DFTHR: A Distributed Framework for Trajectory Similarity Query Based on HBase and Redis
Abstract
:1. Introduction
- We propose a segment-based data model for storing and indexing trajectory data, which ensures efficient pruning and querying capacity.
- We introduce a bulk-based partitioning model and certain optimization strategies to alleviate the cost of adjusting data distribution, so that an incremental dataset can be supported efficiently.
- We design certain strategies to ensure the co-location between each partition and its corresponding index, and implement node-locality-based parallel query algorithms to reduce the inter-worker cost overhead when querying.
- We evaluate our scheme with the dataset of ship trajectory, and the experimental results verify the efficiency.
2. Related Work
3. Preliminary
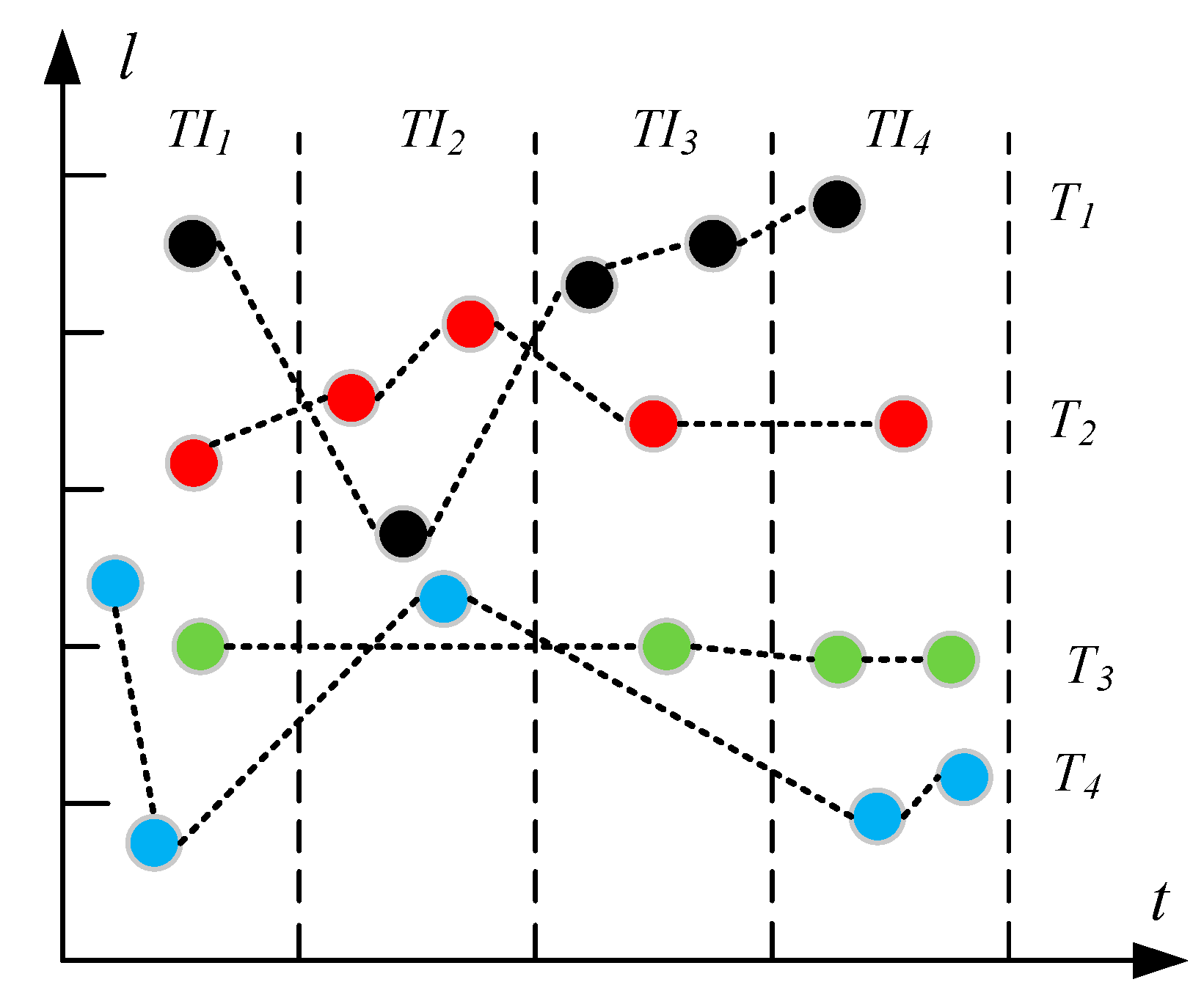
3.1. Problem Formulation
3.2. HBase
3.3. Redis
4. Distributed Framework Based on HBase and Redis (DFTHR) Structure
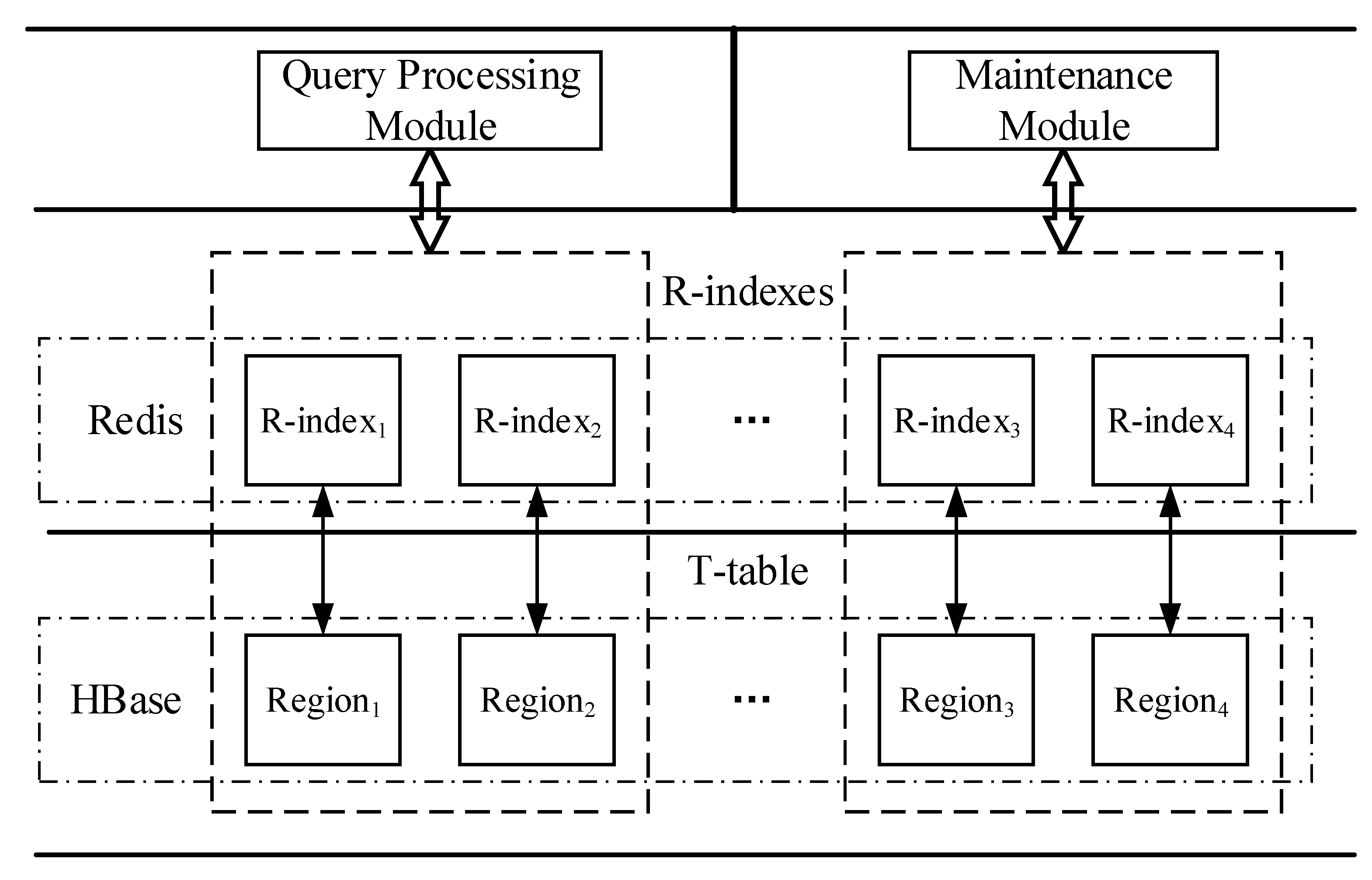
4.1. Overview
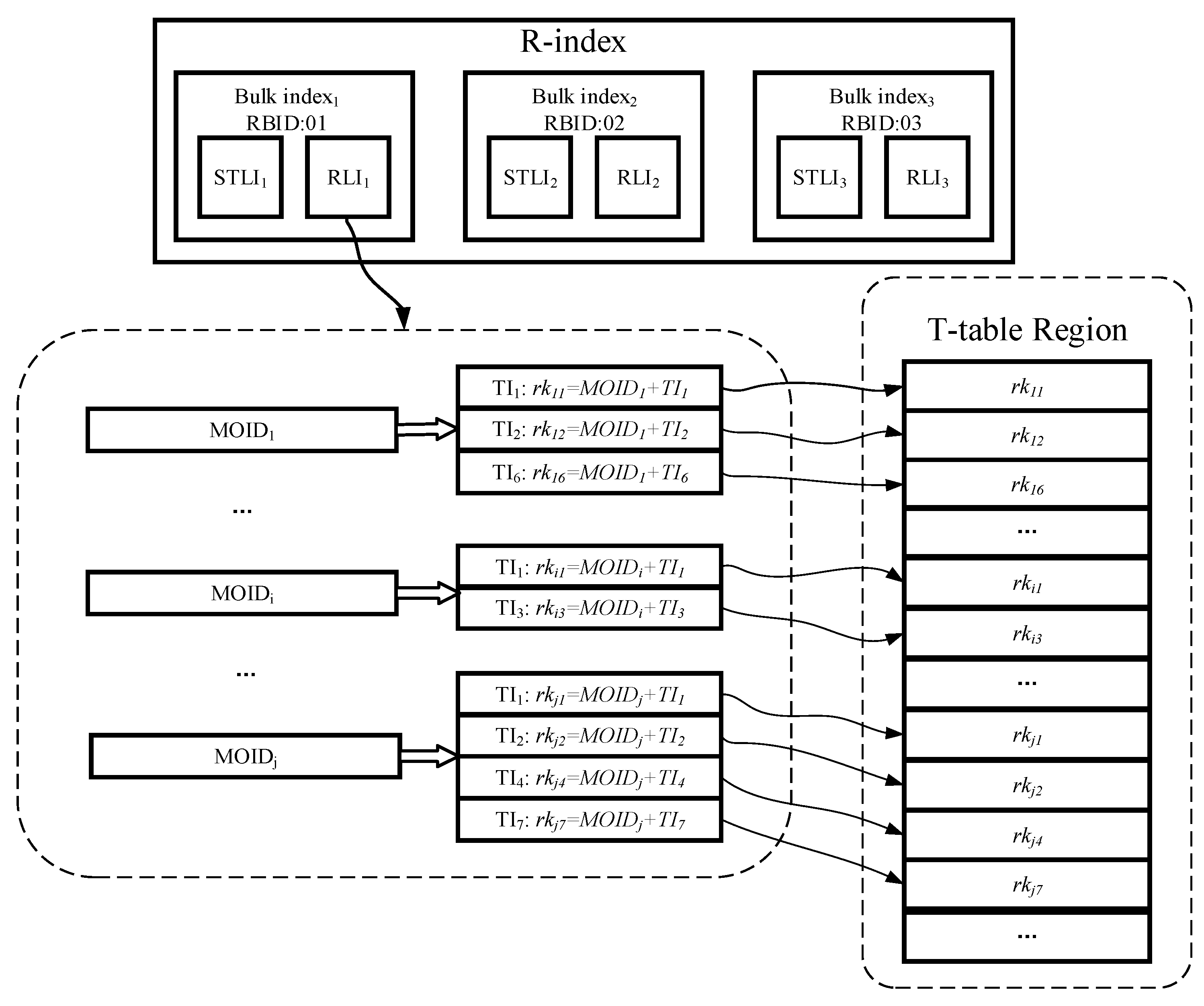
- T-table: used to persist storage trajectory data in HBase cluster. It adopts a segment-based data model to organize the trajectory data, and a bulk-based partitioning model to generate Region boundaries and process splits. This module is detail in Section 4.2.
- R-index: a Region level linear index structure, which stores in Redis so that it is able to process pruning quickly. Each R-index consists of multiple bulk indexes, and each bulk index contains a spatio-temporal linear index (STLI) and a rowkey linear index (RLI); the former is used to process the MBR (minimum bounding rectangle) coverage based pruning, and the latter is used to assist the former to quickly filter out the irrelevant trajectories (detailed in Section 4.3).
- Query processing module: this implements efficient support for two typical trajectory similarity queries based on trajectory segment based pruning method (described in Section 4.4).
- Maintenance module: when inserting data, the maintenance module adopts certain strategies to process the split and migration of Regions and indexes. Especially the processing of split, it uses a bulk-based splitting strategy to greatly reduce processing overhead. Maintenance Module ensures the co-location between Region and its corresponding index, thus reducing the communication overhead during query. We give a detailed description in Section 4.5.
4.2. T-Table
4.2.1. Trajectory Segment-Based Storage Model
- Data locality. The trajectory points that belong to the same MO should be assigned to the same Region to avoid the cross-node network overhead for obtaining data of the same MO.
- Filtering. Since the query results only relate to the trajectory data within the time range, we should avoid storing the whole data of the same MO in a single row, which will increase the filtering overhead of post-processing because the default filtering mechanism of HBase does not take effect until the data is accessed from memory or disk.
- R-index footprint. The rowkeys of T-table need to be recorded in the entries of R-indexes to implement index function. So the data granularity of the T-table row should not be too small, which may cause huge amount of memory occupation of R-indexes.
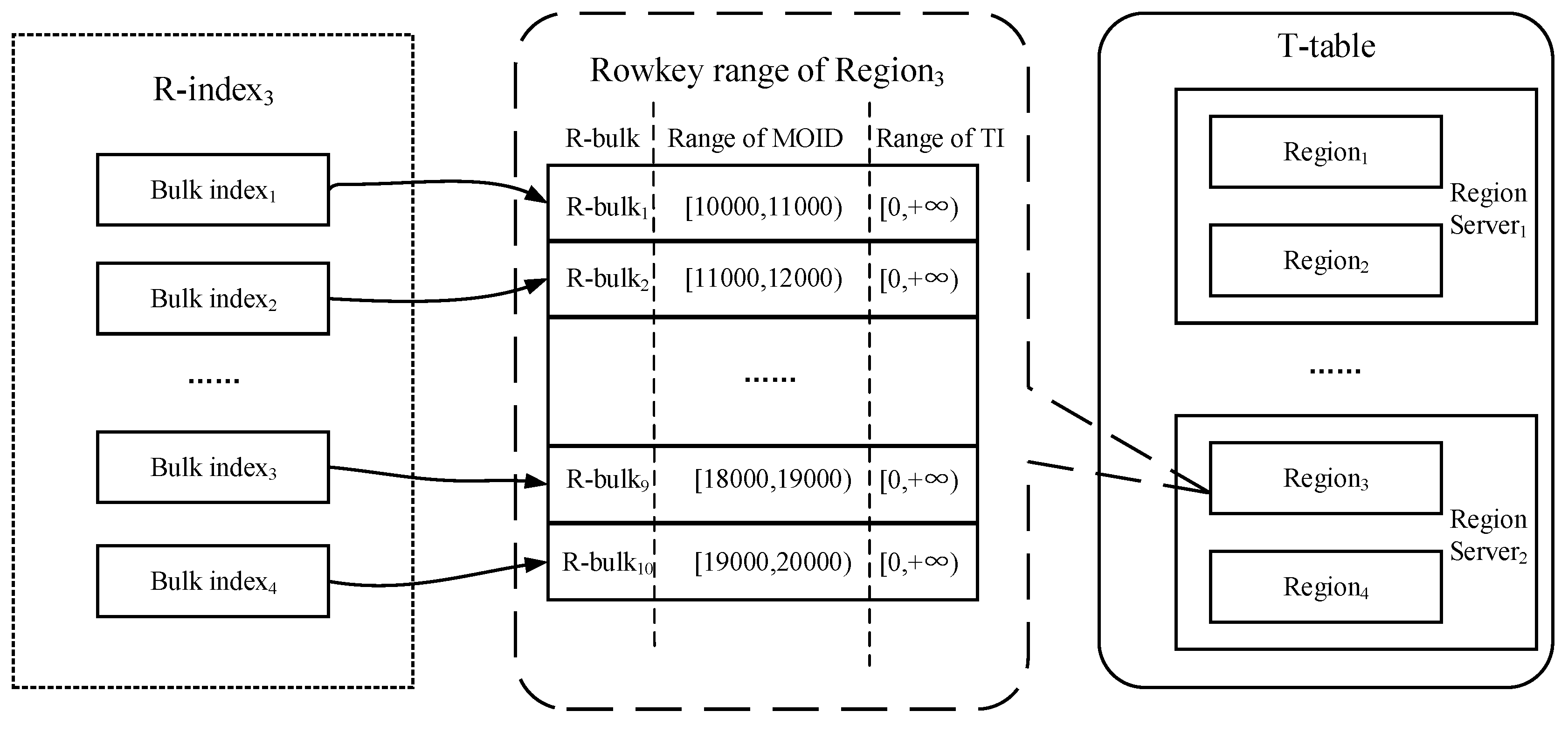
4.2.2. Bulk-Based Partitioning Model
4.3. R-Index
- Access efficiency. The R-index structure does not store actual trajectory point data, so the footprint required is much smaller than the T-table. In order to efficiently access the index data, we implement the deployment of R-indexes through the in-memory database Redis.
- Pruning. Creating a single index entry for the whole trajectory data of an MO may lead to very limited pruning power, because it may result in a large overlap among index entries. Therefore, R-index is also implemented by indexing the trajectory segment, which helps to reduce the overlapping areas.
- Network and maintenance overhead. Based on the consideration of reducing network overhead, we should ensure the co-location between each T-table Region and its corresponding R-index. In the incremental data loading scenario, the state of Region may change due to the increase in data, and the corresponding R-index should be kept up to date (such as split or move). Considering that the update and maintenance cost of linear index is much lower than that of R-Tree, and it can be better applied to the key-value store of Redis, so we use a bulk-based hybrid linear index structure to implement R-index. Furthermore, all Redis instances should be deployed in stand-alone mode, namely each RegionServer deploys an independent Redis instance. Compared with the mode of Redis cluster, the stand-alone mode can implement the co-location between any Region with the corresponding R-index without defining some sort of complex sharding strategy.
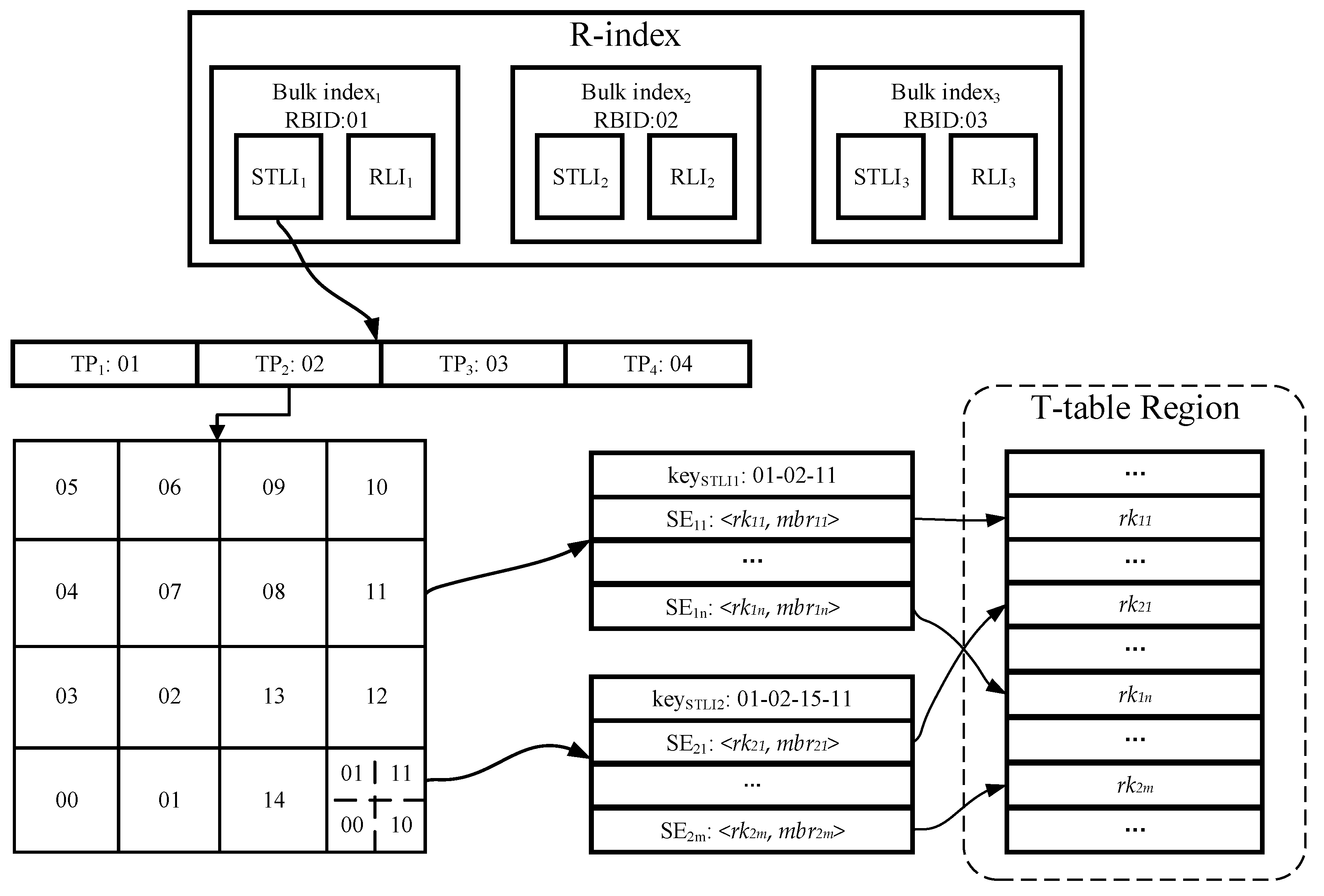
4.3.1. Spatio-Temporal Linear Index (STLI)
- Bico is the RBID value of the corresponding R-bulk, which is used to identify the bulk index to which the current STLI entry belongs.
- In order to index the time attribute, we divide the time dimension into multiple equal-length time periods (TPs) in accordance with time series. Any TI element of trajectory segment must be fully enclosed between the starting and end timestamps of a single TP element to avoid any a trajectory segment being indexed by multiple STLI entries. Since all TPs are of equal length, we can use the starting timestamp as the value of tpco.
- For indexing the spatial attribute, the whole space is first divided into multiple grid cells of the same size as the initial granularity, and next all the grid cells are encoded through adopting a certain encoding technique (such as Z-ordering, Hilbert curve [27]), and each encoding value is treated as the value of gridco to provide index function. However, the grid structure cannot solve data skew, and some grid cells overlaps with too many MBRs of trajectory segments. For such grid cells, we adopt the CIF quad-tree to further divide them to eliminate data skew. In the process of further division, except that the kind of trajectory segment whose MBR intersects with multiple grid cells is still indexed by the intersecting grid cells, other trajectory segments only are indexed by the minimum quad-tree nodes within a defined depth range which surround their MBRs. With quad-tree encoding, we can obtain a quad-tree code cifco for each CIF quad-tree node. Compared with using CIF quad-tree structure alone, our hybrid index structure which combines grid index with CIF quad-tree can effectively reduce the amount of query processing in the massive data, because there is no node with large coverage area in our index structure. Naturally, this index structure also incurs additional storage overheads due to certain data redundancy.
4.3.2. Rowkey Linear Index (RLI)
4.4. Query Processing Module
4.4.1. Trajectory Segment-Based Pruning
4.4.2. Threshold-Based Query
| Algorithm 1 Threshold-Based Query |
| Input: Query time range trq, Query trajectory Tq, Distance Threshold ε; Output: A set of Similar Trajectories; 1 for each T-table Region do in parallel 2 stlisq←getSTLIs(); 3 rlisq←getRLIs(); 4 sess←stlisq.rangeQuery(M(R(Tq)), trq, ε); 5 sess←rlisq.removeElementsBySegment(sess, trq); 6 sess←lowerBoundPruning(sess, ε, Tq); 7 sess←twoSidedPruning(sess, ε, Tq); 8 for each sesi in sess do 9 if distFrom(sesi, Tq)≤ε then 9 TCRq.add(Ti); 11 end if 12 end for 13 end parallel 14 return client.collect(TCRq); |
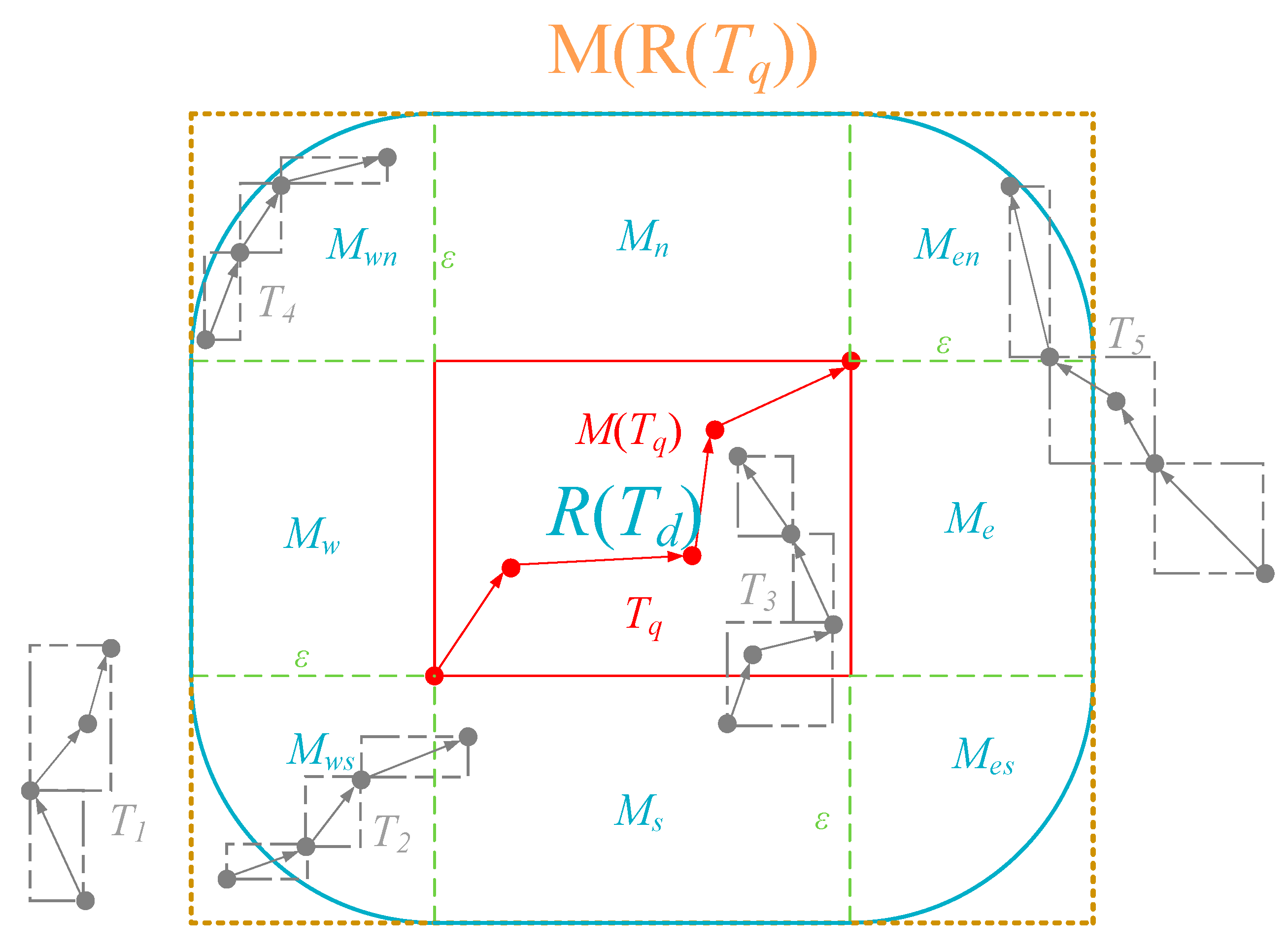
- MBR coverage based pruning (line 4 to 5). We first obtain an extended MBR M(R(Tq)) according to Tq and ε. Next we invoke a spatiotemporal range query in stli sq by M(R(Tq)) and trq (see Section 4.3.1 for query process in STLI). After getting all the candidate SE elements, we group them according to MOID attribute, and obtain a SE set based on MO grouping sess = {ses1, ses2, …, sesn}, where each sesi = {sei1, …, seim} (1 ≤ i ≤ n) is a set of candidate SE elements of an MO. In order to facilitate the description, we define this MO corresponding to sesi element as moi, whose MOID is moidi, and the trajectory generated by moi in trq is Ti. However, each query result sesi can only indicate that Ti have some MBRs of trajectory segment that are fully covered by M(R(Tq)), and according to the requirements of Lemma 3, we need to further check whether all the MBRs of Ti are fully covered by M(R(Tq)). For this, we query rlisq by using trq and each moidi to obtain a rowkey set of the trajectory segments rksi. If at least one rowkey that exists in rksi does not exist in the corresponding sesi, this sesi should be removed from sess.
- Lower bound based pruning (line 6). For each sesi in sess, according to Lemma 4, we first select which SE elements need to further determine the pruning by calculating the lower bound. Next, we calculate the lower bound from each selected SE element to M(Tq), and according to Lemma 1, if there exists a SE element seg = <rkg, mbrg > in sesi, such that HausLB(mbrg, M(Tq))> ε, then we should remove the current sesi from sess.
- Two-sided based pruning (line 7). Considering that the two-sided Hausdorff distance is used as the distance function of the similarity query, for each sesi in sess, we swap Ti to Tq, and repeat the steps of MBR coverage-based pruning and lower bound-based pruning. If there exists an MBR of a trajectory segment in Tq, such that it does not meet the MBR coverage requirement or the lower bound requirement with Ti, then we should remove the corresponding sesi from sess.
- Refining (line 8 to 12). For each sesi in sess, we calculate the Hausdorff distance between Ti and Tq, and check whether the distance value is not beyond ε. Finally, we obtain a collection of trajectories TCRq that satisfy ε and return it to the client. In order to calculate the Hausdorff distance, we need to access the T-table Region through the rowkeys contained in sesi to get the actual trajectory data of Ti. In consideration of reducing the disk overhead, we use the INC-HD (incremental Hausdorff distance) algorithm [28] to calculate the distance. This is because this algorithm can filter more trajectory segments by a more efficient upper bound than others, and the data access overhead is smaller.
4.4.3. k-NN Query
| Algorithm 2 k-NN Query |
| Input: Trajectory number k, Query time range trq, Query trajectory Tq; Output: k most similarity trajectories Tsk to Tq; 1 ε←estimateThreshold(k); 2 repeat 3 for each T-table Region do in parallel 4 TCRq←thresholdQuery (trq, Tq, ε); 5 end parallel 6 ε←ε.increase(); 7 until client.sum(TCRq.size)≥k 8 Tsk←client.collect(TCRq).top(k); 9 return Tsk; |
4.5. Maintenance Module
5. Experiment
5.1. Set Up
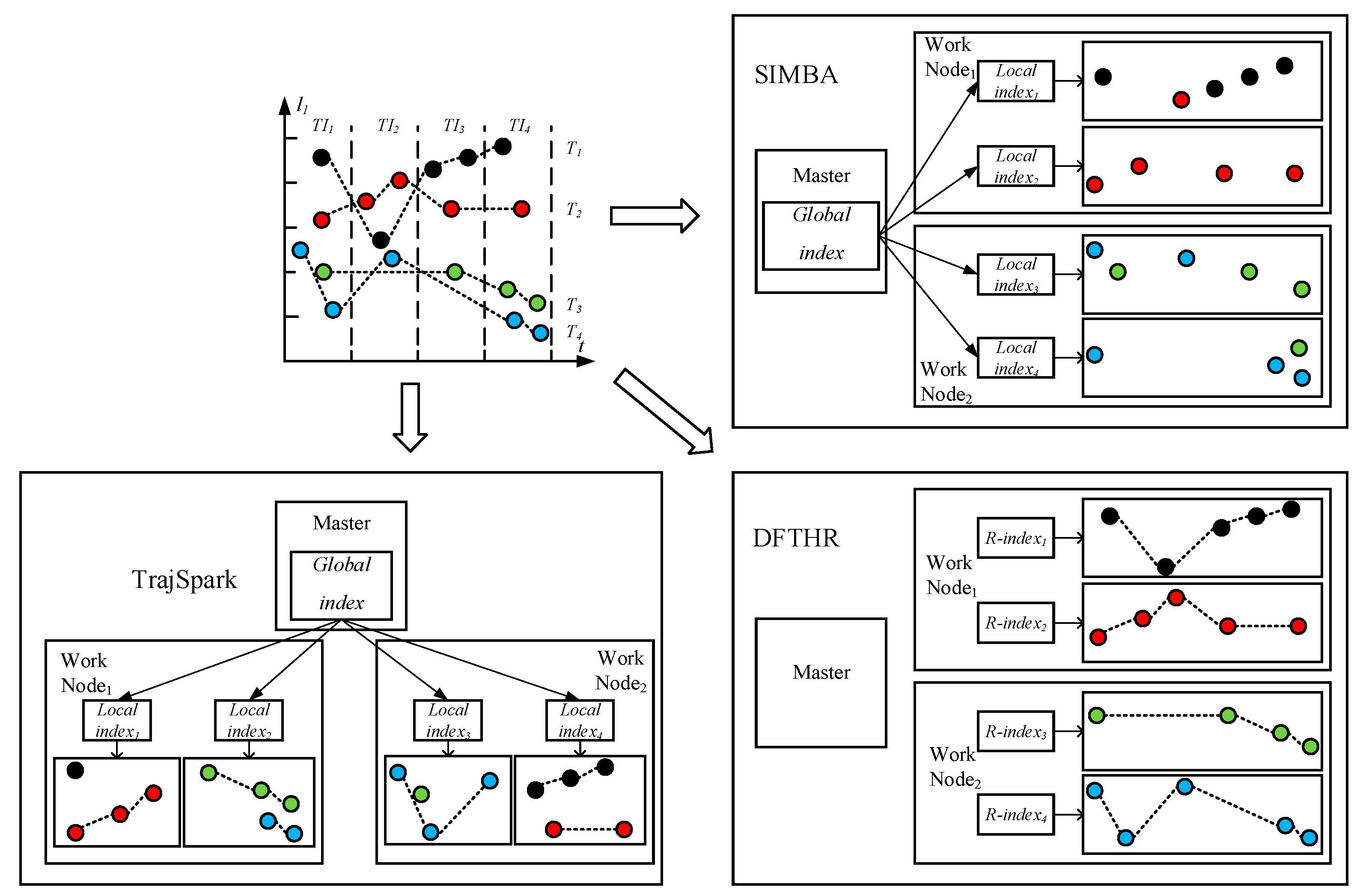
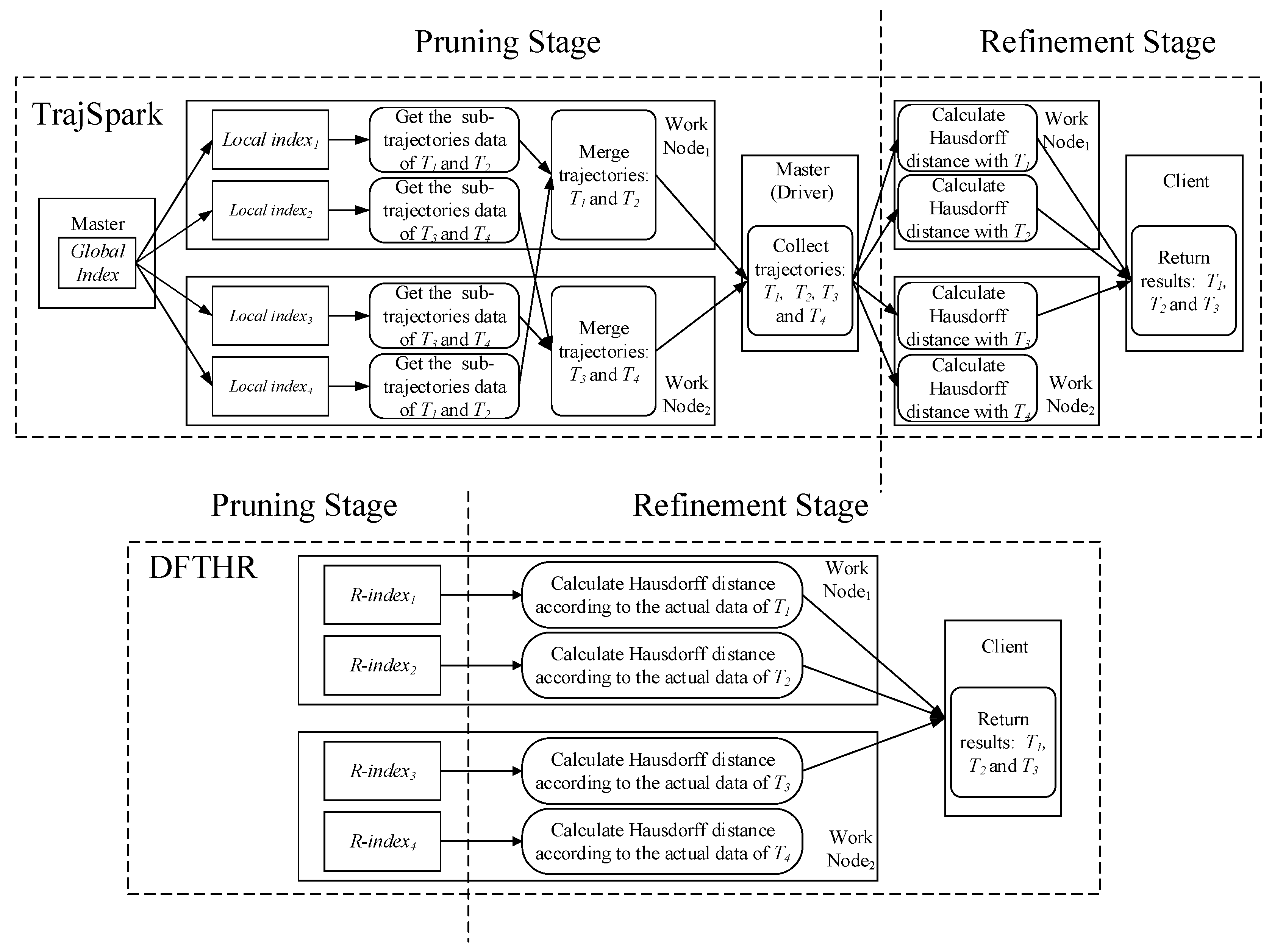
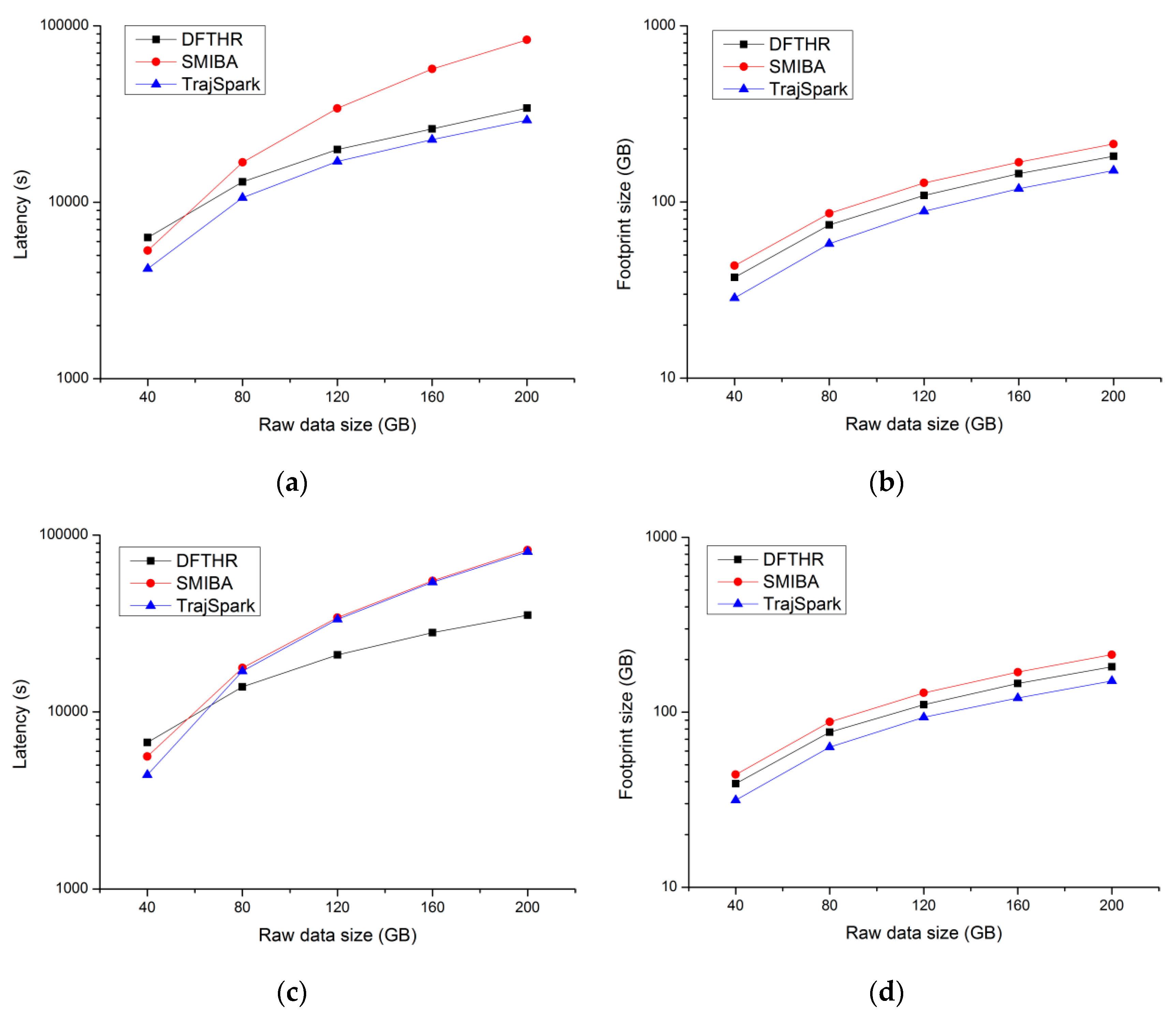
- SIMBA. SIMBA follows a certain spatio-temporal partitioning strategy to distribute the trajectory data on the partitions of the work nodes, and use trajectory point as the basic data unit. As can be seen from Figure 7, it does not store the data of the same MO on the same node or even the same partition. When each batch of data arrives, it needs to repartition all data. For each data partition, this implements a local index structure to support spatio-temporal queries, and maintains a global index structure on the master node for coarse pruning.
- TrajSpark. The structure of TrajSpark is similar to SIMBA, which also includes local indexes and global index, and does not organize the data of the same MO on the same node. The differences are that, first, when data batches are inserted in chronological order, TrajSpark can process only the new data without affecting the historical data, as can be seen from Figure 7, and the second batch inserted is distributed in the different partitions from the first batch. Second, the trajectory data is compressed and stored in the partition in the form of a sub-trajectory.
- DFTHR. The biggest difference between DFTHR and the above two schemes is that it always constrains the data of the same MO to the same partition in the form of trajectory segment. In addition, since it uses HBase instead of Spark, it supports incremental inserting and does not need to adjust all partitions to accommodate data distribution changes. When the state of a partition needs to change, it can alleviate the processing overhead based on the bulk-based maintenance strategy.
5.2. Performance of Data Insertion
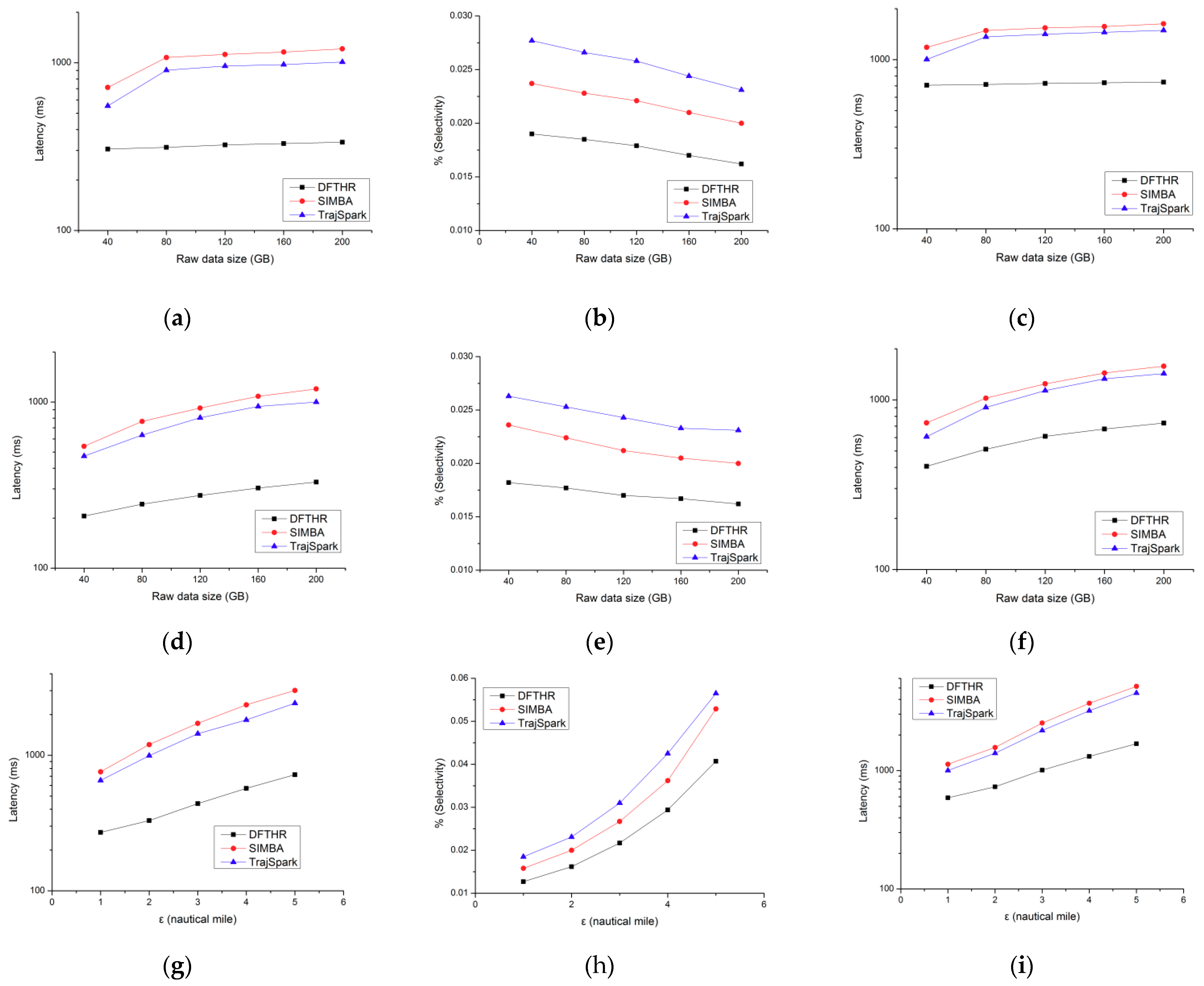
5.3. Performance of Threshold-Based Query
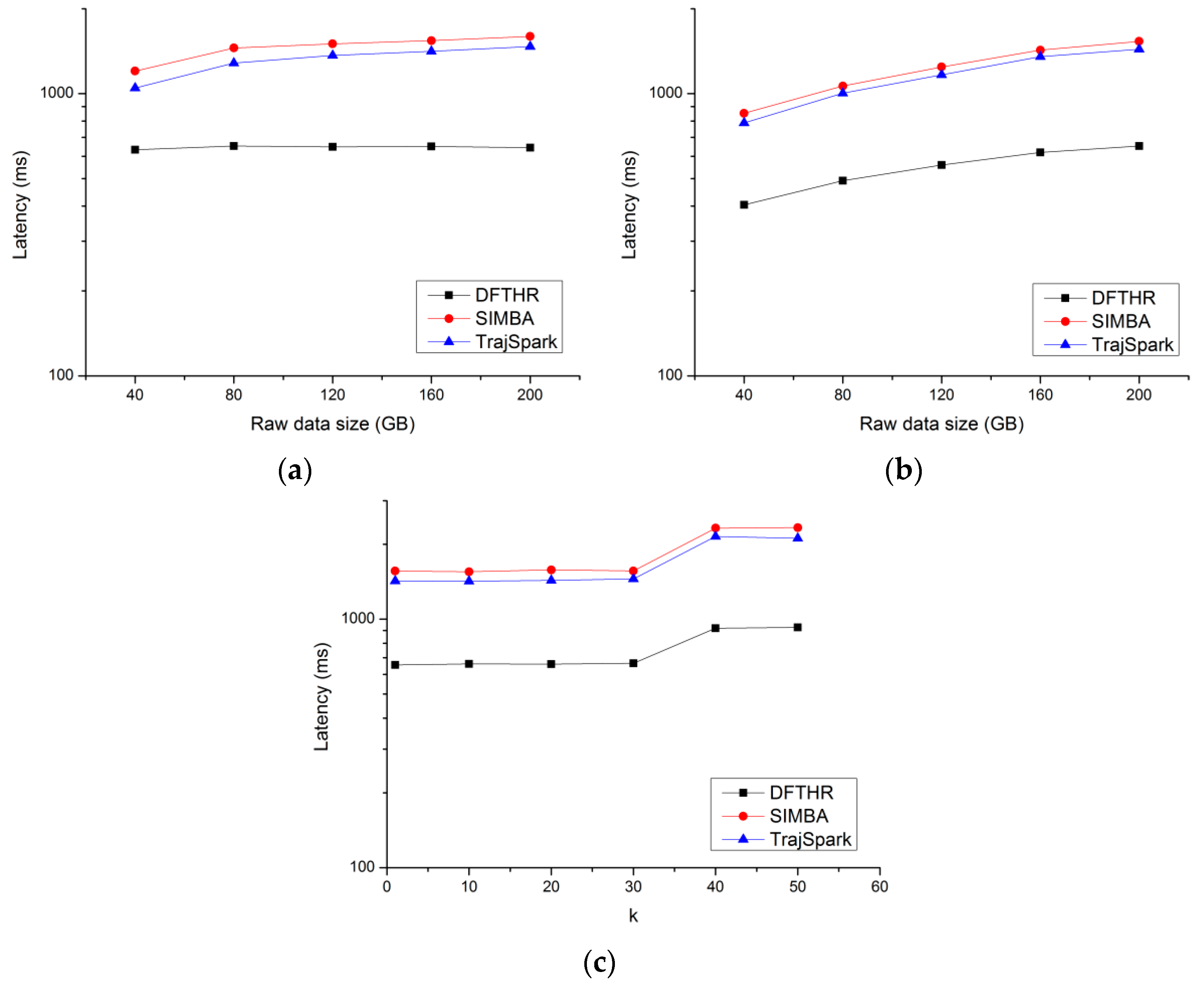
5.4. Performance of k-NN Query
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Salmon, L.; Ray, C. Design principles of a stream-based framework for mobility analysis. GeoInformatica 2017, 21, 237–261. [Google Scholar] [CrossRef]
- He, Z.; Ma, X. Distributed Indexing Method for Timeline Similarity Query. Algorithms 2018, 11, 41. [Google Scholar] [CrossRef]
- Morse, M.D.; Patel, J.M. An efficient and accurate method for evaluating time series similarity. In Proceedings of the 2007 ACM SIGMOD International Conference on Management of Data, Beijing, China, 11–14 June 2007. [Google Scholar]
- Bai, Y.B.; Yong, J.H.; Liu, C.Y.; Liu, X.M.; Meng, Y. Polyline approach for approximating Hausdorff distance between planar free-form curves. Comput. Aided Des. 2011, 43, 687–698. [Google Scholar] [CrossRef]
- Zhou, M.; Wong, M.H. Boundary-Based Lower-Bound Functions for Dynamic Time Warping and Their Indexing. In Proceedings of the 2007 IEEE 23rd International Conference on Data Engineering, Istanbul, Turkey, 15–20 April 2007. [Google Scholar]
- Zhu, Y.; Shasha, D. Warping indexes with envelope transforms for query by humming. In Proceedings of the 2003 ACM SIGMOD International Conference on Management of Data, San Diego, CA, USA, 9–12 June 2003. [Google Scholar]
- Ranu, S.; Deepak, P.; Telang, A.D.; Deshpande, P.; Raghavan, S. Indexing and matching trajectories under inconsistent sampling rates. In Proceedings of the 2015 IEEE 31st International Conference on Data Engineering (ICDE), Seoul, Korea, 13–17 April 2015. [Google Scholar]
- Zheng, B.; Wang, H.; Zheng, K.; Su, H.; Liu, K.; Shang, S. SharkDB: An in-memory column oriented storage for trajectory analysis. WWW 2018, 21, 455–485. [Google Scholar] [CrossRef]
- Peixoto, D.A.; Hung, N.Q.V. Scalable and Fast Top-k Most Similar Trajectories Search Using MapReduce In-Memory. In Proceedings of the Australasian Database Conference, Sydney, Australia, 28–30 September 2016. [Google Scholar]
- Zhang, Z.; Jin, C.; Mao, J.; Yang, X.; Zhou, A. TrajSpark: A Scalable and Efficient In-Memory Management System for Big Trajectory Data. In Proceedings of the Asia-Pacific Web (APWeb) and Web-Age Information Management (WAIM) Joint Conference on Web and Big Data, Beijing, China, 7–9 July 2017. [Google Scholar]
- Xie, D.; Li, F.; Phillips, J.M. Distributed trajectory similarity search. Proc. VLDB Endow. 2017, 10, 1478–1489. [Google Scholar] [CrossRef]
- Shang, Z.; Li, G.; Bao, Z. DITA: A Distributed In-Memory Trajectory Analytics System. In Proceedings of the 2018 International Conference on Management of Data, Houston, TX, USA, 10–15 June 2018. [Google Scholar]
- Alt, H.; Scharf, L. Computing the hausdorff distance between curved objects. JCG Appl. 2008, 18, 307–320. [Google Scholar] [CrossRef]
- Apache HBase. Available online: https://hbase.apache.org/ (accessed on 8 January 2019).
- Redis. Available online: https://redis.io/ (accessed on 8 January 2019).
- Alt, H.; Godau, M. Computing the Fréchet distance between two polygonal curves. IJCGA 1995, 5, 75–91. [Google Scholar] [CrossRef]
- Müller, M. Information Retrieval for Music and Motion; Springer: Berlin/Heidelberg, Germany, 2007; pp. 69–84. [Google Scholar]
- Yang, K.; Shahabi, C. A PCA-based similarity measure for multivariate time series. In Proceedings of the 2nd ACM International Workshop on Multimedia Databases, Washington, DC, USA, 13 November 2004. [Google Scholar]
- Vlachos, M.; Kollios, G.; Gunopulos, D. Discovering similar multidimensional trajectories. In Data Engineering, 2002. In Proceedings of the 18th International Conference, San Jose, CA, USA, 26 February–1 March 2002. [Google Scholar]
- Chen, L.; Özsu, M.T.; Oria, V. Robust and fast similarity search for moving object trajectories. In Proceedings of the 2005 ACM SIGMOD International Conference on Management of Data, New York, NY, USA, 14–16 June 2005. [Google Scholar]
- Xie, D.; Li, F.; Yao, B.; Zhou, L.; Guo, M. Simba: Efficient in-memory spatial analytics. In Proceedings of the 2016 International Conference on Management of Data, San Francisco, CA, USA, 26 June–1 July 2016. [Google Scholar]
- Vora, M.N. Hadoop-HBase for large-scale data. In Proceedings of the 2011 International Conference on Computer Science and Network Technology (ICCSNT), Harbin, China, 24–26 December 2011. [Google Scholar]
- Vashishtha, H.; Stroulia, E. Enhancing query support in hbase via an extended coprocessors framework. In Proceedings of the European Conference on a Service-Based Internet, Poznan, Poland, 26–28 October 2011. [Google Scholar]
- Han, J.; Haihong, E.; Le, G.; Du, J. Survey on NoSQL database. In Proceedings of the 2011 6th International Conference, Pervasive Computing and Applications (ICPCA), Port Elizabeth, South Africa, 26–28 October 2011. [Google Scholar]
- Zaharia, M.; Chowdhury, M.; Franklin, M.J.; Shenker, S.; Stoica, I. Spark: Cluster computing with working sets. In Proceedings of the Usenix Conference on Hot Topics in Cloud Computing, Boston, MA, USA, 22–25 June 2010. [Google Scholar]
- Samet, H. The quadtree and related hierarchical data structures. CSUR 1984, 16, 187–260. [Google Scholar] [CrossRef]
- Lawder, J.K.; King, P.J.H. Querying multi-dimensional data indexed using the Hilbert space-filling curve. SIGMOD 2001, 30, 19–24. [Google Scholar] [CrossRef]
- Nutanong, S.; Jacox, E.H.; Samet, H. An Incremental Hausdorff Distance Calculation Algorithm. Proc. VLDB Endow. 2011, 4, 506–517. [Google Scholar] [CrossRef]
- Harati-Mokhtari, A.; Wall, A.; Brooks, P.; Wang, J. Automatic Identification System (AIS): Data reliability and human error implications. J. Navig. 2007, 60, 373–389. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, J.; Ma, L.; Liu, Q. DFTHR: A Distributed Framework for Trajectory Similarity Query Based on HBase and Redis. Information 2019, 10, 77. https://doi.org/10.3390/info10020077
Qin J, Ma L, Liu Q. DFTHR: A Distributed Framework for Trajectory Similarity Query Based on HBase and Redis. Information. 2019; 10(2):77. https://doi.org/10.3390/info10020077
Chicago/Turabian StyleQin, Jiwei, Liangli Ma, and Qing Liu. 2019. "DFTHR: A Distributed Framework for Trajectory Similarity Query Based on HBase and Redis" Information 10, no. 2: 77. https://doi.org/10.3390/info10020077
APA StyleQin, J., Ma, L., & Liu, Q. (2019). DFTHR: A Distributed Framework for Trajectory Similarity Query Based on HBase and Redis. Information, 10(2), 77. https://doi.org/10.3390/info10020077



