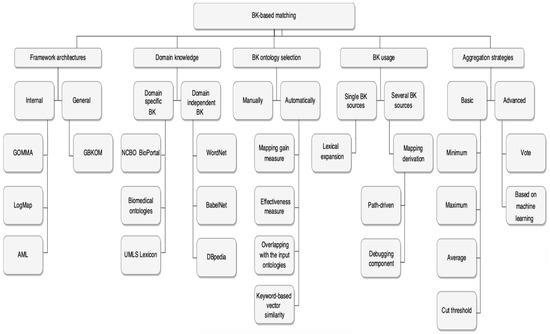
Figure 2.
BK based matching overview.
It is suggested that weighted aggregation be used for the aggregation process. The weighted aggregation technique analyzes each basic matcher’s correspondences differently, taking into account the overall quality of the results provided by each matcher. The most challenging problem is determining an individual basic matcher’s weighting factor or the quality of matching results produced by a specific basic matcher. According to Peukert et al. [44], advanced combination approaches can perform well on some matching tasks, while basic strategies, such as utilizing Average aggregation, are more robust. Some of the most effective matching systems, such as AML [6] and COMA [27], combine the results of individual matchers using relatively simple methods.
4. BK Ontology Matching: A Multimatcher Model
4.1. Overview of Our Approach
We present a BK multimatcher model to combine and aggregate the different mapping alignments created by several automatic matchers, notably, LogMap, LogMapLt, and AML, to enhance the final alignment. Matchers can indeed identify candidate correspondences, which must be confirmed and corrected by human experts. Automatic matchers might miss some correspondences. Moreover, relying on a single matcher to improve calculated ontology mappings and reduce the manual effort required to fix them is insufficient; therefore, various matchers must be combined. In this register, our model is built on the GBKOM architecture presented in [14]. However, significant improvements and changes to the previous approach have been made. The system architecture has been changed to combine and aggregate different alignments obtained by several matcher alignments for different tasks (building the global graph, anchoring, and direct matching). A new aggregation strategy component is created, including Minimum, Maximum, Average, and Vote, and a novel algorithm for path driven inferencing.
The algorithm for final mapping judgment has been improved to its current version by considering the matcher path confidence measure and the false mapping repository. Our model also includes additional features that allow various settings and may be easily integrated into any current matcher. This model is valuable for conducting experiments.
Our proposed model consists of three major components, as shown in Figure 3. We provide matcher aggregation strategies (Algorithm 1), BK path driven inferencing, combing paths and applying the final selection method (Algorithm 2). The model begins by employing various automatic matchers to align the manually chosen BK ontologies. The alignments that each matcher generates are temporarily saved in a processing folder. Then, the model aggregates and determines the final combination based on the model aggregation strategy. After that, several matchers will match the source ontology with the BK ontologies, and the final mapping will be selected using the same aggregation strategy.
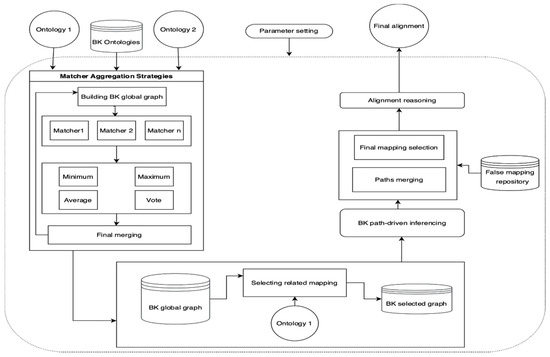
Figure 3.
BK ontology matching: a multimatcher model.
The BK global graph is filtered using the source ontology to build a specific graph (BK selected graph) aligned with the target ontology. In the second component, our model adapted a path driven inferencing method. First, the paths between the source and the target ontologies is established, including the matchers’ names. Then, our suggested measure establishes the matcher confidence value for the created paths, which the final mapping judgment algorithm uses to assist in determining whether the pathways are effective or not. Finally, the third component selects the final mapping judgment among several paths based on the confidence value of the matchers. In addition, post-processing techniques can be used to select only the most appropriate correspondences. Thus, we provide our model with false mappings that start of the art matchers cannot recover, to improve the quality of direct matching (F-measure).
4.2. Matcher Aggregation Strategies
This module is the foundation of our approach. In this work, we apply simple but effective aggregation algorithms. The matching process includes an alignment aggregation step that seeks to combine the best correspondences from the alignments created by the various matchers to produce the final alignment. The final alignment quality can be improved by combining the findings of the individual matchers. Four different alignment combination strategies have been established to combine alignments created by the individual matchers. Three of these strategies represent basic approaches (Minimum, Maximum, and Average) and Vote as a more advanced combination method. In this section, simple and advanced combination methods will be presented. Nonetheless, some more advanced combination approaches that involve machine learning techniques exist. However, these techniques are not explained further because they require training data aligned with the ground truth that is usually unavailable.
The matcher aggregation strategies are as follows: three alignments are expressed in RDF format, one with the matcher LogMap (Table 1), another with the matcher LogMapLt (Table 2), and a third with the matcher AML (Table 3). This article only discusses equivalence mappings. However, our methodology might be expanded to other types of mapping relationships if a mechanism for composing diverse relationships on the same path is developed [45]. Given two ontologies, namely, MA and UBERON, an alignment consists of a collection of correspondences ⟨e1, e2, r, s, m⟩, where r denotes a relationship between e1 and e2, such as equivalence. Where s is a confidence score in (0, 1), indicating how likely it is that e1 and e2 are related to one another. The composition of the confidence value is performed in one of four ways (Table 4, Table 5, Table 6 and Table 7) where:

Table 1.
Part of the alignment between MA and Uberon ontologies using the LogMap matcher.

Table 2.
Part of the alignment between MA and Uberon ontologies using the LogMapLt matcher.

Table 3.
Part of the alignment between MA and Uberon ontologies using the AML matcher.

Table 4.
Part of the final alignment between MA and Uberon ontologies using the minimum aggregation strategy.

Table 5.
Part of the final alignment between MA and Uberon ontologies using the maximum aggregation strategy.

Table 6.
Part of the final alignment between MA and Uberon ontologies using the average aggregation strategy.

Table 7.
Part of the final alignment between MA and Uberon ontologies using the vote aggregation strategy.
Such as equivalence. Where s is a confidence score in (0, 1) indicating how likely it is that e1 and e2 are related to one another. The composition of the confidence value is performed in one of four ways where:
Minimum: The minimization combination method returned the lowest score value for e1 and e2.
Maximum: The maximization combination method returned the highest score value for e1 and e2.
Average: The average combination method returned the average score value for e1 and e2.
Vote: The vote combination method returned majority of the correspondences with the highest score value.
| Algorithm 1. Aggregation Strategies | |
| 1 | Input: ontology 1 (source ontology) and ontology 2 (target ontology) |
| 2 | matchers: matcher 1, matcher 2, matcher 3, and matcher n |
| 3 | Output: Aggregated alignment |
| 4 | if source and target ontologies exist then |
| 5 | for i:= 1 to matcher(n) do |
| 6 | set matcherName to matcher (i) |
| 7 | createAlignment (ontology 1, ontology 2, matcher (i)) |
| 8 | saveAlignmentToList (Matcher(i)) |
| 9 | end for |
| 10 | end if |
| 11 | for A:= 1 to AlignmentsList do |
| 12 | addAllMappingsMaster() |
| 13 | end for |
| 14 | for line:= 1 to allMappingsMaster do |
| 15 | for lineCompare: = 1 to allMappingsMaster do |
| 16 | if(masterLineCompare.equals(lineCompare)) then |
| 17 | addFinalMappings() |
| 18 | end if |
| 19 | end for |
| 20 | if FinalMappings greater than one then |
| 21 | for line:= 1 to FinalMappings do |
| 22 | scoresList = add(score); |
| 23 | if mappingAggregationStrategy = Min then |
| 24 | AggreagatedScore = Min (scoresList) |
| 25 | end if |
| 26 | if mappingAggregationStrategy = Max then |
| 27 | AggreagatedScore = Max (scoresList) |
| 28 | end if |
| 29 | if mappingAggregationStrategy = Avg then |
| 30 | AggreagatedScore = Avg (scoresList) |
| 31 | end if |
| 32 | if mappingAggregationStrategy = Vote then |
| 33 | AggreagatedScore = Vote (scoresList) |
| 34 | end if |
| 35 | end for |
| 36 | end if |
| 37 | end for |
| 38 | if AggreagatedScore > thresholdAggregationSelection then |
| 39 | return finalAggregatedAlignment (AggreagatedScore) |
| 40 | end if |
| 41 | end |
4.3. BK Path Driven Inferencing
A path is a triple composed of three entities: two equivalent entities, and a link entity. After a global graph in the primary component has been created, we use the selected graph to link the source and target concepts. The mappings derived from these paths are applied to form new mappings as illustrated in Figure 4. The paths connecting the concepts within this graph are utilized to generate further mappings. Accordingly, the number of pathways to investigate during derivation and the final returned paths are reduced [14]. The pathways in this graph can lead to the discovery of new mappings. A significant issue with obtaining all pathways is that it is resource intensive, because discovering all the paths between two nodes is impractical in massive graphs. To address this issue, we limit the length of pathways between entity pairs to four intermediate edges (links). The maximum path length exploited had previously been found following extensive tests published in [46] and had also been used in [14]. In light of the results produced by prior solutions, this procedure is assumed to be already addressed.
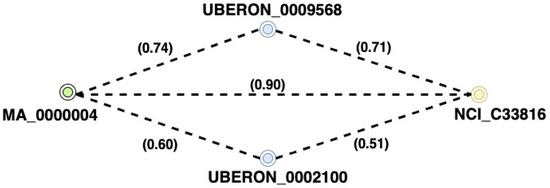
Figure 4.
Example of paths that include scores only.
Another essential feature is the introduction of a new measure called the Matcher Path Confidence Measure. This measure can assist in the process of determining the correct mappings by considering the matcher’s confidence. This metric is only suggested for selecting a single target concept from a set of candidates for a given source concept. Paths are labeled with their matchers. Automatic mapping paths that several matchers have produced can be more significant than single matcher pathways. The identified mappings are explained in Figure 5, to provide a more precise score. We apply weights to various path types between entities based on the matcher that they represent. The present module launches the subsequent phase, which is responsible for path merging and final mapping selection.
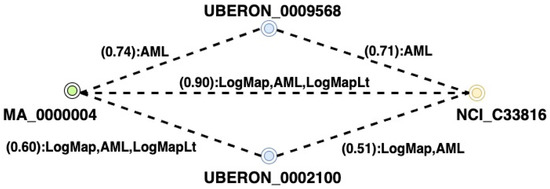
Figure 5.
Example of paths that include scores and matchers.
4.4. Final Mapping Selection
After the aggregated correspondences between all the compared ontologies are determined, a suitable subset of the correspondences must be chosen and included in the final alignment. The paths connecting the source concepts to the target ontology entities should be examined to identify which entities correlate. Several different pathways may represent a single candidate mapping. Thus, related work proposed using algebraic functions, such as multiplication and maximum, to obtain the final score to assemble distinct mapping scores [47]. Furthermore, we present a new algorithm (Algorithm 2) to choose the most relevant mappings from the candidates based on the Matcher Path Confidence Measure and the false mapping repository.
| Algorithm 2. Final Mapping Selection | |
| 1 | Input: foundPaths, |
| 2 | sourceConcepts, targetConcepts |
| 3 | Output: Final alignment |
| 4 | for P:= 1 to foundPaths do |
| 5 | matcherslist = get matchers (linePath) |
| 6 | if matcherslist > 1 then |
| 7 | score:= 1.0 |
| 8 | end if |
| 9 | if refAlignFalseMapping > 0, then |
| 10 | if refAlignFalseMapping equal to |
| 11 | (sourceConcept, targetConcept) then |
| 12 | stopPathFlag=stop |
| 13 | end if |
| 14 | end if |
| 15 | if stopPathFlag not equal to stop, then |
| 16 | if allCandidates (sourceConcept) do not exist then |
| 17 | addCandidate (sourceConcept, score, matcher, pathNo) |
| 18 | else |
| 19 | if allCandidates (targetConcept) not exsit then |
| 20 | addCandidate (targetConcept, score, matcher, pathNo) |
| 21 | else |
| 22 | updateCandidate (maxScore, matcher, pathNo) |
| 23 | end if |
| 24 | end if |
| 25 | end if |
| 26 | end for |
| 27 | for S:= 1 to allCandidates (sourceConcept) do |
| 28 | for T:= 1 to allCandidates (targetConcept) do |
| 29 | if S.pathNo greater than one then |
| 30 | addFinalAlignment(mapping) |
| 31 | stopFlag = true |
| 32 | end if |
| 33 | if (S.maxScore > maxCandidateScore) then |
| 34 | maxCandidateScore = S.maxScore |
| 35 | maxCandidate = sourceConcept |
| 36 | uriCandidate = targetConcept |
| 37 | end if |
| 38 | end for |
| 39 | if stopFlag not true then |
| 40 | addFinalAlignment(mapping) |
| 41 | end if |
| 42 | end for |
| 43 | return (finalAlignment) |
| 44 | end |
5. Experimental and Result Analysis
This section introduces the experimental step and the Anatomy and Large Biomed tracks, which are used to evaluate the performance of our model. The outcomes of various aggregating methods are then reported and compared. Finally, the results of the final alignments are compared with four state of the art matching systems in terms of performance (precision, recall, and F-measure).
5.1. Experimental Setup and Datasets
In this section, we will go over the experimental setup and the data sets. Table 8 summarizes all of the parameter settings. The bold parameter values were leveraged in the tests carried out for this research investigation. The OAEI (2020) Anatomy and Large Biomed tracks are used to measure the overall performance of our model. The Anatomy track consists of two ontologies (one task), namely, the AMA ontology (2744 classes) and a section of the NCI that describes human anatomy (3304 classes). The alignment of classes is the most critical work in this track. The Large biome track (six tracks), consisting of 78,989, 122,464, and 66,724 classes, seeks to find alignments between FMA, SNOMED CT, and NCI. Large biomedical tracks are mainly divided into three related problems: FMA-NCI, FMA-SNOMED, and SNOMED-NCI, each involving various parts of the input ontology.

Table 8.
List of the model parameters.
5.2. Experimental Results and Analysis
The experimental evaluation of our proposed model is presented in this part. Our approach is predicated on the notion that BK based matching can be accomplished by employing many matchers. According to the OAEI findings, some matchers find the correct mappings, whereas others find different ones. In addition, none of them can achieve good results in all matching tasks. Accordingly, it would be more successful in combining alignments produced by several matchers. This experiment investigates many aggregation strategies to confirm our assumption: Minimum, Maximum, Average, and Vote.
5.2.1. Building the Graphs Using Multi Matchers
The most straightforward method of obtaining mappings between ontologies is to employ an automatic matcher. We saw a wide range of outcomes produced by several different matchers, including LogMap, LogMapLt, and AML, as illustrated in Figure 6. We extracted all potential mappings between the preselected ontologies BK1(DOID) and BK2(UBERON) to construct mappings across some intermediate ontologies. According to our experiments, various aggregation procedures resulted in a wide variety of correspondences. LogMap yielded (159) correspondences, whereas AML (62) and LogMapLt created only (6). We arrived at the following result by combining all of the correspondences (227). Different aggregation strategies resulted in a variety of final alignments, namely, Min (194), Max (195), Avg (195), and Vote (19). The Vote method achieved the most precise final alignment. Meanwhile, the recall rate was relatively low. There were just 19 retrieved correspondences. The reason is that LogMapLt only retrieved six matches. Then, the source ontology was matched against the preselected ontologies (SBK1) and (SBK2). Then the constructed graph was compared with the target ontology. The Min (BKTM), Max (BKTX), Avg (BKTA), and Vote (BKTV) strategies produced comparable outcomes throughout the tests. The purpose of BK based matching is to supplement, not to replace, direct matching as defined by (DST). Direct matching may reveal mappings that BK based matching misses, and vice versa.
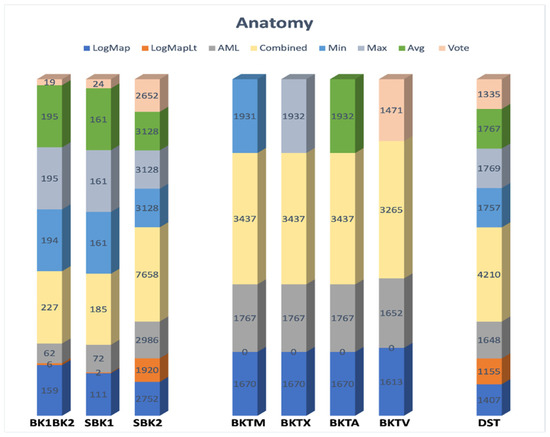
Figure 6.
Applying several matchers and different aggregation strategies on the Anatomy track.
Similar test cases of the Large Biomed tracks were organized to demonstrate the validity of our model in different versions across various matching situations. These six test cases include ontologies where the different aggregation strategies are applied, as shown in Figure 7, Columns (a–f). The voting technique comprised at least two matches to generate the mapping. Meanwhile, Min, Max, and Avg considered all mappings and altered the score’s value. According to these statistics, harvesting multiple matchers is a viable option. We believe that the strength and competency of the final alignment are in using a single aggregation technique and the use of distinct ones across various ontologies based on the preconfiguration process rather than utilizing a single aggregation method. In such a scenario, when vast ontologies are matched, it would be difficult and time consuming to apply Min, Max, and Avg aggregation methods as long as the results are comparable. The F-measure results show that the Max techniques were the most effective because the recall rate is high. The retrieved correspondences have a much higher confidence value than those found by other aggregation methods.
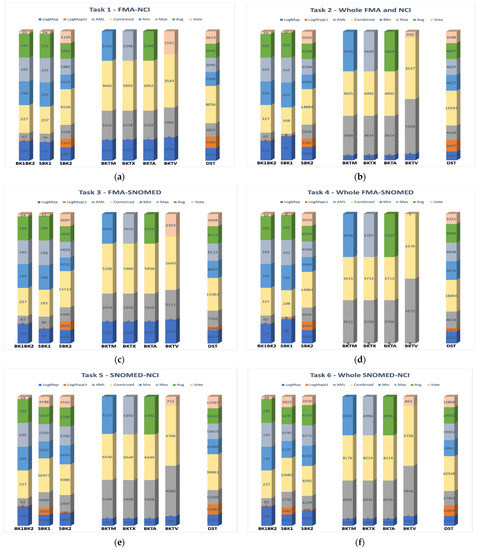
Figure 7.
Applying several matchers and different aggregation strategies as: (a) Task 1—FMA-NCI (b) Task 2—Whole FMA and NCI (c) Task 3—FMA-SNOMED (d) Task 4—Whole FMA-SNOMED (e) Task 5—SNOMED-NCI (f) Task 6—Whole SNOMED-NCI.
5.2.2. BK Path-Driven Inferencing
Pathways between the source and the target entities were searched to derive possible mappings. One or more matchers could define each detected path. The path contains some intermediate concepts that are members of the ontologies that have been preselected. Our research shows that additional mappings and pathways are generated when deriving mappings that include multiple matchers. The candidate mappings returned by many paths and matchers are more likely to be accurate than those returned by a small number of paths and matchers. Pathways with various matchers are more relevant than paths with only one matcher. One of the advantages of taking a multipath method to identify correspondences is that it may return several alternative mappings between two entities, which is helpful in various situations. Such relationships may affirm or contradict one another, which must be considered when determining the final alignment.
Our findings revealed that different aggregation methods resulted in a range of path numbers. The test result shows that the Vote technique returned the smallest number of paths because it only contains the paths established by a minimum of two matchers. The Max and Avg techniques yielded nearly identical path counts throughout the experiments. Meanwhile, the Max method has a higher confidence value. Table 9 illustrates that paths returned by many matchers have a higher confidence positive value. Examples include paths that all matchers have confirmed in the Anatomy Track, Task 1—FMA-NCI, Task 3—FMA-SNOMED, and Task 5—SNOMED-NCI, all of which have positive values greater than 0.900. The other tasks were given lower values because all the matchers did not perform well in large fragment tests as they did in small fragment testing. Another example is Task 6—whole SNOMED-NCI. LogMap and AML matchers created 7519 paths, of which only 2827 are correct, and 4692 are incorrect, and a low positive value (0.374). The paths created with three matchers within the same track have a positive value up to 0.824. Therefore, we used our proposed measure to guide the final rules algorithm to eliminate mappings with low positive values.

Table 9.
Comparison of the correct paths produced by different matchers with the reference alignment.
Moreover, the experiment shows that AML was the most active matcher across all paths, particularly for the Min, Max, and Avg aggregation methods. When the Vote method was used, LogMap generated more candidate paths. In contrast with the previous finding, LogMapLt is the least occurring matcher in all experiments because it has lower actual alignment results than AML and LogMap across all tests. Furthermore, LogMapLt does not generate any unique paths in all tests due to AML, and LogMap produces better actual alignment results as single matchers. The Min, Max, and Avg versions of paths derived by one matcher generated nearly identical results. For example, AML generated more unique paths using the Min version. More incorrect pathways were retrieved in Tasks 2, 4, and 6. In Task 2, 1651 out of the 2132 paths are incorrect due to their size and difficulty. LogMap generated (96) paths for the Anatomy track, but (65) are incorrect.
In the case of paths derived by two matchers, LogMapLt and AML did not create any paths throughout all tests. Meanwhile, LogMap and LogMapLt generated paths in all tasks. Concerning the results that they obtained in Task 6, 1109 out of the 1755 paths are wrong. In Task 2, only 17 out of the 195 paths are correct. Finally, more correct correspondences were found once all matchers formulated a path, as shown in Table 9.
5.2.3. Our Model with Different Direct Matchers and GBKOM
This work aims to compare the results obtained by four versions of our model based on aggregation strategies with state of the art matching systems. We use traditional precision, recall, and F-measure to evaluate our model. More correct correspondences were obtained when the recall value is high. Meanwhile, the number of successfully discovered correspondences is limited when the recall value is low. Considering the measure of precision, less false matching occurs when its value is high.
The number of false correspondences discovered by the system must be kept to a minimum to maintain a high precision value. If the F-Measure value is significant, then the expert’s additional work to correct derived correspondences is reduced. The matching system aims to reach the best possible recall and precision values to make less work correcting results. Our proposed algorithm assists us in excluding the possibility of false mapping. Our results are illustrated in Table 10, Table 11 and Table 12. The findings of each test case in the Anatomy and Large Biomed tracks generated by four versions of our model and cutting-edge matching methods are shown. The overall results of these four versions of our model are nearly the same. However, several test cases provided by the Vote approach produced quite different outcomes, demonstrating that our hypothesis still has potential for improvement in matching Large Biomed tracks. In addition, it serves as justification for carrying out this research’s overall goal of developing a novel aggregation method.

Table 10.
Compare our model with GBKOM and different direct matchers using the precision measure.

Table 11.
Compare our model with GBKOM and different direct matchers using the recall measure.

Table 12.
Compare our model with GBKOM and different direct matchers using the f-measure measure.
To demonstrate our model’s quality, we compared it against the LogMap, LogMapLt, AML, and GBKOM systems in various matching scenarios. According to these seven separate test scenarios, we can compare four versions of our model and other systems. Table 10 compares the findings for several test case groups using the precision measure. In this case, our model’s (Vote) version outperformed other versions and systems in terms of precision across all test groups. However, the findings for other versions are nearly identical for all sets of test cases. Hence, the other versions yield satisfactory results for the Anatomy and Large Biomed tracks. The recall result is shown in Table 11. Our model achieves better outcomes for all groups of test cases in recall measures. However, no significant variation in recall results is observed between our three versions, namely, Min, Max, and Avg.
Table 12 shows a more detailed look at F-Measure, which evaluates the matching process. Our model’s (Max) version achieved marginally better outcomes than other systems in all test instances except for the Anatomy Track and Task 2. A possible explanation for this situation is the usage of the preselected ontology UBERON. By contrast, the Vote version produced the best results. AML and GBKOM have also shown positive results. Furthermore, the values of our model in different versions are interesting to observe, and the overall matching results are nearly as good as other systems for the majority of test groups. The evaluation findings reveal that, while the number of correct correspondences found by the three versions of our model is nearly identical, our model finds more trustworthy correspondences because it incorporates the BK false mappings repository. The time results are not comparable because the matchers were not launched under the same conditions and with different BKs.
Finally, we can conclude that there is no optimal ontology matching strategy. The user’s requirements for precision, recall, and computation time are considered while selecting a particular approach or the system that implements that strategy. When possible, we believe it would be helpful to report participant findings with and without specialized BK resources. On the one hand, this provides a more accurate assessment of the advantage of utilizing BK resources in matching results and calculation time on the other side. On the other hand, systems that do not use BK resources can be compared.
6. Conclusions and Future Work
We present a BK multimatcher approach in this work and demonstrate how to combine and aggregate distinct mapping alignments generated by several automatic matchers. We presented an aggregation model consisting of four aggregation methods to establish the final alignment between the compared ontologies: Min, Max, Avg, and Vote. The experimental findings reveal that the Max version discovers more dependable correspondences because the values are significantly higher than those of correspondences found by other versions. Accordingly, the recall is high. According to the experiments, the voting process provides the most precise final alignment, but low recall rate. Another essential feature is the addition of a new measure known as the matcher path confidence measure. This measure can aid in identifying the correct mappings by taking the matcher’s confidence into account. The names of the matchers are also placed in the paths.
We also proposed the final mapping selection algorithm to decide the final alignment. The results show that our matching model demonstrated effectiveness throughout many test cases within the Anatomy and Large Biomed tracks, as our system’s performance is the best. Our system performed remarkably well due to the higher recall levels obtained besides utilizing the false mapping repository and the guidance of the proposed matcher path confidence measure. In future work, we will enhance our proposed final mapping selection algorithm to identify more false mappings. Moreover, it would be difficult and time consuming in such a scenario, when vast ontologies are matched to use all matchers simultaneously. Therefore, we intend to select some matchers from the matchers library to employ for a specific task. Then, the matchers can be arranged in a parallel composition. Furthermore, future studies will aim to demonstrate the model outside the biomedical domain to overcome the limitation of domain dependence in our study. In general, our model may serve as a first step toward supporting domain independent solutions by applying hybrid matchers. Finally, exploiting unstructured BK sources will be attractive to investigate as our model only exploits ontologies as BK sources.
Author Contributions
Conceptualization, S.A.-Y., W.-W.G., E.-X.T., N.Z.J. and P.B.; Methodology, S.A.-Y., W.-W.G., E.-X.T., N.Z.J. and P.B; software, S.A.-Y.; formal analysis, S.A.-Y.; writing—original draft preparation, S.A.-Y.; writing—review and editing, S.A.-Y., W.-W.G., E.-X.T., N.Z.J. and P.B.; supervision, W.-W.G., E.-X.T., N.Z.J. and P.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used to support this study’s findings are available from OAEI and are available online which can be accessed on http://oaei.ontologymatching.org/2020/ (accessed on 21 November 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| AMA | Adult Mouse Anatomy |
| AML | AgreementMakerLight |
| BK | Background Knowledge |
| COMA | Combination of Schema Matching Approaches |
| DOID | Human Disease Ontology |
| FMA | Foundational Model of Anatomy |
| GBKOM | A Generic framework for BK Based Ontology Matching |
| GOMMA | Generic Ontology Matching and Mapping Management |
| LogMap | Logic Based and Scalable Ontology Matching |
| LogMapBio | LogMap BioPortal |
| LogMapLt | LogMap Lightweight |
| MA | Mouse Anatomy |
| NCI | National Cancer Institute Thesaurus |
| NCBO | The National Center for Biomedical Ontology |
| OAEI | Ontology Alignment Evaluation Initiative |
| SNOMED CT | SNOMED Clinical Terms |
| UBERON | The Uber Anatomy Ontology |
| UMLS | The Unified Medical Language System |
| YAM++ | Yet Another Matcher for Ontology Matching |
References
- Al-Yadumi, S.; Xion, T.E.; Wei, S.G.W.; Boursier, P. Review on Integrating Geospatial Big Datasets and Open Research Issues. IEEE Access 2021, 9, 10604–10620. [Google Scholar] [CrossRef]
- El Hajjamy, O.; Alaoui, L.; Bahaj, M. Semantic integration of heterogeneous classical data sources in ontological data warehouse. In Proceedings of the International Conference on Learning and Optimization Algorithms: Theory and Applications, Rabat, Morocco, 2–5 May 2018; pp. 1–8. [Google Scholar]
- Euzenat, J.; Shvaiko, P. Ontology Matching, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2013; Available online: http://book.ontologymatching.org/ (accessed on 3 February 2021).
- Tudorache, T. Ontology engineering: Current state, challenges, and future directions. Semant. Web 2020, 11, 125–138. [Google Scholar] [CrossRef]
- Pesquita, C. Towards Semantic Integration for Explainable Artificial Intelligence in the Biomedical Domain. In Proceedings of the ACM SIGMOD International Conference on Management of Data, 14 June 2005; Baltimore, MD, USA; pp. 906–908. [Google Scholar] [CrossRef]
- Faria, D.; Pesquita, C.; Mott, I.; Martins, C.; Couto, F.M.; Cruz, I.F. Tackling the challenges of matching biomedical ontologies. J. Biomed. Semant. 2018, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Zhu, Y.; Song, J. Progress and Challenges on Entity Alignment of Geographic Knowledge Bases. ISPRS Int. J. Geo-Inf. 2019, 8, 77. [Google Scholar] [CrossRef]
- Portisch, J.P. Towards Matching of Domain-Specific Schemas Using General-Purpose External Background Knowledge. In Proceedings of the European Semantic Web Conference, Heraklion, Greece, 31 May–4 June 2020; 12124 LNCS. pp. 270–279. [Google Scholar] [CrossRef]
- Nkisi-Orji, I.; Wiratunga, N.; Massie, S.; Hui, K.-Y.; Heaven, R. Ontology alignment based on word embedding and random forest classification. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Dublin, Ireland, 10 September 2018; pp. 557–572. [Google Scholar] [CrossRef]
- Karimi, H.; Kamandi, A. Ontology alignment using inductive logic programming. In Proceedings of the 2018 4th International Conference on Web Research, ICWR 2018, Tehran, Iran, 25 April 2018; pp. 118–127. [Google Scholar] [CrossRef]
- Pesquita, C.; Santos, E.; Palmonari, M.; Cruz, I.F.; Couto, F.M. The AgreementMakerLight ontology matching system. In Proceedings of the On the Move to Meaningful Internet Systems (OTM 2013), Graz, Austria, 9–13 September 2013; pp. 527–541. [Google Scholar] [CrossRef]
- Aumueller, D.; Do, H.-H.; Massmann, S.; Rahm, E. Schema and ontology matching with COMA++. In Proceedings of the ACM SIGMOD International Conference on Management of Data, Baltimore, MD, USA, 14 June 2005; pp. 906–908. [Google Scholar] [CrossRef]
- Ren, F.; Deng, J. Background Knowledge Based Multi-Stream Neural Network for Text Classification. Appl. Sci. 2018, 8, 2472. [Google Scholar] [CrossRef]
- Annane, A.; Bellahsene, Z. GBKOM: A generic framework for BK-based ontology matching. J. Web Semant. 2020, 63, 100563. [Google Scholar] [CrossRef]
- Locoro, A.; David, J.; Euzenat, J. Context-Based Matching: Design of a Flexible Framework and Experiment. J. Data Semant. 2013, 3, 25–46. [Google Scholar] [CrossRef][Green Version]
- Annane, A.; Bellahsene, Z.; Azouaou, F.; Jonquet, C. Selection and combination of heterogeneous mappings to enhance biomedical ontology matching. In Proceedings of the European Knowledge Acquisition Workshop, Bologna, Italy, 19–23 November 2016; pp. 19–33. [Google Scholar] [CrossRef]
- Portisch, J.; Hladik, M.; Paulheim, H. Background Knowledge in Schema Matching. Semant. Web J. 2020, 1, 1–5. Available online: http://www.semantic-web-journal.net/system/files/swj2645.pdf (accessed on 10 January 2021).
- Real, F.J.Q.; Bella, G.; McNeill, F.; Bundy, A. Using domain lexicon and grammar for ontology matching. In Proceedings of the 15th International Workshop on Ontology Matching, Online. Athens, Greece, 2–3 November 2020; Volume 2788, pp. 1–12. [Google Scholar]
- Annane, A.; Bellahsene, Z.; Azouaou, F.; Jonquet, C. Building an effective and efficient background knowledge resource to enhance ontology matching. J. Web Semant. 2018, 51, 51–68. [Google Scholar] [CrossRef]
- Gherbi, S.; Khadir, M.T. Inferred Ontology Concepts Alignment Using Instances and an External Dictionary. Procedia Comput. Sci. 2016, 83, 648–652. [Google Scholar] [CrossRef]
- Yousfi, A.; Hafid, M.; Zellou, A. xMatcher: Matching Extensible Markup Language Schemas using Semantic-based Techniques. Int. J. Adv. Comput. Sci. Appl. 2020, 11, 655–665. [Google Scholar] [CrossRef]
- Destro, J.M.; Vargas, J.A.; dos Reis, J.C.; Torres, R.D.S. EVOCROS: Results for OAEI 2019. CEUR Workshop Proc. 2019, 2536, 131–137. [Google Scholar]
- Schmidt, D.; Trojahn, C.; Vieira, R.; Kamel, M. Validating Top-Level and Domain Ontology Alignments Using WordNet. In Proceedings of the Brazilian Seminar Ontology (ONTOBRAS 2016), Curitiba, Brazil, 3–6 October 2016. [Google Scholar]
- Jiménez-Ruiz, E. LogMap family participation in the OAEI 2020. In Proceedings of the 15th International Workshop on Ontology Matching (OM 2020), Athens, Greece, 2–6 November 2020; Volume 2788, pp. 201–203. [Google Scholar]
- Kachroudi, M.; Diallo, G.; Ben Yahia, S. On the composition of large biomedical ontologies alignment. In Proceedings of the 7th International Conference on Web Intelligence, Mining and Semantics, Amantea, Italy, 19–22 June 2017; pp. 1–10. [Google Scholar] [CrossRef]
- Nikooie Pour, M.A.; Algergawy, A.; Amini, R.; Faria, D.; Fundulaki, I.; Harrow, I.; Hertling, S.; Jimenez-Ruiz, E.; Jonquet, C.; Karam, N.; et al. Results of the ontology alignment evaluation initiative 2020. City Res. Online 2020, 37, 1591–1601. [Google Scholar]
- Kirsten, T.; Gross, A.; Hartung, M.; Rahm, E. GOMMA: A component-based infrastructure for managing and analyzing life science ontologies and their evolution. J. Biomed. Semant. 2011, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Ruiz, E.; Cuenca Grau, B. LogMap: Logic-based and scalable ontology matching. In Proceedings of the 10th International Semantic Web Conference, Bonn, Germany, 23–27 October 2011; pp. 273–288. [Google Scholar] [CrossRef]
- Groß, A.; Hartung, M.; Kirsten, T.; Rahm, E. Mapping composition for matching large life science ontologies. In Proceedings of the International Conference on Biomedical Ontology: ICBO 2011, Buffalo, NY, USA, 26 July 2011; Volume 833, pp. 109–116. [Google Scholar]
- Hartung, M.; Groß, A.; Rahm, E. Composition methods for link discovery. In Proceedings of the Datenbanksysteme für Business, Technologie und Web (BTW), Magdeburg, Germany, 11–15 March 2013; pp. 261–277. [Google Scholar]
- Chen, X.; Xia, W.; Jiménez-Ruiz, E.; Cross, V.V. Extending an ontology alignment system with BIOPORTAL: A preliminary analysis. In Proceedings of the ISWC 2014 Posters & Demonstrations Track a Track within the 13th International Semantic Web Conference, Riva del Garda, Italy, 21 October 2014; Volume 1272, pp. 313–316. [Google Scholar]
- Geometry, R.; Analysis, G. Automatic Background Knowledge Selection for Matching Biomedical Ontologies. PLoS ONE 2014, 11, e111226. [Google Scholar]
- Hartung, M.; Gross, A.; Kirsten, T.; Rahm, E. Effective composition of mappings for matching biomedical ontologies. In Proceedings of the Extended Semantic Web Conference, Bethlehem, PA, USA, 11–15 October 2015; Volume 7540, pp. 176–190. [Google Scholar] [CrossRef]
- Tigrine, A.N.; Bellahsene, Z.; Todorov, K. Selecting optimal background knowledge sources for the ontology matching task. In Proceedings of the European Knowledge Acquisition Workshop, Bologna, Italy, 19–23 November 2016; pp. 651–665. [Google Scholar] [CrossRef]
- Quix, C.; Roy, P.; Kensche, D. Automatic selection of background knowledge for ontology matching. In Proceedings of the International Workshop on Semantic Web Information Management, SWIM 2011, Athens, Greece, 12–16 June 2011; Volume 5, pp. 1–7. [Google Scholar] [CrossRef]
- Rahm, E. Towards Large-Scale Schema and Ontology Matching. In Schema Matching and Mapping; Bellahsene, Z., Bonifati, A., Rahm, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 3–27. [Google Scholar] [CrossRef]
- Gulić, M.; Vrdoljak, B.; Banek, M. CroMatcher: An ontology matching system based on automated weighted aggregation and iterative final alignment. J. Web Semant. 2016, 41, 50–71. [Google Scholar] [CrossRef]
- Duchateau, F.; Bellahsene, Z. YAM: A step forward for generating a dedicated schema matcher. In Transactions on Large-Scale Data- and Knowledge-Centered Systems XXV; Springer: Berlin/Heidelberg, Germany, 2016; Volume 9620, pp. 150–185. [Google Scholar] [CrossRef]
- Cardoso, S.D.; Da Silveira, M.; Lin, Y.-C.; Christen, V.; Rahm, E.; Reynaud-Delaître, C.; Pruski, C. Combining semantic and lexical measures to evaluate medical terms similarity. In Proceedings of the International Conference on Data Integration in the Life Sciences, Hannover, Germany, 20–21 November 2018; pp. 17–32. [Google Scholar] [CrossRef]
- Gulić, M.; Vrdoljak, B.; Vuković, M. An Iterative Automatic Final Alignment Method in the Ontology Matching System. J. Inf. Organ. Sci. 2018, 42, 39–61. [Google Scholar] [CrossRef]
- Gross, A.; Hartung, M.; Kirsten, T.; Rahm, E. On matching large life science ontologies in parallel. In Proceedings of the International Conference on Data Integration in the Life Sciences, Gothenburg, Sweden, 25–27 August 2010; pp. 35–49. [Google Scholar] [CrossRef]
- Wang, S.; Schlobach, S.; Takens, J.; Van Atteveldt, W. Mapping-chains for studying concept shift in political ontologies. In Proceedings of the 4th International Workshop on Ontology Matching (OM-2009), Fairfax, VA, USA, 25 October 2009; Volume 551, pp. 13–24. [Google Scholar]
- rojahn, C.; Moraes, M.; Quaresma, P.; Vieira, R. A cooperative approach for composite ontology mapping. In Journal on Data Semantics X; Springer: Berlin, Germany, 2008; pp. 237–263. [Google Scholar] [CrossRef]
- Peukert, E.; Maßmann, S.; König, K. Comparing similarity combination methods for schema matching. INFORMATIK 2010. Serv. Sci. Neue Perspekt. Für Die Inform. 2020, 1, 692–701. [Google Scholar]
- Euzenat, J. Algebras of ontology alignment relations. In Proceedings of the International Semantic Web Conference, Karlsruhe, Germany, 26–30 October 2008; pp. 387–402. [Google Scholar] [CrossRef]
- Nunes, B.P.; Dietze, S.; Casanova, M.A.; Kawase, R.; Fetahu, B.; Nejdl, W. Combining a co-occurrence-based and a semantic measure for entity linking. In Proceedings of the Extended Semantic Web Conference, Montpellier, France, 26–30 May 2013; pp. 548–562. [Google Scholar] [CrossRef]
- Mascardi, V.; Locoro, A.; Rosso, P. Automatic Ontology Matching via Upper Ontologies: A Systematic Evaluation. IEEE Trans. Knowl. Data Eng. 2009, 22, 609–623. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).