HMIC: Hierarchical Medical Image Classification, A Deep Learning Approach
Abstract
1. Introduction and Related Works
2. Data Source
3. Pre-Processing
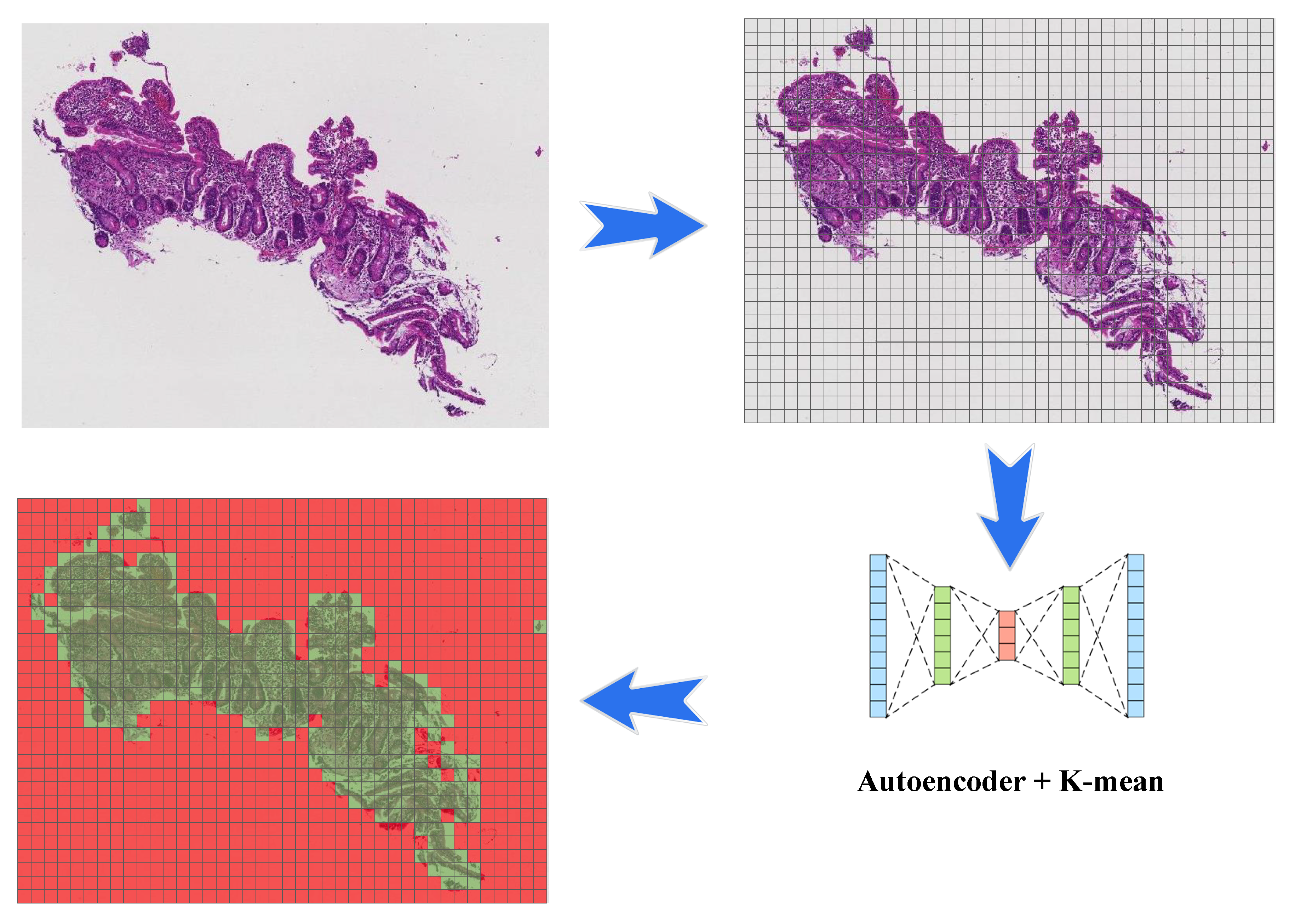
3.1. Image Patching
3.2. Clustering
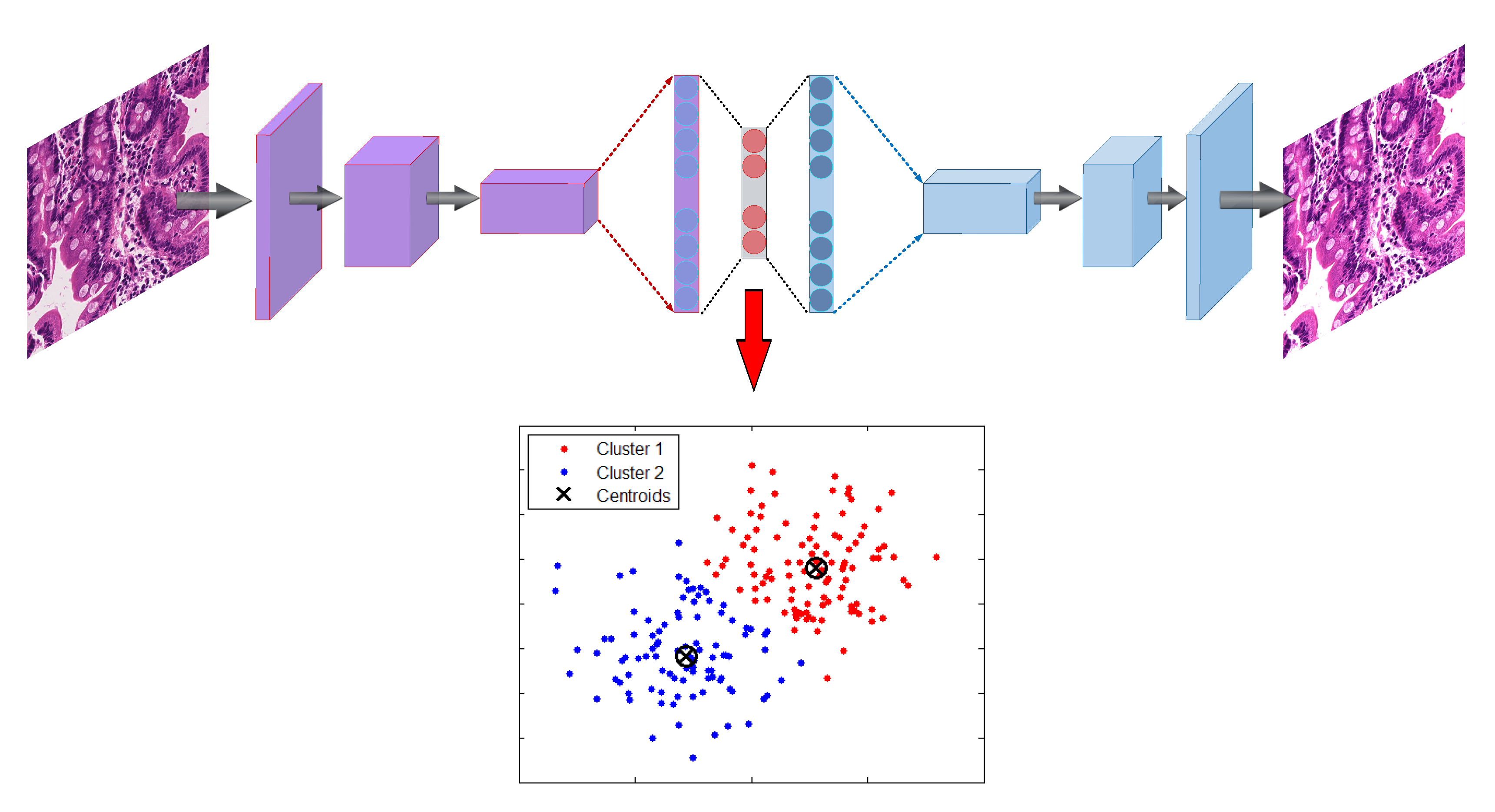
3.2.1. Autoencoder
Encode
Decode
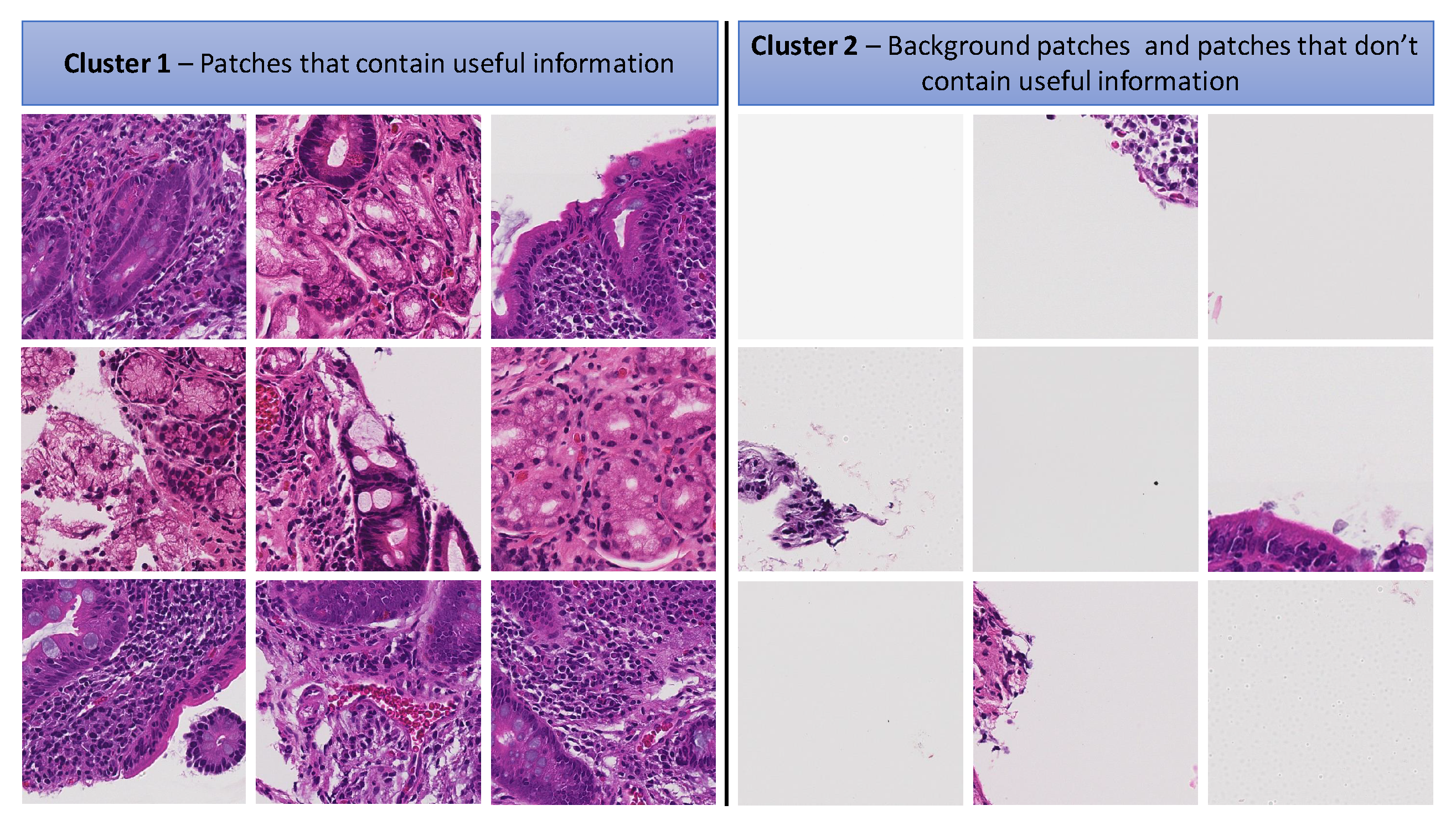
3.2.2. K-Means
| Algorithm 1 K-means algorithm for 2 clusters medical images |
 |
3.3. Medical Image Staining
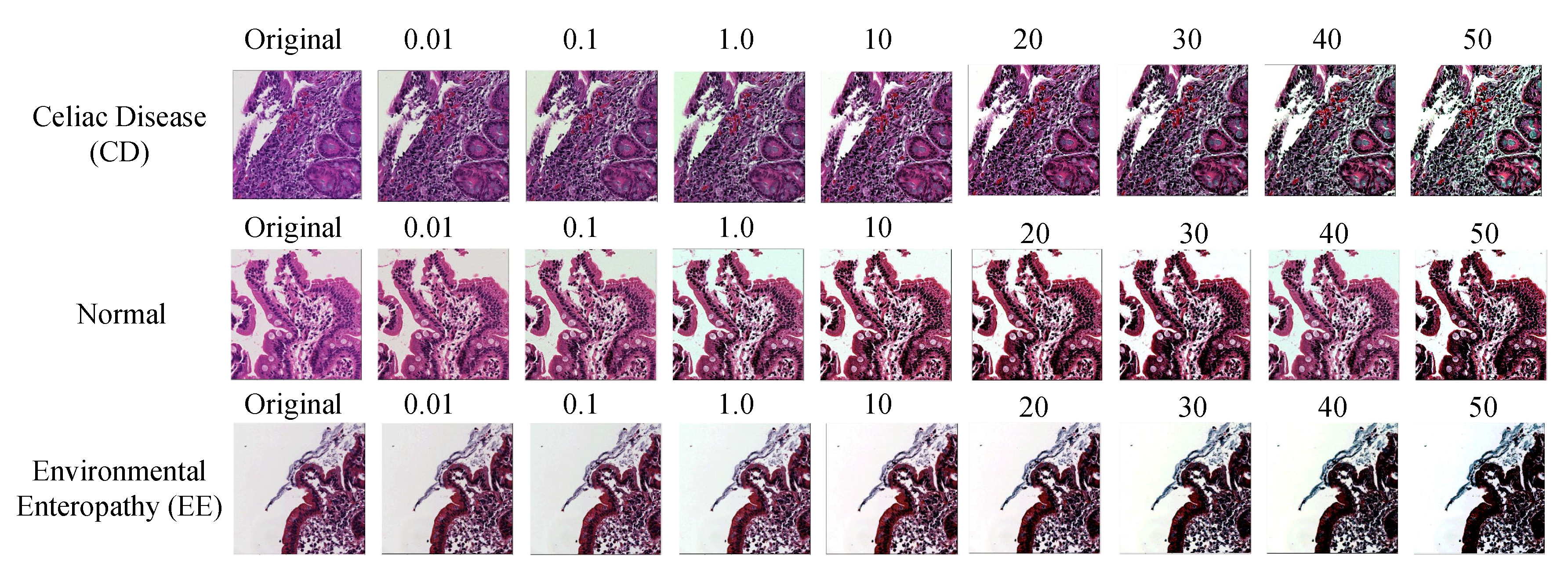
3.3.1. Color Balancing
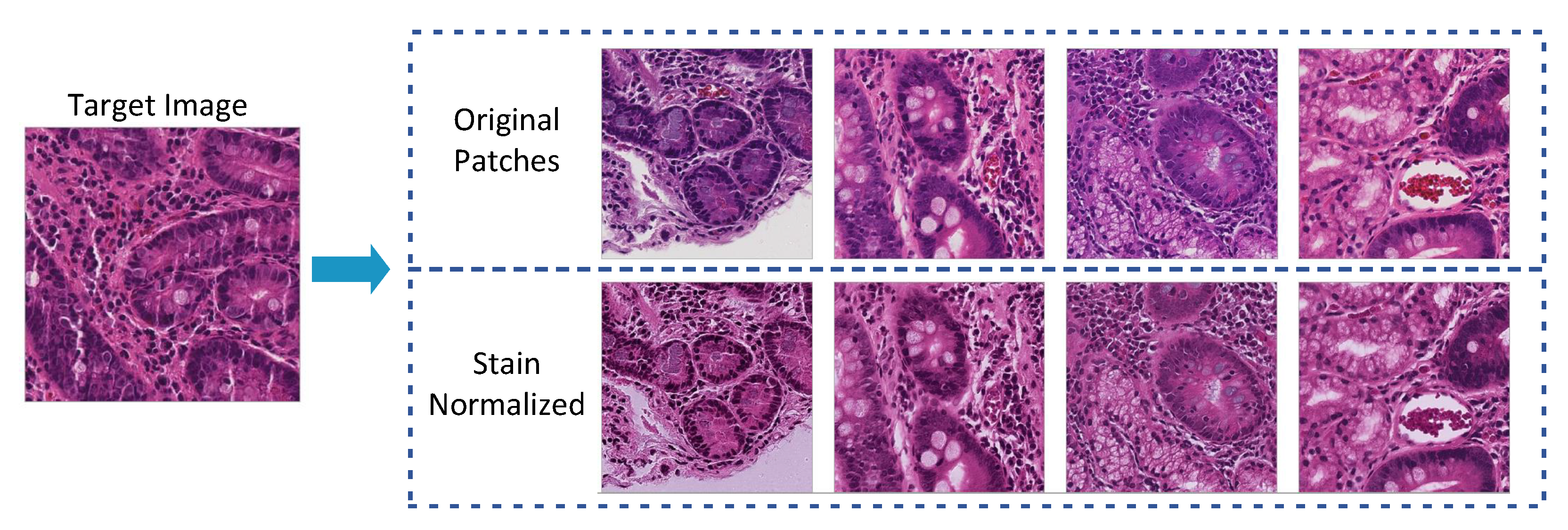
3.3.2. Stain Normalization
4. Baseline
4.1. Deep Convolutional Neural Networks
4.2. Deep Neural Networks
5. Method
5.1. Convolutional Neural Networks
5.1.1. Convolutional Layer
5.1.2. Pooling Layer
5.1.3. Neuron Activation
5.1.4. Optimizer
5.1.5. Network Architecture
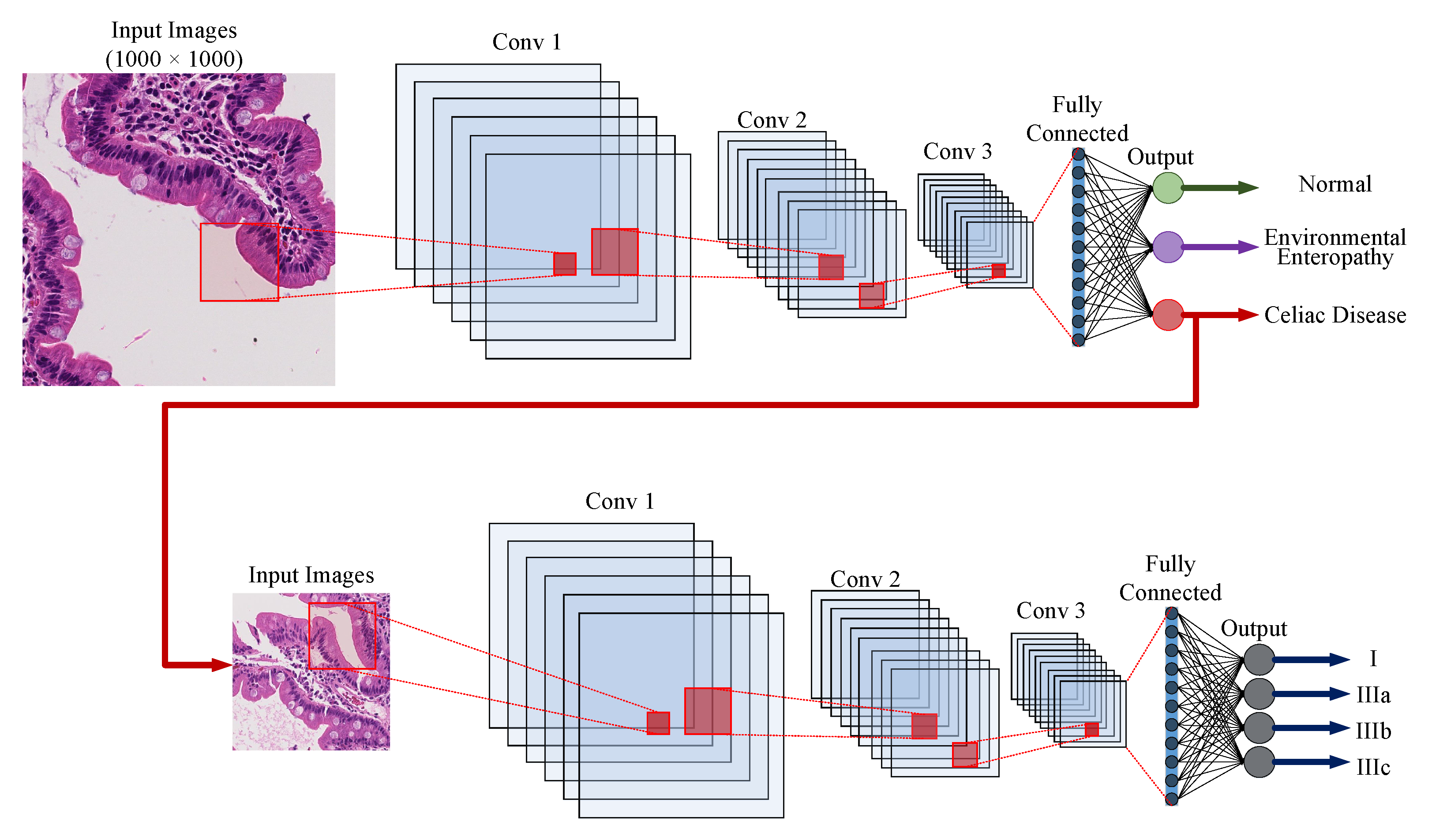
5.2. Whole Slide Classification
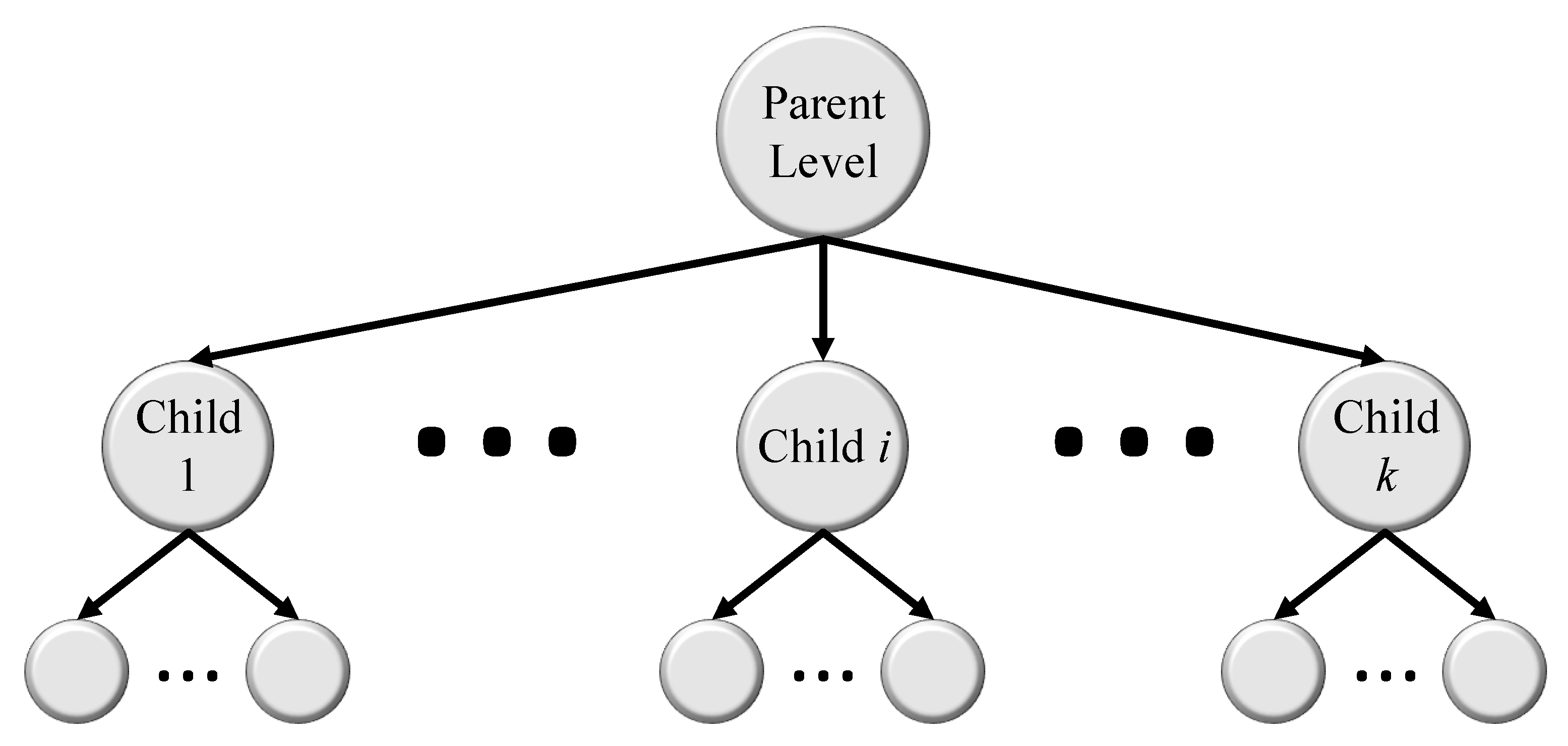
5.3. Hierarchical Medical Image Classification
6. Results
6.1. Evaluation Setup
6.2. Experimental Setup
6.3. Empirical Results
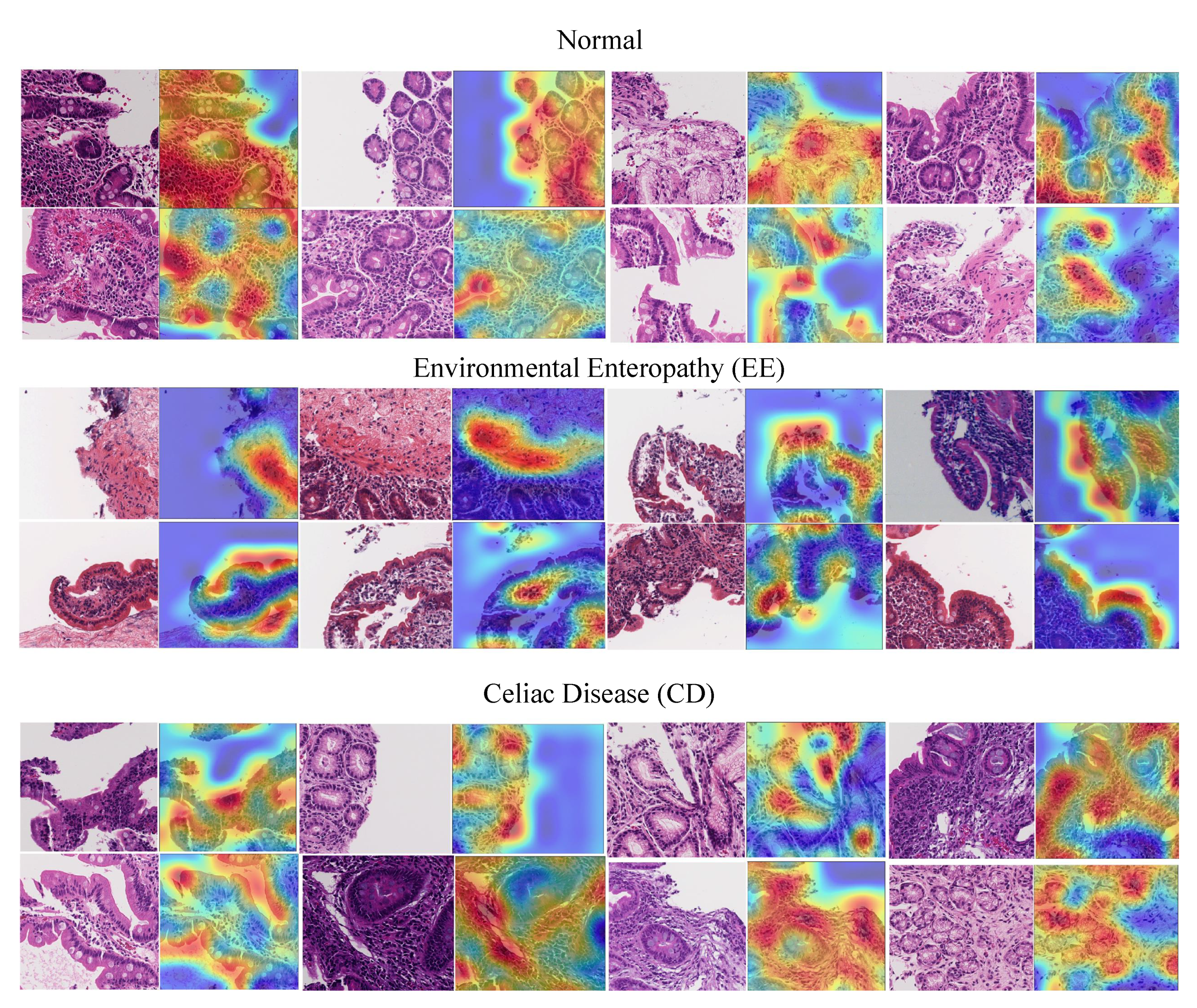
6.4. Visualization
- EE: surface epithelium with IELs and goblet cells was highlighted. Within the lamina propria, the heatmaps also focused on mononuclear cells.
- CD: heatmaps highlighted the edge of crypt cross sections, surface epithelium with IELs and goblet cells, and areas with mononuclear cells within the lamina propria.
- Histologically Normal: surface epithelium with epithelial cells containing abundant cytoplasm was highlighted.
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sali, R.; Ehsan, L.; Kowsari, K.; Khan, M.; Moskaluk, C.A.; Syed, S.; Brown, D.E. CeliacNet: Celiac Disease Severity Diagnosis on Duodenal Histopathological Images Using Deep Residual Networks. arXiv 2019, arXiv:1910.03084. [Google Scholar]
- Kowsari, K.; Sali, R.; Khan, M.N.; Adorno, W.; Ali, S.A.; Moore, S.R.; Amadi, B.C.; Kelly, P.; Syed, S.; Brown, D.E. Diagnosis of celiac disease and environmental enteropathy on biopsy images using color balancing on convolutional neural networks. In Proceedings of the Future Technologies Conference; Springer: Cham, Switzerland, 2019; pp. 750–765. [Google Scholar]
- Kowsari, K. Diagnosis and Analysis of Celiac Disease and Environmental Enteropathy on Biopsy Images using Deep Learning Approaches. Ph.D. Thesis, University of California, Los Angeles, CA, USA, 2020. [Google Scholar] [CrossRef]
- Kowsari, K.; Jafari Meimandi, K.; Heidarysafa, M.; Mendu, S.; Barnes, L.; Brown, D. Text Classification Algorithms: A Survey. Information 2019, 10, 150. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Nobles, A.L.; Glenn, J.J.; Kowsari, K.; Teachman, B.A.; Barnes, L.E. Identification of imminent suicide risk among young adults using text messages. In Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems, Montreal, QC, Canada, 21–26 April 2018; ACM: New York, NY, USA, 2018; p. 413. [Google Scholar]
- Zhai, S.; Cheng, Y.; Zhang, Z.M.; Lu, W. Doubly convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 1082–1090. [Google Scholar]
- Hegde, R.B.; Prasad, K.; Hebbar, H.; Singh, B.M.K. Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images. Biocybern. Biomed. Eng. 2019, 39, 382–392. [Google Scholar] [CrossRef]
- Zhang, J.; Kowsari, K.; Harrison, J.H.; Lobo, J.M.; Barnes, L.E. Patient2Vec: A Personalized Interpretable Deep Representation of the Longitudinal Electronic Health Record. IEEE Access 2018, 6, 65333–65346. [Google Scholar] [CrossRef]
- Pavik, I.; Jaeger, P.; Ebner, L.; Wagner, C.A.; Petzold, K.; Spichtig, D.; Poster, D.; Wüthrich, R.P.; Russmann, S.; Serra, A.L. Secreted Klotho and FGF23 in chronic kidney disease Stage 1 to 5: A sequence suggested from a cross-sectional study. Nephrol. Dial. Transplant. 2013, 28, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Kowsari, K.; Brown, D.E.; Heidarysafa, M.; Meimandi, K.J.; Gerber, M.S.; Barnes, L.E. Hdltex: Hierarchical deep learning for text classification. In Proceedings of the 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA), Cancun, Mexico, 18–21 December 2017; pp. 364–371. [Google Scholar]
- Dumais, S.; Chen, H. Hierarchical classification of web content. In Proceedings of the 23rd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, Athens, Greece, 24–28 July 2000; pp. 256–263. [Google Scholar]
- Yan, Z.; Piramuthu, R.; Jagadeesh, V.; Di, W.; Decoste, D. Hierarchical Deep Convolutional Neural Network for Image Classification. U.S. Patent 10,387,773, 20 August 2019. [Google Scholar]
- Seo, Y.; Shin, K.S. Hierarchical convolutional neural networks for fashion image classification. Expert Syst. Appl. 2019, 116, 328–339. [Google Scholar] [CrossRef]
- Ranjan, N.; Machingal, P.V.; Jammalmadka, S.S.D.; Thenaknidiyoor, V.; Dileep, A. Hierarchical Approach for Breast cancer Histopathology Images Classification. 2018. Available online: https://openreview.net/forum?id=rJlGvTojG (accessed on 10 January 2019).
- Zhu, X.; Bain, M. B-CNN: Branch convolutional neural network for hierarchical classification. arXiv 2017, arXiv:1709.09890. [Google Scholar]
- Sali, R.; Adewole, S.; Ehsan, L.; Denson, L.A.; Kelly, P.; Amadi, B.C.; Holtz, L.; Ali, S.A.; Moore, S.R.; Syed, S.; et al. Hierarchical Deep Convolutional Neural Networks for Multi-category Diagnosis of Gastrointestinal Disorders on Histopathological Images. arXiv 2020, arXiv:2005.03868. [Google Scholar]
- Syed, S.; Ali, A.; Duggan, C. Environmental enteric dysfunction in children: A review. J. Pediatr. Gastroenterol. Nutr. 2016, 63, 6. [Google Scholar] [CrossRef]
- Naylor, C.; Lu, M.; Haque, R.; Mondal, D.; Buonomo, E.; Nayak, U.; Mychaleckyj, J.C.; Kirkpatrick, B.; Colgate, R.; Carmolli, M.; et al. Environmental enteropathy, oral vaccine failure and growth faltering in infants in Bangladesh. EBioMedicine 2015, 2, 1759–1766. [Google Scholar] [CrossRef] [PubMed]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A.; Catassi, C. Current approaches to diagnosis and treatment of celiac disease: An evolving spectrum. Gastroenterology 2001, 120, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Samaras, D.; Kurc, T.M.; Gao, Y.; Davis, J.E.; Saltz, J.H. Patch-based convolutional neural network for whole slide tissue image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2424–2433. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Volume 1. [Google Scholar]
- Wang, W.; Huang, Y.; Wang, Y.; Wang, L. Generalized autoencoder: A neural network framework for dimensionality reduction. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Columbus, OH, USA, 23–28 June 2014; pp. 490–497. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning Internal Representations by Error Propagation; Technical Report; California Univ San Diego La Jolla Inst for Cognitive Science: La Jolla, CA, USA, 1985. [Google Scholar]
- Liang, H.; Sun, X.; Sun, Y.; Gao, Y. Text feature extraction based on deep learning: A review. EURASIP J. Wirel. Commun. Netw. 2017, 2017, 211. [Google Scholar] [CrossRef]
- Masci, J.; Meier, U.; Cireşan, D.; Schmidhuber, J. Stacked convolutional auto-encoders for hierarchical feature extraction. In International Conference on Artificial Neural Networks; Springer: Berlin/Heidelberg, Germany, 2011; pp. 52–59. [Google Scholar]
- Chen, K.; Seuret, M.; Liwicki, M.; Hennebert, J.; Ingold, R. Page segmentation of historical document images with convolutional autoencoders. In Proceedings of the 2015 13th International Conference on Document Analysis and Recognition (ICDAR); IEEE: Washington, DC, USA, 2015; pp. 1011–1015. [Google Scholar]
- Geng, J.; Fan, J.; Wang, H.; Ma, X.; Li, B.; Chen, F. High-resolution SAR image classification via deep convolutional autoencoders. IEEE Geosci. Remote. Sens. Lett. 2015, 12, 2351–2355. [Google Scholar] [CrossRef]
- Jain, A.K. Data clustering: 50 years beyond K-means. Pattern Recognit. Lett. 2010, 31, 651–666. [Google Scholar] [CrossRef]
- Gao, Q.; Xu, H.X.; Han, H.G.; Guo, M. Soft-sensor Method for Surface Water Qualities Based on Fuzzy Neural Network. In Proceedings of the 2019 Chinese Control Conference (CCC), Guangzhou, China, 27–30 July 2019; pp. 6877–6881. [Google Scholar]
- Kowsari, K.; Yammahi, M.; Bari, N.; Vichr, R.; Alsaby, F.; Berkovich, S.Y. Construction of fuzzyfind dictionary using golay coding transformation for searching applications. arXiv 2015, arXiv:1503.06483. [Google Scholar] [CrossRef]
- Kowsari, K.; Alassaf, M.H. Weighted unsupervised learning for 3d object detection. arXiv 2016, arXiv:1602.05920. [Google Scholar] [CrossRef]
- Alassaf, M.H.; Kowsari, K.; Hahn, J.K. Automatic, real time, unsupervised spatio-temporal 3d object detection using rgb-d cameras. In Proceedings of the 2015 19th International Conference on Information Visualisation, Barcelona, Spain, 22–24 July 2015; pp. 444–449. [Google Scholar]
- Manning, C.D.; Raghavan, P.; Schutze, H. Introduction to Information Retrieval; Cambridge University Press: Cambridge, UK, 2008; Volume 20, pp. 405–416. [Google Scholar]
- Mahajan, M.; Nimbhorkar, P.; Varadarajan, K. The Planar k-Means Problem is NP-Hard. In WALCOM: Algorithms and Computation; Das, S., Uehara, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 274–285. [Google Scholar]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb. Protoc. 2008, 2008, pdb–prot4986. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J. An introduction to Routine and special staining. Retrieved August 2011, 18, 2014. [Google Scholar]
- Khan, A.M.; Rajpoot, N.; Treanor, D.; Magee, D. A nonlinear mapping approach to stain normalization in digital histopathology images using image-specific color deconvolution. IEEE Trans. Biomed. Eng. 2014, 61, 1729–1738. [Google Scholar] [CrossRef] [PubMed]
- Bianco, S.; Cusano, C.; Napoletano, P.; Schettini, R. Improving CNN-Based Texture Classification by Color Balancing. J. Imaging 2017, 3, 33. [Google Scholar] [CrossRef]
- Bianco, S.; Schettini, R. Error-tolerant color rendering for digital cameras. J. Math. Imaging Vis. 2014, 50, 235–245. [Google Scholar] [CrossRef]
- Vahadane, A.; Peng, T.; Sethi, A.; Albarqouni, S.; Wang, L.; Baust, M.; Steiger, K.; Schlitter, A.M.; Esposito, I.; Navab, N. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med Imaging 2016, 35, 1962–1971. [Google Scholar] [CrossRef]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Kowsari, K.; Heidarysafa, M.; Brown, D.E.; Meimandi, K.J.; Barnes, L.E. Rmdl: Random multimodel deep learning for classification. In Proceedings of the 2nd International Conference on Information System and Data Mining, Lakeland, FL, USA, 9–11 April 2018; pp. 19–28. [Google Scholar]
- Li, Q.; Cai, W.; Wang, X.; Zhou, Y.; Feng, D.D.; Chen, M. Medical image classification with convolutional neural network. In Proceedings of the 2014 13th International Conference on Control Automation Robotics & Vision (ICARCV), Singapore, 10–12 December 2014; pp. 844–848. [Google Scholar]
- Heidarysafa, M.; Kowsari, K.; Brown, D.E.; Jafari Meimandi, K.; Barnes, L.E. An Improvement of Data Classification Using Random Multimodel Deep Learning (RMDL). arXiv 2018, arXiv:1808.08121. [Google Scholar] [CrossRef]
- Scherer, D.; Müller, A.; Behnke, S. Evaluation of pooling operations in convolutional architectures for object recognition. In Proceedings of the Artificial Neural Networks–ICANN 2010, Thessaloniki, Greece, 15–18 September 2010; pp. 92–101. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Chollet, F. Keras: Deep Learning Library for Theano and Tensorflow. 2015. Available online: https://keras.io/ (accessed on 19 August 2019).
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Yang, Y. An evaluation of statistical approaches to text categorization. Inf. Retr. 1999, 1, 69–90. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Altman, N. Points of significance: Classification evaluation. Nat. Methods 2016, 13, 603–604. [Google Scholar] [CrossRef]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
| Total Population | Pakistan | Zambia | US | ||
|---|---|---|---|---|---|
| Data | 150 | EE (n = 10) | EE (n = 16) | Celiac (n = 63) | Normal (n = 61) |
| Biopsy Images | 461 | 29 | 19 | 239 | 174 |
| Age, median (IQR), months | 37.5 (19.0 to 121.5) | 22.2 (20.8 to 23.4) | 16.5 (9.5 to 21.0) | 130.0 (85.0 to 176.0) | 25.0 (16.5 to 41.0) |
| Gender, n (%) | M = 77 (%51.3) F = 73 (%48.7) | M = 5 (%50) F = 5 (%50) | M = 10 (%62.5) F = 6 (%37.5) | M = 29 (%46) F = 34 (%54) | M = 33 (%54) F = 28 (%46) |
| LAZ/ HAZ, median (IQR) | −0.6 (−1.9 to 0.4) | −2.8 (−3.6 to -2.3) | −3.1 (−4.1 to −2.2) | −0.3 (−0.8 to 0.7) | ( to 0.5) |
| Data | Train | Test | Total | ||||
|---|---|---|---|---|---|---|---|
| Normal | 22,676 | 9717 | 32,393 | ||||
| Environmental Enteropathy | 20,516 | 8792 | 29,308 | ||||
| Celiac Disease | Parent | Child | Parent | Child | Parent | Child | |
| I | 21,140 | 4988 | 9058 | 2137 | 30,198 | 7125 | |
| IIIa | 4790 | 2052 | 6842 | ||||
| IIIb | 5684 | 2436 | 8120 | ||||
| IIIc | 5678 | 2433 | 8111 | ||||
| Precision | Recall | F1-Score | |
|---|---|---|---|
| Normal | 89.97 ± 0.59 | 89.35 ± 0.61 | 89.66 ± 0.60 |
| Environmental Enteropathy | 94.02 ± 0.49 | 97.30 ± 0.33 | 95.63 ± 0.42 |
| Celiac Disease | 91.12 ± 0.32 | 88.71 ± 0.35 | 89.90 ± 1.27 |
| Model | Precision | Recall | F1-Score | |
|---|---|---|---|---|
| Baseline | CNN | 76.76 ± 0.49 | 80.18 ± 0.47 | 78.43 ± 0.48 |
| Multilayer perceptron | 76.19 ± 0.50 | 79.40 ± 0.47 | 77.76 ± 0.49 | |
| Deep CNN | 82.95 ± 0.44 | 87.28 ± 0.39 | 85.06 ± 0.42 | |
| HMIC | Non Whole slide | 84.13 ± 0.37 | 93.56 ± 0.29 | 88.61 ± 0.37 |
| Whole slide | 88.01 ± 0.38 | 93.98 ± 0.28 | 90.89 ± 0.38 | |
| Model | Precision | Recall | F1-Score | |||
|---|---|---|---|---|---|---|
| Baseline | CNN | Normal | 87.83 ± 0.57 | 90.77 ± 0.65 | 89.28 ± 0.61 | |
| Environmental Enteropathy | 90.93 ± 0.61 | 82.48 ± 0.79 | 86.50 ± 0.71 | |||
| Celiac Disease | I | 68.37 ± 1.98 | 68.62 ± 1.96 | 68.50 ± 1.96 | ||
| IIIa | 56.26 ± 1.01 | 56.26 ± 2.21 | 59.29 ± 1.95 | |||
| IIIb | 65.28 ± 0.97 | 98.28 ± 2.01 | 66.64 ± 1.87 | |||
| IIIc | 62.66 ± 1.99 | 66.83 ± 1.99 | 64.68 ± 2.02 | |||
| Multilayer perceptron | Normal | 87.97 ± 0.76 | 81.87 ± 0.76 | 84.81 ± 0.71 | ||
| Environmental Enteropathy | 87.25 ± 0.69 | 90.18 ± 0.62 | 88.69 ± 0.66 | |||
| Celiac Disease | I | 57.92 ± 2.07 | 60.74 ± 2.07 | 59.30 ± 2.09 | ||
| IIIa | 62.58 ± 2.09 | 62.18 ± 2.09 | 60.89 ± 2.11 | |||
| IIIb | 65.00 ± 1.89 | 66.09 ± 1.87 | 65.56 ± 1.88 | |||
| IIIc | 67.97 ± 1.85 | 74.85 ± 1.72 | 71.24 ± 1.78 | |||
| DCNN | Normal | 95.14 ± 0.42 | 94.91 ± 0.43 | 95.14 ± 0.42 | ||
| Environmental Enteropathy | 92.22 ± 0.55 | 90.62 ± 0.60 | 91.52 ± 0.58 | |||
| Celiac Disease | I | 75.41 ± 1.82 | 72.63 ± 1.89 | 73.99 ± 1.85 | ||
| IIIa | 70.81 ± 1.92 | 72.47 ± 1.93 | 71.63 ± 1.79 | |||
| IIIb | 81.08 ± 0.81 | 74.67 ± 1.84 | 77.74 ± 1.65 | |||
| IIIc | 75.07 ± 1.83 | 76.37 ± 1.81 | 75.71 ± 1.81 | |||
| HMIC | Non Whole Slide | Normal | 89.97 ± 0.59 | 89.35 ± 0.61 | 89.66 ± 0.61 | |
| Environmental Enteropathy | 94.02 ± 0.49 | 97.30 ± 0.33 | 95.63 ± 0.33 | |||
| Celiac Disease | I | 83.25 ± 1.58 | 80.91 ± 1.66 | 82.06 ± 1.62 | ||
| IIIa | 80.34 ± 1.62 | 80.46 ± 1.71 | 80.40 ± 1.57 | |||
| IIIb | 85.35 ± 1.49 | 81.77 ± 1.67 | 83.52 ± 1.47 | |||
| IIIc | 85.54 ± 1.49 | 82.71 ± 1.60 | 84.10 ± 1.55 | |||
| Whole Slide | Normal | 90.64 ± 0.57 | 90.06 ± 0.57 | 90.35 ± 0.58 | ||
| Environmental Enteropathy | 94.08 ± 0.49 | 97.33 ± 0.42 | 98.68 ± 0.42 | |||
| Celiac Disease | I | 88.73 ± 1.34 | 85.07 ± 1.51 | 86.86 ± 1.43 | ||
| IIIa | 81.19 ± 1.65 | 81.19 ± 1.65 | 82.44 ± 1.51 | |||
| IIIb | 90.51 ± 1.24 | 90.48 ± 1.27 | 90.49 ± 1.16 | |||
| IIIc | 89.26 ± 1.31 | 90.18 ± 1.26 | 89.72 ± 1.28 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowsari, K.; Sali, R.; Ehsan, L.; Adorno, W.; Ali, A.; Moore, S.; Amadi, B.; Kelly, P.; Syed, S.; Brown, D. HMIC: Hierarchical Medical Image Classification, A Deep Learning Approach. Information 2020, 11, 318. https://doi.org/10.3390/info11060318
Kowsari K, Sali R, Ehsan L, Adorno W, Ali A, Moore S, Amadi B, Kelly P, Syed S, Brown D. HMIC: Hierarchical Medical Image Classification, A Deep Learning Approach. Information. 2020; 11(6):318. https://doi.org/10.3390/info11060318
Chicago/Turabian StyleKowsari, Kamran, Rasoul Sali, Lubaina Ehsan, William Adorno, Asad Ali, Sean Moore, Beatrice Amadi, Paul Kelly, Sana Syed, and Donald Brown. 2020. "HMIC: Hierarchical Medical Image Classification, A Deep Learning Approach" Information 11, no. 6: 318. https://doi.org/10.3390/info11060318
APA StyleKowsari, K., Sali, R., Ehsan, L., Adorno, W., Ali, A., Moore, S., Amadi, B., Kelly, P., Syed, S., & Brown, D. (2020). HMIC: Hierarchical Medical Image Classification, A Deep Learning Approach. Information, 11(6), 318. https://doi.org/10.3390/info11060318






