Abstract
A study on the application of surface-enhanced Raman spectroscopy (SERS) in detecting biological threats is here reported. Simulants of deadly Bacillus anthracis endospores were used. This study proposes an automated device where SERS is used as a fast, pre-alarm technique of a two-stage sensor equipped with a real-time polymerase chain reaction (PCR). In order to check the potentialities of SERS in terms of sensitivity and specificity for on-site, real-time, automatic detection and identification of biological agents, two strains of genetically and harmless closely B. anthracis-related spores, Bacillus thuringiensis and Bacillus atrophaeus, were used as simulants. In order to assure the selectivity of the SERS substrate against B. thuringiensis spores, the substrate was functionalized by specific peptides. The obtained SERS measurements are classified as positive or negative hits by applying a special data evaluation based on the Euclidian distance between each spectrum and a reference spectrum of blank measurement. Principal component analysis (PCA) was applied for discriminating between different strains representing dangerous and harmless spores. The results show that the SERS sensor is capable of detecting a few tenths of spores in a few minutes, and is particularly sensitive and fast for this purpose. Post-process analysis of the spectra allowed for discrimination between the contaminated and uncontaminated SERS sensors and even between different strains of spores, although not as clearly. For this purpose, the use of a non-functionalized SERS substrate is suggested.
1. Introduction
The great worry of the presence of microorganisms in air and the multiplying of related epidemics has strongly lead to the search for more rapid methods for their identification, not only because of their common pathogenesis, but also because they could be employed as dangerous and devastating weapons for terroristic attacks as well as chemical or nuclear arms [1].
A potential attack with biological weapons, even if basic knowledge of biology and/or handling skills is required, can be easily conducted in public crowded areas such as metro stations, airports, and commercial centers. Measures by traditional sensors are, generally, time-consuming and expensive due to the cost of the necessary chemicals. The main problem related to the use of this kind of weapon is its noiseless lag time from infection to expression of the disease symptoms. In addition, their effectiveness is assured by their capability to both, spread throughout the air and to be dangerous in a relatively low amount [2]. Bio-agents include microorganisms such as bacteria, fungi, yeasts, and viruses and can infect people, animals, and plants, causing both sickness and death. These biological weapons can be easily spread through the contamination of water and food, and as aerosols suspended in wet or dry formulations [3]. A recent classification made by the Centers for Disease Control and Prevention (CDC) in conjunction with military intelligence, as well as medical and public health agencies, [4] divides bio-agents into different categories according to their dangerousness and potential for exploitation as a biological weapon. The reaction of the first responders to such an attack is crucial and must be rapid in order to strongly reduce any impact on human lives [5]. The loss of time connected to culturing suspected cells and their successive identification allows for pathogen propagation. Therefore, the adoption of a sensor able to identify the biological agent with sufficient reliability in real time, directly at the scene, is crucial for incisive emergency management [6]. The capability of continuous unattended monitoring is a feature not actually available for commercial biological agent sensors, but it would improve the emergency management in cases of large events, particularly in critical infrastructures (airports, underground stations, etc.).
Raman and the related surface-enhanced Raman spectroscopy (SERS) are already used for the rapid screening and identification of biological samples [7,8,9,10]. In particular, SERS has also already been demonstrated as being capable of differentiating between bacteria and hoax materials [11] among different bacteria species [8,12,13] and even between species of the same kind of bacteria [14,15].
SERS is useful for the label-free detection of microorganisms, thanks to its ability to identify single molecules by using their intrinsic vibrational fingerprint and by amplifying the inherently weak Raman signal nonlinearly by several orders of magnitude [16]. Gold and/or silver are usually employed to enhance the signal. In order to make SERS substrates more efficient in the selective capture of biological targets, these substrates can be functionalized by means of specific receptors, such as phages, peptides, and aptamers [17].
In the framework of the RAMBO (rapid-air monitoring particles against biological threats) project, the application of the SERS technique for the rapid identification of a few units of Bacillus atrophaeus (SBA) and Bacillus thuringiensis endospores (SBT) have been carried out. These bacteria are generally employed as simulants of deadly Bacillus anthracis because, although harmless, they are genetically and bodily similar [18]. Bacillus anthracis have been used since ancient times and is even used today as an infectious agent in terrorist attacks based on biological weapons. The design of the final RAMBO system foresees the use of two miniaturized sensors: a SERS providing an early warning of the bio-agent, and a DNA-based polymerase chain reaction (PCR) for a selective revelation for the final confirmation of positive detection [19]. The combination of SERS + PCR in a microfluidic cell enhances the reliability of the sensor detection. For the SERS sensor, substrates functionalized with specific peptides [20,21] are used to selectively bind the SBT spores. Before measurements, the interfering particles (not captured by the receptors) can be washed out by rinsing the sample with sterile water.
In this work, SBA and SBT spores are characterized by Raman spectroscopy on SERS substrates functionalized by peptides specific to SBT and integrated in a microfluidic cell. The real differential affinity between receptors and the two spore species is assessed in order to evaluate the effectiveness of the method in the early detection of harmful bio-agents only. Scanning electron microscopy (SEM) is used to characterize the samples and to infer the average number of spores under detection. Principal component analysis evidences characteristic spectral features of the microorganisms, useful for discriminating among them. Finally, a routine based on Euclidean distance, developed for our purpose, is followed to rapidly evaluate the membership of the Raman spectra to the reference cluster.
2. Materials and Methods
2.1. Instrument Set-Up and SERS Substrates
The SERS measurements were performed with a table-top, portable BW&Tek iRaman system characterized by a laser source at 785 nm, a spectral resolution of 3 cm−1, a range from 0 to 4000 cm−1, and a CCD array of 2048 pixels (Figure 1). In order to improve the conditions for detection (images and spectra), preliminary measurements were performed with different objectives: 4x, 10x, 20x, 40x, 80x, and 100x. The experimental conditions chosen consisted of a laser power of 150 mW, an integration time of 20 s, and an objective of 40x (with a spot size of ~40 μm).
Figure 1.
Raman apparatus measuring the surface-enhanced Raman spectroscopy (SERS) sensor in the microfluidic cell (MFC).
The laser light was focused onto the sample through a Raman probe connected to a spectrometer with a 1.5 m collection/excitation fiber. The spot size at the sample surface was 85 μm, while the focusing optics had a working distance of 5.4 mm and were protected from accidental contamination from the surface via a metallic distance regulator. The covered spectral range was between 75 and 3200 cm−1 (Raman shift) with a resolution of 3 cm−1.
Figure 1 shows the apparatus and the details of the Raman probe focused on the SERS microfluidic cell.
Klarite® SERS substrates from Renishaw Diagnostics Inc. (Wotton-under-Edge, UK) were used. They are composed of regular arrays of inverted pyramidal pits that deposit a sputtered gold layer on a silicon substrate with an ordered nanostructure produced via electron beam lithography [22]. The regularity of the nanostructures guarantees a uniform enhancement of the weak Raman signal over the entire excited area. The full chip active area was 4 × 4 mm2. Each pit had an aperture of 1.5 × 1.5 μm2 and a total pitch size (aperture + distance to the next microcavity) of 2 μm with a fixed apex pit angle of 70.5°. The cavity depth was 1.06 μm.
In order to enhance the selectivity and to assure the tie with the substrates, some of these were functionalized with specific peptides of Bacillus thuringiensis [21] at University Claude Bernard (Lyon, France). The peptides (SLLPGLPGGGC. βAla-βAla -βAla K.K.K code MC25) were deposited as a thin, uniform layer on the SERS sensor by using a standardized protocol [23,24].
2.2. Bacillus Endospores
Bacillus thuringiensis (SBT) (ATCC® 10792TM) and Bacillus atrophaeus (SBA) (ATCC® 9372TM) endospore suspensions were provided by the University of Technology, Institute of Optoelectronics (Warsaw, Poland). The SBT was considered a simulant of the deadly Bacillus anthracis, whereas SBA were considered a harmless interfering sample. The concentrations utilized were both 109 CFU/mL in sterile water. For each sample, 40 μL of these suspensions were injected through the injection valves in two different cartridges containing functionalized SERS sensors. Then, after 10 minutes of allowing bonding with the peptides, they were rinsed with pure sterile water (MilliQ, Millipore) to remove unlinked spores and finally measured.
To obtain a rough estimation of the spore density on the sensor after the entire procedure, the active SERS surface was investigated with a Phenom Pro SEM (scanning electron microscope) (Phenom, Eindhoven, The Netherlands ).
2.3. SERS Sensor in Microfluidic Cartridge
The SERS sensor was embedded in a microfluidic cartridge provided by Microfluidic ChipShop (Jena, Germany). The microfluidic chamber consists of a sealed chamber that contains the sensing surface with the functionalized SERS array to which the target bio-molecules are stuck. The cartridge is made of a transparent polymer and has the dimensions of a standard microscope glass. It implements connections to two valves and two 100-μm-wide micro-channels in order to inject and remove samples in suspension of sterile water, to flush the substrate with rinsing water, and, if necessary, to inject the solution with the SERS labels.
The sensor surface is covered with a thin inspection window, (100 μm in thickness) made of a ciclic olefin polymer (COP) such as PMMA, through which the SERS plate can be interrogated.
Finally, the cartridge is implemented on a two-dimension scanning stage, to enable sequential analysis of different spots over the SERS plate. Figure 2 shows the SERS included in the microfluidic cell.
Figure 2.
The microfluidic cell with the SERS sensor.
2.4. Data Collection
SERS spectra were collected, and, in order to clearly identify the interfering blank signal coming from the clean cartridge, before collecting the spectra of SBT and SBA, the spectra of the clean cartridge were acquired. Then, on 20 different points of the whole inoculated area (with a sensing area of 4 × 4 mm2), tree spectra were collected. The spectral interval between 350 and 2200 cm−1 was considered for the characterization of the spectra, mainly because the signal out of this interval was often recorded saturated (near the laser excitation at low Raman shifts) or very weak (far from the laser excitation at higher Raman shifts) and featureless. Spectra were baseline-corrected and normalized between 0 and 1 prior to performing any multivariate statistics or classification.
2.5. Data Processing
In order to extract more detailed spectral information from such complex datasets and assume possible discrimination criteria, principal component analysis (PCA) [25,26,27] was applied. The multivariate statistical approach uses linear transformations to reduce the dimensionality of the dataset by projecting the original measures into a subspace along the directions of their maximum variance. The new uncorrelated variables are called principal components (PCs) [25,26,27]. PCA was applied to sets of blank and positive measurements. In order to immediately distinguish whether a spectrum corresponds to SBT or not and measure the sensitivity of the SERS technique in the cartridge, the normalized Euclidean distance [28] were calculated. The calculation was performed according to Equation (1) between each single spectrum and the blank spectrum (internal standard), obtained as the average of all the spectra of the clean cartridge:
where , are the two spectra, and si is the standard deviation. Thus, we achieved a measure of the similarity of each spectrum to this standard spectrum in units of the standard deviation of a set of blank measurements. Spectra with a normalized Euclidean distance over 1 standard deviation band (1sB) are considered positive identifications (hits), whereas shorter differences are considered negative identifications.
3. Results
Raman spectra of the clean system (constituted by the clean SERS substrate and functionalized with peptides, which are supposed to be selective towards SBT), inserted in the MFC and covered by the above-mentioned PMMA window, were acquired by means of a 40X microscope objective and then compared with the spectra acquired (in the same conditions and with the same equipment) after the SBT inoculation. As evident from Figure 3, the intense signals due to the whole clean system (blue line) highly affect the relatively weak Raman signal of the SBT (red line), so that the SBT spectral signatures can be barely individuated.
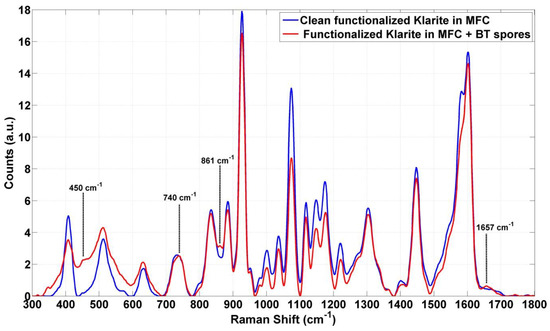
Figure 3.
The average of the SERS spectra acquired on the clean cartridge (blue lines) compared to substrate inoculated with Bacillus thuringiensis endospores (SBT) (red lines).
Nevertheless, the presence of SBT can be argued from the slight variation of the intensities; in particular, in the range from 450 to 900 and 1657 cm−1, some weak peaks can be recognized (Figure 3).
In order to clarify if those peaks can be conferred to SBT and to separate SBT from the clean system, reducing their dimensionality, the spectral data were also processed by Euclidian distance defined in Equation (1), and PCA.
Figure 4 shows the obtained results. Every point corresponds to an average of each spectrum (x-axis) for which, on the y-axis, the Euclidean distance, calculated with respect to the average spectrum of the clean substrate (brown line), is given.
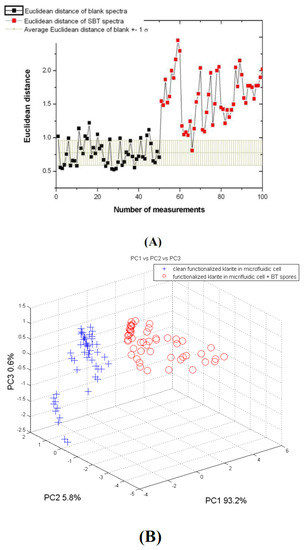
Figure 4.
(A) Euclidean distance for the blank spectra (black dots) and SBT spectra (red dots) compared to the average value of the Euclidian distance obtained for the blank spectra (the brown line shows the range individuated by mean value ± 1sB). (B) First three principal component (PC) score plots of the SERS spectra acquired on the clean cartridge (blue crosses) and inoculated with SBT (red dots).
Both Euclidian distance analysis and the principal component (PC) score plots show two groups of spectra well represented by the substrate and the spores of SBT likely captured by peptides (Figure 4). With reference to the results obtained from the Raman spectra acquired on SBA in the same experimental conditions described above, unexpected results were observed. In fact, for the Euclidian distance, the mean values and the standard deviation (sd) relative to the SERS spectra of the spores of SBA and the clean substrate are respectively 4.00 sd ± 0.46 and 1.6 sd ± 0.23. The distance between the two groups is even higher than the one obtained for SBT, which are tied by specific peptides receptors. Further experiments were carried out with a Raman probe on Klarite substrates, which were functionalized as in the previous tests. The tests with SBT were performed in the same experimental conditions as above, while the tests with SBA were performed out of the MFC in order to simplify the measurement system. The results are shown in Figure 5 and Figure 6.
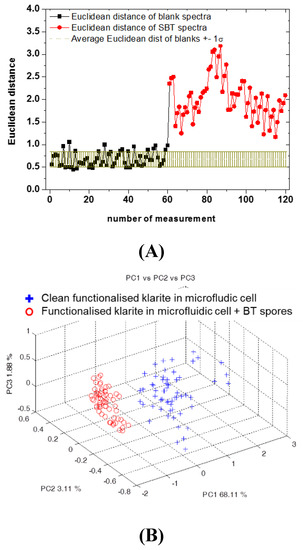
Figure 5.
(A) Euclidean distance for the blank spectra (black dots) and SBT spectra (red dots) compared to the average value of the Euclidian distance obtained for the blank spectra (the brown line shows the range individuated by mean value ± 1sB). (B) First three principal component (PC) score plots of the SERS spectra acquired on the clean cartridge (blue crosses) and inoculated with SBT (red dots).
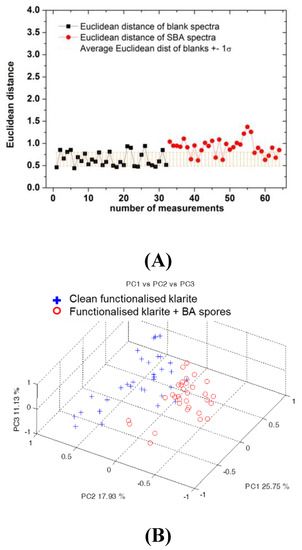
Figure 6.
(A) Euclidean distance for the blank spectra (black dots) and Bacillus atrophaeus (SBA) spectra (red dots) compared to the average value of the Euclidian distance obtained for the blank spectra (the brown line shows the range individuated by mean value ± 1sB). (B) First three principal component (PC) score plots of the SERS spectra acquired on the clean cartridge (blue crosses) and inoculated with SBT (red dots).
In the case of the SBT samples, the previous results were confirmed by the Euclidian distance that well represents the identification of spores compared to the blank (Figure 5A), and by the PCA (Figure 5B).
As regards SBA, the results obtained previously were confirmed (Figure 6). In fact, even if the majority of points are close to the range individuated by the mean value of the Euclidian distance of the blank spectra ± 1sB, as expected after rinsing, almost all the points are out of this range (Figure 6A). The PCA results are in line with this observation (Figure 6B): the total variance explained by the first three PCs is lower, and an overlapping of the two groups is present, but a clear separation is still evident.
This result seems to be the proof of the permanence, for both Bacillus spores, of the functionalized SERS substrate, even after rinsing.
Thus, to give an estimation of the SBA spore (not the expected spores bound to the peptides) density on the sensor after the entire procedure, the active SERS surface was inspected with a Phenom Pro SEM microscope. Figure 7 shows three SEM images of the SERS sensor contaminated with SBA at different magnifications. It can be observed that, a few tenths of spores (~80) remain linked to the sensor after rinsing over an area of about 150 × 150 μm2. This area is approximately four times larger than the one usually investigated in a single Raman probe measurement (80 μm in diameter from the technical specifications of the Raman probe).
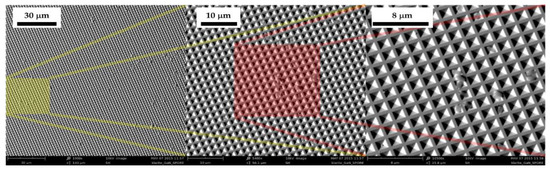
Figure 7.
SEM pictures of the SERS sensor contaminated with SBA after the whole deposition-rinsing procedure. The coloured areas represent areas zoomed at different magnifications.
4. Discussions
The study for the rapid detection and identification of Bacillus spores in a MFC highlighted that the main signal in the spectrum shown in Figure 3 (red line) seems to be due to the whole cartridge. In particular, the two layers constituted by the polymeric coverslip and the thin layer of peptides, which are in close contact with the SERS surface, interfere with the Raman signal of the spores. However, by the comparison between the signals from the clean substrate and the substrate with spores, a few weak signals, most likely due to the presence of spores of Bacillus thuringiensis (SBT) bound selectively to the specific peptides, can be noticed (marked in Figure 3).
According to the literature, [29] the spectral features at 740, 861, and 1650 cm−1 could be assigned to adenine and phenylalanine [10], or, as a glycosidic ring, associated to the peptidoglican, a principal component of the cell wall that is present in the endospores coat (at 740 cm−1) [29,30,31]. It could be explained by the possibility of having some components of spores, as well as the functional group of the main component of Bacillus spores, in solution. [29,32,33] In particular, the signal at 1650 cm−1 can be attributed to the amide II combination modes and the amide I mode (C = O stretch) [17]. Nevertheless, for both kinds of spores, statistical analysis demonstrates the ability to discriminate between Raman spectra before and after spore inoculation and surface rinsing. In fact, the results obtained for measurements by Micro Raman (40x objective) are particularly evident for SBT as shown in Figure 4 and for SBA as discussed in Section 3. Similarly, in the case of measurements with the probe, applying statistical approaches (Figure 5 and Figure 6), Raman signals on substrates and samples for both SBA and SBT can be discriminated. In particular, by measuring the Euclidean distance from a proper reference spectrum, it was possible to classify each spectrum as a positive (hit) or negative (no-hit) identification of dangerous spores (Figure 5 and Figure 6).
In addition, for SBA, several points are far from the blank spectra according to Euclidian distance (Figure 6), as is also the case for SBT (only for SBT are expected to be joined by their specific peptides), leading to the conclusion that peptides are not as selective as foreseen. In fact, several SBA remain on the substrate, as confirmed by SEM (Figure 7). In accordance with the results obtained, the use of a substrate not functionalized with peptides is suggested.
5. Conclusions
Concerning the realization of an automated device for the rapid detection of the biological threat of aerosol, the SERS technique, in combination with chemometric analysis, as well as normalized Euclidian distance and PCA, can be proposed mainly as a rapid pre-alarm system for fast continuous monitoring sensing. Higher selectivity can be achieved at the second stage of the device, e.g., performing PCR measurements. The functionalization of the SERS with the peptides not only enhances the selectivity of the substrate for SBT, but also, despite being very thin, strongly affects the signal from the spores, because it is very close to the SERS active surface. Additional studies with SERS substrates not functionalized with peptides, as well as studies on more biologically interfering particles that can be commonly found in air, pollen, and the spores of fungi and bacteria, are ongoing to confirm these results.
Acknowledgments
The project RAMBO is funded by the EDA (European Defence Agency) Joint Investment Programme on CBRNE protection under grant agreement A-1152-RT-GP.
Author Contributions
All authors conceived, performed, and designed the experiments and analyzed the data. Antonia Lai wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hess, G. Biosecurity: An evolving challenge. Chem. Eng. News 2012, 90, 30–32. [Google Scholar]
- Pedrero, M.; Campuzano, S.; Pingarron, J.M. Magnetic Beads-Based Electrochemical Sensors Applied to the Detection and Quantification of Bioterrorism/Biohazard Agents. Electroanal 2012, 24, 470–482. [Google Scholar] [CrossRef]
- Katz, R. Biological Weapons: A National Security Problem That Requires a Public Health Response, Working Paper 2001–2004. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.680.5464&rep=rep1&type=pdf (accessed on 19 December 2016).
- Kortepeter, M.G.; Parker, G.W. Potential biological weapons threats. Emerg. Infect. Dis. 1999, 5, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.M.; Kozarsky, P.E.; Stephens, D.S. Clinical issue in the prophylaxis diagnosis and treatment of anthrax. Emerg. Infect. Dis. 2002, 8, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Kellogs, M. Detection of biological agents used for terrorism: Are we ready? Clin. Chem. 2010, 56, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Félix-Rivera, H.; Hernández-Rivera, S.P. Raman Spectroscopy Techniques for the Detection of Biological Samples in Suspensions and as Aerosol Particles: A Review. Sens. Imaging 2012, 13, 1–25. [Google Scholar] [CrossRef]
- Jarvis, R.M.; Goodacre, R. Discrimination of bacteria using sers. Anal. Chem. 2004, 76, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Cowcher, D.P.; Xu, Y.; Goodacre, R. Portable quantitative detection of bacillus bacteria spores using sers. Anal. Chem. 2013, 85, 3297–3302. [Google Scholar] [CrossRef] [PubMed]
- Shende, C.; Inscore, F.; Huang, H.; Farquharson, S.; Sengupta, A. Detection of Bacillus spores within 15 minutes by surface-enhanced Raman spectroscopy. In Proceedings of the SPIE Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XIII, Baltimore, MD, USA, 1 May 2012. [CrossRef]
- Farquharson, S.; Smith, W.W. Differentiating bacterial spores from hoax materials by Raman spectroscopy. In Proceedings of the SPIE Chemical and Biological Point Sensors for Homeland Defense, Providence, RI, USA, 8 March 2004. [CrossRef]
- Mungroo, N.A.; Oliveira, G.; Neethirajan, S. SERS based point-of-care detection of food-borne pathogens. Microchim. Acta 2016, 183, 697–707. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Moir, D.T.; Klempner, M.S.; Krieger, N.; Jones, G., II; Ziegler, L.D. Characterization of the surface Enhanced Raman Scattering (SERS) of Bacteria. J. Phys. Chem. B 2005, 109, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Guicheteau, J.; Argue, L.; Emge, D.; Hyre, A.; Jacobson, M.; Christesen, S. Bacillus Spore Classification via Surface-Enhanced Raman Spectroscopy and Principal Component Analysis. Appl. Spectrosc. 2008, 62, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, R.M.; Brooker, A.; Goodrace, R. Surface-enhanced Raman scattering for the rapid discrimination of bacteria. Faraday Discuss. 2006, 132, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Pieczonkaa, N.P.W.; Arocaa, R.F. Single molecule analysis by surfaced-enhanced Raman scattering. Chem. Soc. Rev. 2008, 37, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Shende, C.; Farquharson, S.; Inscore, F. Detection of Bacillus anthracis Spores Using Peptide Functionalized SERS-Active Substrates. Int. J. Spectrosc. 2012, 2012. [Google Scholar] [CrossRef]
- Ash, C.; Farrow, J.A.E.; Wallbanks, S.; Collins, M.D. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 1991, 13, 202–206. [Google Scholar] [CrossRef]
- Innis, M.A.; Gelfand, D.H.; Sninsky, J.J.; White, T.J. PCR Protocols: A Guide to Methods and Applications; Academic Press: Waltham, MA, USA, 2012. [Google Scholar]
- Beer, M.; Liu, C.Q. Panning of a Phage Display Library against a Synthetic Capsule for Peptide Ligands that Bind to the Native Capsule of Bacillus anthracis. PLoS ONE 2012, 7, e45472. [Google Scholar] [CrossRef] [PubMed]
- Knurr, J.; Benedek, O.; Heslop, J.; Vinson, R.B.; Boydston, J.A.; McAndrew, J.; Kearney, J.F.; Turnbough, C.L., Jr. Peptide ligands that bind selectively to spores of Bacillus subtilis and closely related species. Appl. Environ. Microbiol. 2003, 69, 6841–6847. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.A.; Le, D.M. Characterization of a commercialized SERS-active substrate and its application to the identification of intact Bacillus endospores. Appl. Opt. 2007, 46, 3878–3890. [Google Scholar] [CrossRef] [PubMed]
- Corgier, B.P.; Marquette, C.A.; Blum, L.J. Diazonium-Protein Adducts for Graphite Electrode Microarrays Modification: Direct and Addressed Electrochemical Immobilization. J. Am. Chem. Soc. 2005, 127, 18328–18332. [Google Scholar] [CrossRef] [PubMed]
- Mandon, C.A.; Blum, L.J.; Marquette, C.A. Aryl diazonium for biomolecules immobilization onto SPRi chips. Chem. Phys. Chem. 2009, 10, 3273–3277. [Google Scholar] [CrossRef] [PubMed]
- Jolliffe, I.T. Principal Component Analysis, 2nd ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Serber, G.A.F. Multivariate Observations; John Wiley and Sons: New York, NY, USA, 1984. [Google Scholar] [CrossRef]
- Al-Kandari, N.M.; Jolliffe, I.T. Variable selection and interpretation in correlation principal components. Environmetrics 2005, 16, 659–672. [Google Scholar] [CrossRef]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D.L. The Mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Morrowa, J.B.; Almeidab, J.; Colec, K.D.; Reipad, V. Raman spectroscopy of Bacillus thuringiensis physiology and inactivation. Eur. Phys. J. Plus 2012, 127. [Google Scholar] [CrossRef]
- Félix-Rivera, H.; Gonzalez, R.; Del Mar Rodrıguez, G.; Primera-Pedrozo, O.M.; Rıos-Velazquez, C.; Hernández-Rivera, S.P. Improving SERS Detection of Bacillus thuringiensis Using Silver Nanoparticles Reduced with Hydroxylamine and with Citrate Capped Borohydride. Int. J. Spectrosc. 2011, 2011. [Google Scholar] [CrossRef]
- Atrih, A.; Zollner, P.; Allmaier, G.; Foster, J.A. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J. Bacteriol. 1996, 178, 6173–6183. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Bergman, N.H.; Thomason, B.; Shallom, S.; Hazen, A.; Crossno, J.; Rasko, D.A.; Ravel, J.; Read, T.D.; Peterson, S.N.; et al. Formation and composition of the Bacillus anthracis endospore. J. Bacteriol. 2004, 186, 164–178. [Google Scholar] [CrossRef] [PubMed]
- Farquharson, S.; Grigely, L.; Khitrov, V.; Smith, W.W.; Sperry, J.F.; Fenerty, G. Detecting Bacillus cereus spores on a mail sorting system using Raman spectroscopy. J. Raman Spectrosc. 2004, 35, 3582–3586. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).