Multiomics and Systems Biology Are Needed to Unravel the Complex Origins of Chronic Disease
Abstract
1. Introduction
2. Perinatal Influences on Immune Development and Chronic Disease
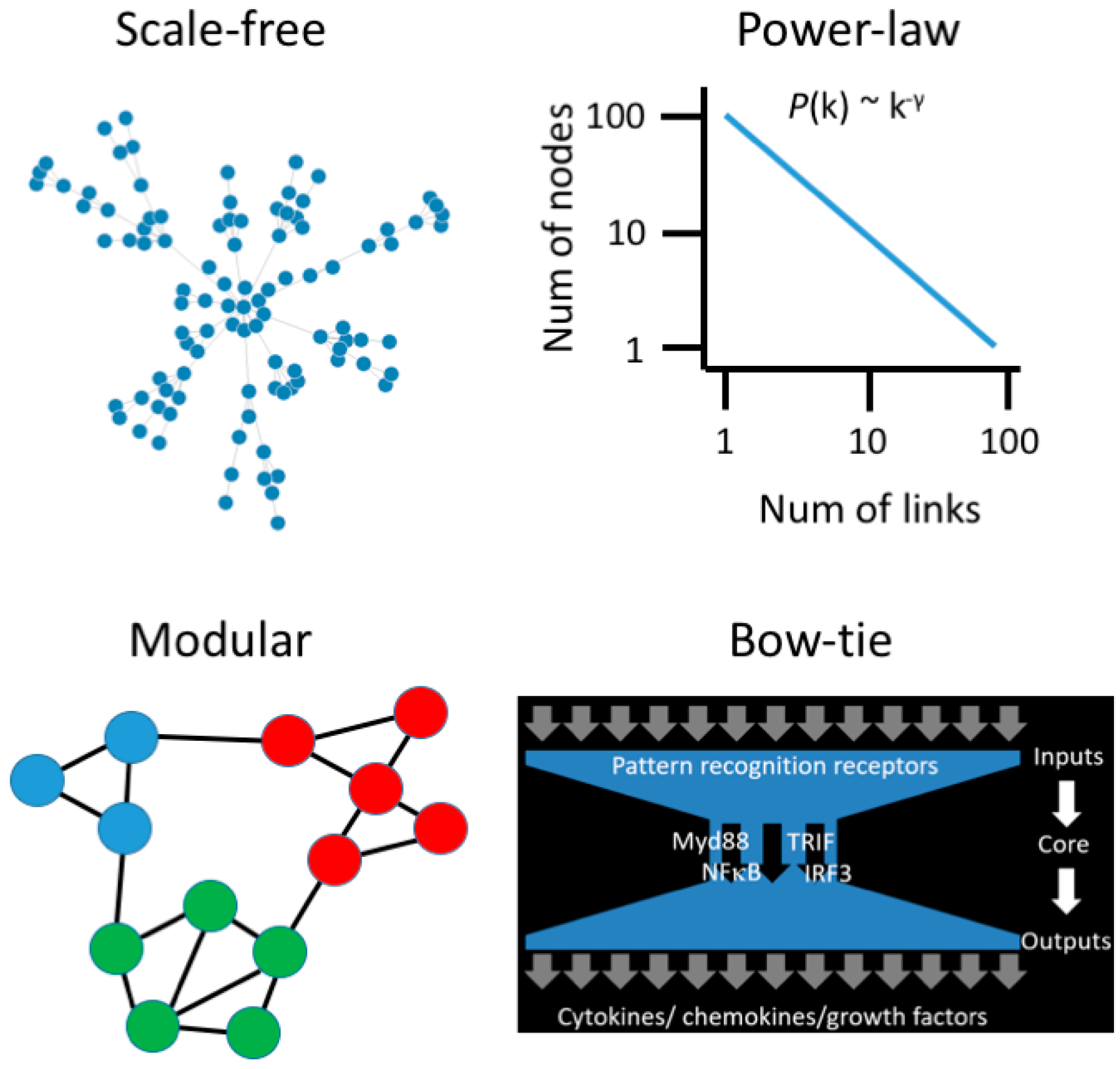
3. Moving beyond Reductionist Biology: Systems-Level Understanding of Immune Dysregulation
4. Utilizing Systems Biology Approaches for Very Early Prediction and Intervention for Immune-Mediated Diseases
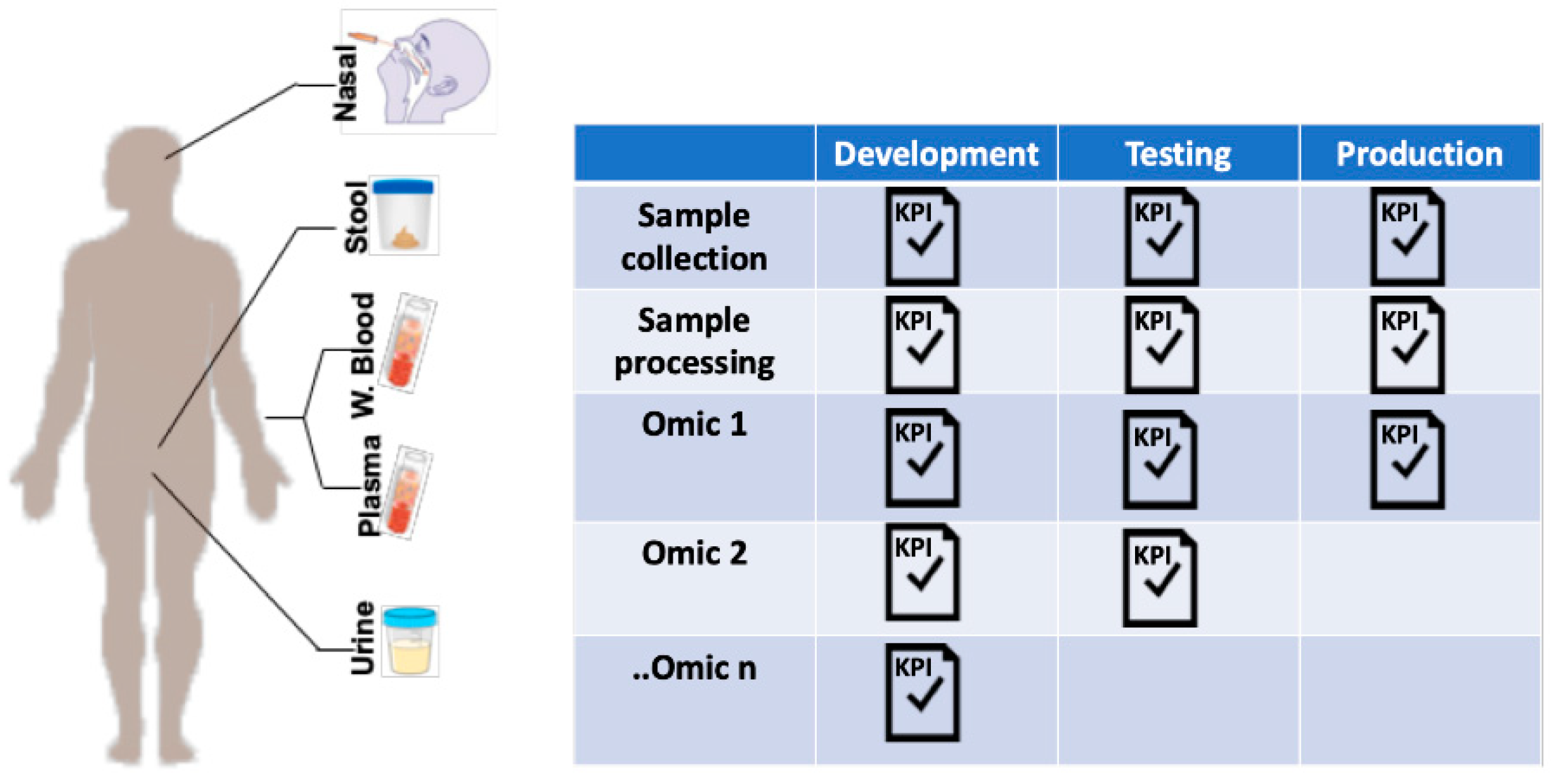
5. Multiomic Studies: Challenges and Opportunities
5.1. Sample Collection
5.2. Data Collection
5.3. Data Management
5.4. Data Analysis
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Renz, H.; Holt, P.G.; Inouye, M.; Logan, A.C.; Prescott, S.L.; Sly, P.D. An exposome perspective: Early-life events and immune development in a changing world. J. Allergy Clin. Immunol. 2017, 140, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Corbett, S.; Courtiol, A.; Lummaa, V.; Moorad, J.; Stearns, S. The transition to modernity and chronic disease: Mismatch and natural selection. Nat. Rev. Genet. 2018, 19, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Abajobir, A.A.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abera, S.F.; Aboyans, V.; Adetokunboh, O.; Afshin, A.; Agrawal, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef]
- Bilbo, S.D.; Schwarz, J.M. The immune system and developmental programming of brain and behavior. Front. Neuroendocrinol. 2012, 33, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-X.; Wheelock, C.E.; Sköld, C.M.; Wheelock, Å.M. Integration of multi-omics datasets enables molecular classification of COPD. Eur. Respir. J. 2018, 51, 1701930. [Google Scholar] [CrossRef] [PubMed]
- Moore, T.; Arefadib, N.; Deery, A.; Keyes, M.; West, S. The First Thousand Days: An Evidence Paper; Centre for Community Child Health, Murdoch Children’s Research Institute: Parkville, VIC, Australia, 2017. [Google Scholar]
- Gollwitzer, E.S.; Marsland, B.J. Impact of Early-Life Exposures on Immune Maturation and Susceptibility to Disease. Trends Immunol. 2015, 36, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Olin, A.; Henckel, E.; Chen, Y.; Lakshmikanth, T.; Pou, C.; Mikes, J.; Gustafsson, A.; Bernhardsson, A.K.; Zhang, C.; Bohlin, K.; et al. Stereotypic Immune System Development in Newborn Children. Cell 2018, 174, 1277–1292. [Google Scholar] [CrossRef]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, Å.; Hagopian, W.A.; Rewers, M.J.; She, J.-X.; Toppari, J.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef]
- West, C.E.; Renz, H.; Jenmalm, M.C.; Kozyrskyj, A.L.; Allen, K.J.; Vuillermin, P.; Prescott, S.L. The gut microbiota and inflammatory noncommunicable diseases: Associations and potentials for gut microbiota therapies. J. Allergy Clin. Immunol. 2015, 135, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.M.; Hrusch, C.L.; Gozdz, J.; Igartua, C.; Pivniouk, V.; Murray, S.E.; Ledford, J.G.; Marques dos Santos, M.; Anderson, R.L.; Metwali, N.; et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. NEJM 2016, 375, 411–421. [Google Scholar] [CrossRef]
- Amenyogbe, N.; Kollmann, T.R.; Ben-Othman, R. Early-Life Host–Microbiome Interphase: The Key Frontier for Immune Development. Front. Pediatr. 2017, 5, 343. [Google Scholar] [CrossRef] [PubMed]
- Bateson, P.; Gluckman, P.; Hanson, M. The biology of developmental plasticity and the Predictive Adaptive Response hypothesis. J. Physiol. 2014, 592, 2357–2368. [Google Scholar] [CrossRef] [PubMed]
- Fleming, T.P.; Watkins, A.J.; Velazquez, M.A.; Mathers, J.C.; Prentice, A.M.; Stephenson, J.; Barker, M.; Saffery, R.; Yajnik, C.S.; Eckert, J.J.; et al. Origins of lifetime health around the time of conception: Causes and consequences. Lancet 2018, 391, 1842–1852. [Google Scholar] [CrossRef]
- Hanson, M.A.; Gluckman, P.D. Early Developmental Conditioning of Later Health and Disease: Physiology or Pathophysiology? Physiol. Rev. 2014, 94, 1027–1076. [Google Scholar] [CrossRef] [PubMed]
- Martino, D.; Allen, K. Meeting the challenges of measuring human immune regulation. J. Immunol. Methods 2015, 424, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wang, X.; Li, X.; Inlora, J.; Wang, T.; Liu, Q.; Snyder, M. Dynamic Human Environmental Exposome Revealed by Longitudinal Personal Monitoring. Cell 2018, 175, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Niedzwiecki, M.M.; Walker, D.I.; Vermeulen, R.; Chadeau-Hyam, M.; Jones, D.P.; Miller, G.W. The Exposome: Molecules to Populations. Annu. Rev. Pharmacol. Toxicol. 2018, 59, 107–127. [Google Scholar] [CrossRef]
- Ma’ayan, A. Complex systems biology. J. R. Soc. Interface 2017, 14, 20170391. [Google Scholar] [CrossRef]
- Zotenko, E.; Mestre, J.; O’Leary, D.P.; Przytycka, T.M. Why do hubs in the yeast protein interaction network tend to be essential: Reexamining the connection between the network topology and essentiality. PLoS Comput. Biol. 2008, 4, e1000140. [Google Scholar] [CrossRef]
- Chevrier, N.; Mertins, P.; Artyomov, M.N.; Shalek, A.K.; Iannacone, M.; Ciaccio, M.F.; Gat-Viks, I.; Tonti, E.; DeGrace, M.M.; Clauser, K.R.; et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell 2011, 147, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Bosco, A.; Ehteshami, S.; Panyala, S.; Martinez, F.D. Interferon regulatory factor 7 is a major hub connecting interferon-mediated responses in virus-induced asthma exacerbations in vivo. J. Allergy Clin. Immunol. 2012, 129, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef]
- Kitano, H.; Oda, K. Robustness trade-offs and host-microbial symbiosis in the immune system. Mol. Syst. Biol. 2006, 2. [Google Scholar] [CrossRef]
- Price, N.D.; Magis, A.T.; Earls, J.C.; Glusman, G.; Nature, R.L. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 2017, 35, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Vassos, E. Prospects for using risk scores in polygenic medicine. Genome Med. 2017, 9, 96. [Google Scholar] [CrossRef] [PubMed]
- Dinalankara, W.; Ke, Q.; Xu, Y.; Ji, L.; Pagane, N.; Lien, A.; Matam, T.; Fertig, E.J.; Price, N.D.; Younes, L.; et al. Digitizing omics profiles by divergence from a baseline. Proc. Natl. Acad. Sci. USA 2018, 115, 4545–4552. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.D. Early-Life Origins of Chronic Obstructive Pulmonary Disease. NEJM 2016, 375, 871–878. [Google Scholar] [CrossRef]
- Torkamani, A.; Wineinger, N.E.; Topol, E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018, 19, 581–590. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Tang, H.H.; Teo, S.M.; Belgrave, D.C.; Evans, M.D.; Jackson, D.J.; Brozynska, M.; Kusel, M.M.; Johnston, S.L.; Gern, J.E.; Lemanske, R.F.; et al. Trajectories of childhood immune development and respiratory health relevant to asthma and allergy. Elife 2018, 7, 28. [Google Scholar] [CrossRef]
- Piening, B.D.; Zhou, W.; Contrepois, K.; Röst, H.; Gu Urban, G.J.; Mishra, T.; Hanson, B.M.; Bautista, E.J.; Leopold, S.; Yeh, C.Y.; et al. Integrative Personal Omics Profiles during Periods of Weight Gain and Loss. Cell Syst. 2018, 6, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Peakman, T.; Elliott, P. Current standards for the storage of human samples in biobanks. Genome Med. 2010, 2, 72. [Google Scholar] [CrossRef] [PubMed]
- Kolker, E.; Özdemir, V.; Martens, L.; Hancock, W.; Anderson, G.; Anderson, N.; Aynacioglu, S.; Baranova, A.; Campagna, S.R.; Chen, R.; et al. Toward more transparent and reproducible omics studies through a common metadata checklist and data publications. OMICS 2014, 18, 10–14. [Google Scholar] [CrossRef]
- Gomez-Cabrero, D.; Abugessaisa, I.; Maier, D.; Teschendorff, A.; Merkenschlager, M.; Gisel, A.; Ballestar, E.; Bongcam-Rudloff, E.; Conesa, A.; Tegnér, J. Data integration in the era of omics: Current and future challenges. BMC Syst. Biol. 2014, 8. [Google Scholar] [CrossRef] [PubMed]
- Chiang, G.-T.; Clapham, P.; Qi, G.; Sale, K.; Coates, G. Implementing a genomic data management system using iRODS in the Wellcome Trust Sanger Institute. BMC Bioinform. 2011, 12, 361. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.; Rujan, T.; Genomics, J.H.A.T. A collaborative approach to develop a multi-omics data analytics platform for translational research. Appl. Transl. Genomics 2014, 3, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Hernández-de-Diego, R.; Tarazona, S.; Martínez-Mira, C.; Balzano-Nogueira, L.; Furió-Tarí, P.; Pappas, G.J.; Conesa, A. PaintOmics 3: A web resource for the pathway analysis and visualization of multi-omics data. Nucleic Acids Res. 2018, 46, W503–W509. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Ghandikota, S.; Hershey, G.; Bioinformatics, T.M. GENEASE: Real time bioinformatics tool for multi-omics and disease ontology exploration, analysis and visualization. Bioinformatics 2018, 34, 3160–3168. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Gligorijević, V.; Pržulj, N. Methods for biological data integration: Perspectives and challenges. J. R. Soc. Interface 2015, 12, 20150571. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martino, D.; Ben-Othman, R.; Harbeson, D.; Bosco, A. Multiomics and Systems Biology Are Needed to Unravel the Complex Origins of Chronic Disease. Challenges 2019, 10, 23. https://doi.org/10.3390/challe10010023
Martino D, Ben-Othman R, Harbeson D, Bosco A. Multiomics and Systems Biology Are Needed to Unravel the Complex Origins of Chronic Disease. Challenges. 2019; 10(1):23. https://doi.org/10.3390/challe10010023
Chicago/Turabian StyleMartino, David, Rym Ben-Othman, Danny Harbeson, and Anthony Bosco. 2019. "Multiomics and Systems Biology Are Needed to Unravel the Complex Origins of Chronic Disease" Challenges 10, no. 1: 23. https://doi.org/10.3390/challe10010023
APA StyleMartino, D., Ben-Othman, R., Harbeson, D., & Bosco, A. (2019). Multiomics and Systems Biology Are Needed to Unravel the Complex Origins of Chronic Disease. Challenges, 10(1), 23. https://doi.org/10.3390/challe10010023




