Machine Learning Classification Algorithms for Predicting Karenia brevis Blooms on the West Florida Shelf
Abstract
:1. Introduction
2. Materials and Methods
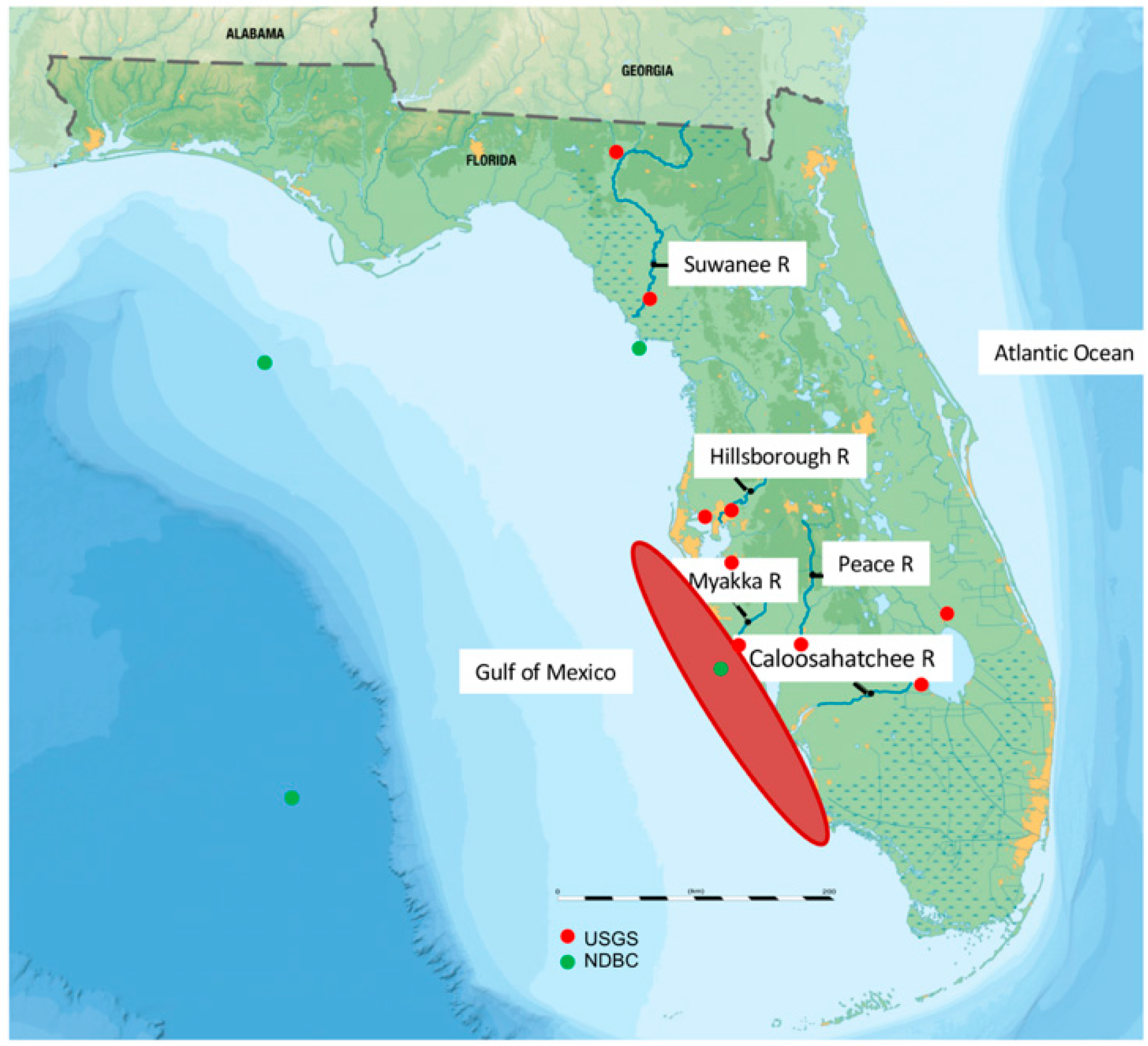
2.1. The Dataset and Preparation of Explanatory and Dependent Variables
2.2. Machine Learning Algorithms
2.2.1. Support Vector Machine
2.2.2. Relevance Vector Machine
2.2.3. Naïve Bayes
2.2.4. Artificial Neural Network
2.3. Model Evaluation and Metrics
2.4. Sensitivity Analysis
2.5. In Silico Experiments
3. Results
3.1. Overall Model Performance
3.2. Role of Wind
3.3. Role of River Flow and Associated Nutrients
3.4. Role of Sea Surface Height
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, D.M. Toxic Algal Blooms and Red Tides: A Global Perspective. In Red Tides: Biology, Environmental Science, and Toxicology; Okaichi, T., Anderson, D., Nemoto, T., Eds.; Elsevier Science Publishing Company: New York, NY, USA, 1989; pp. 11–16. [Google Scholar]
- Hallegraeff, G.M. A review of harmful algal blooms and their apparent global increase. Phycologia 1993, 32, 79–99. [Google Scholar] [CrossRef] [Green Version]
- Glibert, P.M.; Burkholder, J.M. Causes of harmful algal blooms. In Harmful Algal Blooms: A Compendium Desk Reference; Shumway, S., Burkholder, J.M., Morton, S.L., Eds.; Wiley Blackwell: Singapore, 2018; pp. 1–38. [Google Scholar]
- Heisler, J.; Glibert, P.M.; Burkholder, J.; Anderson, D.; Cochlan, W.; Dennison, W.; Dortch, Q.; Heil, C.; Humphries, E.; Lewitus, A.; et al. Eutrophication and harmful algal blooms: A scientific consensus. Harmful Algae 2008, 8, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Fu, F.X.; Tatters, A.O.; Hutchins, D.A. Global change and the future of harmful algal blooms in the ocean. Mar. Ecol. Progr. Ser. 2012, 470, 207–233. [Google Scholar] [CrossRef] [Green Version]
- Wells, M.L.; Trainer, V.L.; Smayda, T.J.; Karlson, B.S.; Trick, C.G.; Kudela, R.M.; Ishikawa, A.; Bernard, S.; Wulff, A.; Anderson, D.A.; et al. Harmful algal blooms and climate change: Learning from the past and present to forecast the future. Harmful Algae 2015, 49, 68–93. [Google Scholar] [CrossRef] [Green Version]
- Glibert, P.M.; Burford, M.A. Globally changing nutrient loads and harmful algal blooms: Recent advances, new paradigms and continuing challenges. Oceanography 2017, 30, 44–55. [Google Scholar] [CrossRef] [Green Version]
- Glibert, P.M. Harmful algal at the complex nexus of eutrophication and climate change. Harmful Algae 2020, 9. [Google Scholar] [CrossRef]
- Steidinger, K.A. Historical perspective on Karenia brevis red tide research in the Gulf of Mexico. Harmful Algae 2009, 8, 549–561. [Google Scholar] [CrossRef]
- Brand, K.; Compton, A. Long-term increase in Karenia brevis abundance along the southwest Florida coast. Harmful Algae 2007, 6, 232–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glibert, P.M. Why were the water and beaches in west Florida so gross in summer 2018? Red tides! Front. Young Minds 2019, 7. [Google Scholar] [CrossRef] [Green Version]
- Fears, D.; Rozsa, L. Florida’s Unusually Long Red Tide Is Killing Wildlife, Tourism and Businesses. The Washington Post. Available online: https://www.washingtonpost.com/national/health-science/floridas-unusually-long-red-tide-is-killing-wildlife-tourism-and-businesses/2018/08/28/245fc8da-aad5-11e8-8a0c-70b618c98d3c_story.html (accessed on 12 December 2018).
- Monuz, C.R. Red tide episode kills record number of sea turtles. Herald Tribune, 15 January 2019. [Google Scholar]
- Heil, C.A.; Bronk, D.A.; Dixon, L.K.; Hitchcock, G.L.; Kirkpatrick, G.J.; Mulholland, M.R.; O’Neil, J.M.; Walsh, J.J.; Weisberg, R.H.; Garrett, M. The Gulf of Mexico ECOHAB: Karenia program 2006–2012. Harmful Algae 2014, 38, 3–7. [Google Scholar] [CrossRef]
- Weisberg, R.H.; He, R. Local and deep-ocean forcing contributions to anomalous water properties on the West Florida Shelf. J. Geophys. Res. 2003, 108, 3184. [Google Scholar] [CrossRef]
- Liu, Y.; Weisberg, R.H. Seasonal variability on the West Florida Shelf. Progr. Oceanogr. 2012, 104, 80–98. [Google Scholar] [CrossRef]
- Mayer, D.A.; Weisberg, R.H.; Zheng, L.; Liu, Y. Winds on the West Florida Shelf: Regional comparisons between observations and model estimates. J. Geophys. Res. Oceans 2017, 122, 834–846. [Google Scholar] [CrossRef] [Green Version]
- Weisberg, L.; Zheng, L.; Liu, Y.; Lembke, C.; Lenes, J.M.; Walsh, J.J. Why a red tide was not observed on the west Florida continental shelf in 2010. Harmful Algae 2014, 38, 119–126. [Google Scholar] [CrossRef]
- Liu, Y.; Weisberg, R.H.; Lenes, J.M.; Zheng, L.; Hubbard, K.; Walsh, J.J. Offshore forcing on the “pressure point” of the West Florida Shelf: Anomalous upwelling and its influence on harmful algal blooms. J. Geophys. Res. 2016, 121, 5501–5515. [Google Scholar] [CrossRef]
- Hu, C.; Muller-Karger, F.E.; Swarzenski, P.W. Hurricanes, submarine groundwater discharge, and Florida’s red tides. Geophys. Res. Lett. 2006, 33, L11601. [Google Scholar] [CrossRef]
- Vargo, G.A.; Heil, C.A.; Fanning, K.A.; Dixon, K.L.; Neely, M.B.; Lester, K.; Ault, D.; Murasko, S.; Havens, J.; Walsh, J.; et al. Nutrient availability in support of Karenia brevis blooms on the central West Florida Shelf: What keeps Karenia blooming? Cont. Shelf Res. 2008, 28, 73–98. [Google Scholar] [CrossRef]
- Vargo, G.A. A brief summary of the physiology and ecology of Karenia brevis Davis (G. Hansen and Moestrup comb. nov.) red tides on the West Florida Shelf and of hypotheses posed for their initiation, growth, maintenance, and termination. Harmful Algae 2009, 8, 573–584. [Google Scholar] [CrossRef]
- Lenes, J.M.; Darrow, B.A.; Walsh, J.J.; Prospero, J.M.; He, R.; Weisberg, R.H.; Vargo, G.A.; Heil, C.A. Saharan dust and phosphatic fidelity: A three-dimensional biogeochemical model of Trichodesmium as a nutrient source for red tides on the West Florida Shelf. Cont. Shelf Res. 2008, 28, 1091–1115. [Google Scholar] [CrossRef]
- Glibert, P.M.; Burkholder, J.M.; Kana, T.M.; Alexander, J.A.; Schiller, C.; Skelton, H. Grazing by Karenia brevis on Synechococcus enhances their growth rate and may help to sustain blooms. Aquat. Microb. Ecol. 2009, 55, 17–30. [Google Scholar] [CrossRef] [Green Version]
- O’Neil, J.M.; Heil, C.A. Preface to ECOHAB: Karenia Special Edition of Harmful Algae. Harmful Algae 2014, 38, 1–2. [Google Scholar] [CrossRef]
- Glibert, P.M.; Allen, J.I.; Bouwman, L.; Brown, C.; Flynn, K.J.; Lewitus, A.; Madden, C. Modeling of HABs and eutrophication: Status, advances, challenges. J. Mar. Syst. 2010, 83, 262–275. [Google Scholar] [CrossRef]
- Glibert, P.M.; Beusen, A.H.W.; Bouwman, A.F.; Burkholder, J.M.; Flynn, K.J.; Heil, C.A.; Li, M.; Lin, C.H.; Madden, C.J.; Mitra, A.; et al. Multifaceted climatic and nutrient effects on harmful algae require multifaceted model. In Climate Change and Marine and Freshwater Toxins, 2nd ed.; Botana, L.M., Louzao, C., Vilariño, N., Eds.; DeGruyter Publishers: Berlin, Germany, 2021; pp. 473–518. [Google Scholar]
- McGillicuddy, D.J., Jr.; de Young, B.; Doney, S.; Glibert, P.M.; Stammer, D.; Werner, F.E. Models: Tools for synthesis in international oceanographic research programs. Oceanography 2010, 23, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M. HABs in a changing world: A perspective on harmful algal blooms, their impacts, and research and management in a dynamic era of climatic and environmental change. In Harmful Algae 2012, Proceedings of the 15th International Conference on Harmful Algae: 29 October—2 November 2012; Kim, H.-G., Reguera, B., Hallegraeff, G.M., Lee, C.K., Han, M.S., Choi, J.K., Eds.; CECO: Changwon, Gyeongnam, 2014; pp. 3–17. [Google Scholar]
- Franks, P.J.S. Recent advances in modeling of harmful algal blooms. In Global Ecology and Oceanography of Harmful Algal Blooms; Glibert, P.M., Berdalet, E., Burford, M., Pitcher, G., Zhou, M.J., Eds.; Springer: Cham, Switzerland, 2018; pp. 359–380. [Google Scholar]
- Flynn, K.J.; McGillicuddy, D.J. Modeling marine harmful algal blooms: Current status and future prospects. In Harmful Algal Blooms: A Compendium Desk Reference; Shumway, S., Burkholder, J.M., Morton, S.L., Eds.; Wiley Blackwell: Singapore, 2018; pp. 115–134. [Google Scholar]
- Stumpf, R.P.; Culver, M.E.; Tester, P.A.; Tomlinson, M.; Kirkpatrick, G.J.; Pederson, B.A.; Truby, E.; Ransibrahmanakul, V.; Soracco, M. Monitoring Karenia brevis blooms in the Gulf of Mexico using satellite ocean color imagery and other data. Harmful Algae 2003, 2, 147–160. [Google Scholar] [CrossRef]
- Stumpf, R.P.; Tomlinson, M.C.; Calkins, J.A.; Kirkpatrick, B.; Fisher, K.; Nierenberg, K.; Currier, R.; Wynne, T.T. Skill assessment for an operational algal bloom forecast system. J. Mar. Syst. 2009, 76, 151–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisberg, R.H.; Barth, A.; Alvera-Azcárate, A.; Zheng, L. A coordinated coastal ocean observing and modeling system for the West Florida Shelf. Harmful Algae 2009, 8, 585–598. [Google Scholar] [CrossRef]
- Walsh, J.J.; Weisberg, R.H.; Dieterle, D.A.; He, R.; Darrow, B.P.; Jolliff, J.K.; Lester, K.M.; Vargo, G.A.; Kirkpatrick, G.J.; Fanning, K.A.; et al. Phytoplankton response to intrusions of slope water on the West Florida Shelf: Models and observations. J. Geophys. Res. 2003, 108, 15. [Google Scholar] [CrossRef] [Green Version]
- Walsh, J.J.; Joliff, J.K.; Darrow, B.P.; Lenes, J.M.; Milroy, S.P.; Remsen, A.; Dieterle, D.A.; Carder, K.L.; Chen, F.R.; Vargo, G.A.; et al. Red tides in the Gulf of Mexico: Where, when, and why. J. Geophys. Res. 2006, 111, 1–46. [Google Scholar] [CrossRef]
- Milroy, S.P.; Dieterle, D.A.; He, R.; Kirkpatrick, G.J.; Lester, K.M.; Steidinger, K.A.; Vargo, G.A.; Walsh, J.J.; Weisberg, R.H. A three-dimensional biophysical model of Karenia brevis dynamics on the west Florida shelf: A look at physical transport and potential zooplankton grazing controls. Cont. Shelf Res. 2008, 28, 112–136. [Google Scholar] [CrossRef]
- Lenes, J.M.; Darrow, B.P.; Walsh, J.J.; Jolliff, J.K.; Chen, F.I.; Weisberg, R.W.; Zheng, L. A 1-D simulation analysis of the development and maintenance of the 2001 red tide of the ichthyotoxic dinoflagellate Karenia brevis on the West Florida shelf. Cont. Shelf Res. 2012, 41, 92–110. [Google Scholar] [CrossRef]
- Cruz, R.C.; Reis Costa, P.; Vinga, S.; Krippahl, L.; Lopes, M.B. A review of recent machine learning advances for forecasting harmful algal blooms and shellfish contamination. J. Mar. Sci. Eng. 2021, 9, 283. [Google Scholar] [CrossRef]
- Lee, J.H.W.; Huang, Y.; Dickman, M.; Jayawardena, A.W. Neural network modelling of coastal algal blooms. Ecol. Model. 2003, 159, 179–201. [Google Scholar] [CrossRef]
- Velo-Suarez, L.; Gutierrez-Estrada, J.C. Artificial neural network approaches to one-step weekly prediction of Dinophysis acuminata blooms in Huelva (Western Andalucıa, Spain). Harmful Algae 2007, 6, 361–371. [Google Scholar] [CrossRef]
- Guallar, C.; Delgado, M.; Diogène, J.; Fernández-Tejedo, M. Artificial neural network approach to population dynamics of harmful algal blooms in Alfacs Bay (NW Mediterranean): Case studies of Karlodinium and Pseudo-nitzschia. Ecol. Model. 2016, 338, 37–50. [Google Scholar] [CrossRef]
- Xie, Z.; Lou, I.; Ung, W.K.; Mok, K.M. Freshwater algal bloom prediction by support vector machine in Macau storage reservoirs. Math. Prob. Eng. 2012, 2012, 397473. [Google Scholar] [CrossRef]
- Shen, J.; Qin, Q.; Wang, Y.; Sisson, M. A data-driven modeling approach for simulating algal blooms in the tidal freshwater of James River in response to nutrient loading. Ecol. Model. 2019, 398, 44–54. [Google Scholar] [CrossRef] [Green Version]
- Gokaraju, B.; Durbha, S.S.; King, R.L.; Younan, N.H. A Machine Learning Based Spatio-Temporal Data Mining Approach for Detection of Harmful Algal Blooms in the Gulf of Mexico. IEEE J. Selected Topics Appl. Earth Observ. Rem. Sens. 2011, 4, 710–720. [Google Scholar] [CrossRef]
- Hill, P.R.; Kumar, A.; Temini, M.; Bull, D.R. HABNet: Machine learning, remote sensing based detection and prediction of harmful algal blooms. IEEE J. Selected Topics Appl. Earth Observ. Rem. Sens. 2019, arXiv:1912.02305. [Google Scholar]
- Florida Fish and Wildlife Conservation Commission. Available online: https://myfwc.com/research/redtide/ (accessed on 24 February 2020).
- Maze, G.; Olascoaga, M.J.; Brand, L. Historical analysis of environmental conditions during Florida red tide. Harmful Algae 2015, 50, 1–7. [Google Scholar] [CrossRef] [Green Version]
- US Water Data for the Nation. Available online: https://waterdata.usgs.gov/nwis (accessed on 24 October 2020).
- University of South Florida Water Institute. Available online: http://www.wateratlas.usf.edu (accessed on 6 February 2020).
- National Data Buoy Center. Available online: https://www.ndbc.noaa.gov/ (accessed on 10 October 2020).
- E.U. Copernicus Marine Service Monitoring Service (CMEMS). Available online: http://marine.copernicus.eu/ (accessed on 6 February 2020).
- Sun, Y.; Wong, A.K.C.; Kamel, M.S. Classification of imbalanced data: A review. Int. J. Pattern Recognit. Artif. Intell. 2009, 23, 687–719. [Google Scholar] [CrossRef]
- Japkowicz, N.; Holte, R. Workshop report: AAAI2000 workshop on learning from imbalanced data-sets. AI Mag. 2000, 22, 127–136. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Fernandez, A.; Garcia, S.; Herrera, F.; Chawla, N.V. SMOTE for learning from imbalanced data: Progress and challenges, marking the 15-year anniversary. J. Art. Intel. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- Haibo, H.; Garcia, E.A. Learning from Imbalanced Data. IEEE Trans. Knowl. Data. Engin. 2009, 21, 1263–1284. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Nello, C.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Basak, D.; Pal, S. Patranabis. Support vector regression. Neural Info. Process Letts Rev. 2007, 11, 203–224. [Google Scholar]
- Vapnik, V. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995. [Google Scholar]
- Boser, B.; Guyon, I.; Vapnik, V. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, COLT ’92, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Schölkopf, B.; Smola, A. Support Vector Machines and Kernel Algorithms. In Encyclopedia of Biostatistics; Armitage, P., Colton, T., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2002; pp. 5328–5335. [Google Scholar]
- Tipping, M.E. Sparse Bayesian Learning and the Relevance Vector Machine. J. Mach. Learn. Res. 2001, 1, 211–244. [Google Scholar] [CrossRef] [Green Version]
- Camps-Valls, G.; Gómez-Chova, L.; Muñoz-Marí, J.; Vila-Francés, J.; Amorós-López, J.; Calpe-Maravilla, J. Retrieval of oceanic chlorophyll concentration with relevance vector machines. Remote Sens. Environ. 2006, 105, 23–33. [Google Scholar] [CrossRef]
- Maron, M.E. Automatic indexing: An experimental inquiry. J. Assoc. Comp. Mach. 1961, 8, 404–417. [Google Scholar] [CrossRef]
- Hand, D.J.; Yu, K. Idiots Bayes—not so stupid after all? Int. Stat. Rev. 2001, 69, 385–398. [Google Scholar]
- Hassoun, M.H. Fundamentals of Artificial Neural Networks; The MIT Press: Massachusetts, MA, USA, 1995; 511p. [Google Scholar]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Werbos, P. Backpropagation through time: What it does and how to do it. Proc. IEEE 1990, 78, 1550–1560. [Google Scholar] [CrossRef] [Green Version]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learn; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Hijmans, R. Raster: Geographic Data Analysis and Modeling. R Package Version 3.0–7. 2017. Available online: https://CRAN.R-project.org/package=raster (accessed on 12 March 2019).
- Calaway, R.; Microsoft Corporation; Weston, S.; Tenenbaum, D. doParallel: Foreach Parallel Adaptor for the ‘Parallel’ Package. 2017. R Package Version 1.0.15. Available online: https://CRAN.R-project.org/package=doParallel (accessed on 12 March 2019).
- Karatzoglou, A.; Smola, A.; Hornik, K.; Zeileis, A. Kernlab. An S4 package for kernel methods in R. J. Stat. Softw. 2004, 11, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Torgo, L. Data Mining Using R: Learning with Case Studies; CRC Press: Boca Raton, FL, USA, 2010; pp. 209–211. [Google Scholar]
- Schnute, J.; Boers, M.; Haigh, R.; Couture-Beil, A.; Chabot, D.; Grandin, C.; Johnson, A.; Wessel, P.; Antonio, F.; Lewin-Koh, N.J.; et al. PBSmapping: Mapping Fisheries Data and Spatial Analysis Tools. R Package Version 2.70.4. 2017. Available online: https://CRAN.R-project.org/package=PBSmapping (accessed on 13 March 2019).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F.; Chang, C.C.; Lin, C.C. e1071: Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien. R Package Version 1.7–2. 2021. Available online: https://CRAN.R-project.org/package=e1071 (accessed on 2 May 2021).
- Fritsch, S.; Guenther, F.; Wright, M.N.; Suling, M.; Mueller, S.M. Training of Neural Networks. R Package Version 1.44.2. 2019. Available online: https://CRAN.R-project.org/package=neuralnet (accessed on 3 October 2019).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. R Package Version 3.3.5. 2020. Available online: https://cran.r-project.org/package=ggplot2 (accessed on 2 January 2021).
- R Core Team. A Language and Environment for Statistical Computing. 2017. R: R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 13 March 2019).
- Anguita, D.; Ghio, A.; Ridella, S.; Sterpi, D. K-Fold cross validation for error rate estimate in support vector machines. In Proceedings of the 2009 International Conference on Data Mining, Miami, FL, USA, 13–16 July 2009; DMIN: Las Vegas, NV, USA, 2009; pp. 1–7. [Google Scholar]
- Cawley, G.C.; Tablot, N.L.C. Fast exact leave-one-out cross validation of sparse least-squared support vector machines. Neural Netw. 2004, 17, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. Roy. Stat. Soc. Ser. B 1974, 36, 111–133. [Google Scholar] [CrossRef]
- Geisser, S. The predictive sample reuse method with applications. J. Amer. Stat. Assoc. 1975, 70, 320–328. [Google Scholar] [CrossRef]
- Bergmeir, C.; Benitez, J.M. On the use of cross-validation for time series predictor evaluation. Inform. Sci. 2012, 191, 192–213. [Google Scholar] [CrossRef]
- Roberts, D.R. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]
- Burman, P.R.; Chow, E.; Nolan, D. A cross-validatory method for dependent data. Biometrika 1994, 81, 351–358. [Google Scholar] [CrossRef]
- Racine, J. Consistent cross-validatory model-selection for dependent data: Hv-block cross-validation. J. Economet. 2000, 99, 39–61. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness & correlation. J. Mach. Learn. Technol. 2011, 2, 37–63. [Google Scholar]
- Platt, J.C. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In Advances in Large Margin Classifiers; Smola, A.J., Bartlett, P., Schölkopf, S., Eds.; MIT Press: Cambridge, MA, USA, 1999; pp. 61–74. [Google Scholar]
- Lin, H.; Lin, C.; Weng, R.C. A note on Platt’s probabilistic outputs for support vector machines. Mach. Learn. 2007, 68, 267–276. [Google Scholar] [CrossRef] [Green Version]
- He, R.; Weisberg, R.H. A Loop Current intrusion case study on the West Florida Shelf. J. Phys. Oceanogr. 2003, 33, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Hadjisolomou, E.; Stefanidis, K.; Herodotou, H.; Michaelides, M.; Papatheodorou, G.; Papastergiadou, E. Modelling freshwater eutrophication with limited limnological data using artificial neural networks. Water 2021, 13, 1590. [Google Scholar] [CrossRef]
- Deng, T.; Chau, K.W.; Duan, H.-F. Machine learning based marine water quality prediction for coastal hydro-environment management. J. Envir. Manag. 2021, 284, 112051. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y. Real-time probabilistic forecasting of river water quality under data missing situation: Depp learning plus post-processing techniques. J. Hydrol. 2020, 589, 125164. [Google Scholar] [CrossRef]
- IPCC (Intergovernmental Panel on Climate Change). Impacts, adaptation, and vulnerability, Summary for Policymakers. In Climate Change; Field, C.B., Barros, V.R., Dokken, D.J., Mach, K.J., Mastrandrea, M.D., Bilir, T.E., Chatterjee, M., Ebi, K.L., Estrada, Y.O., Genova, R.C., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2014; pp. 1–32. [Google Scholar]
- Sillmann, J.; Kharin, V.V.; Zhang, X.; Zwiers, F.W.; Bronaugh, D. Climate extremes indices in the CMIP5 multimodel ensemble: Part 1. Model evaluation in the present climate. J. Geophys. Res. Atmos. 2013, 118, 1716–1733. [Google Scholar] [CrossRef]
- Sillmann, J.; Kharin, V.V.; Zwiers, F.W.; Zhang, X.; Bronaugh, D. Climate extremes indices in the CMIP5 multimodel ensemble: Part 2. Future climate projections. J. Geophys. Res. Atmos. 2013, 118, 2473–2493. [Google Scholar] [CrossRef]
- Russo, S.; Dosio, A.; Graversen, R.G.; Sillmann, J.; Carrao, H.; Dunbar, M.B.; Singleton, A.; Montagna, P.; Barbola, P.; Vogt, J.V. Magnitude of extreme heat waves in present climate and their projection in a warming world. J. Geophys. Res. Atmos. 2014, 119, 12500–12512. [Google Scholar] [CrossRef] [Green Version]








| Model | Performance Metric | k-Fold Cross-Validation (Random Oversampling) | k-Fold Cross-Validation (SMOTE) | Block Cross-Validation (Random Oversampling) | Block Cross-Validation (SMOTE) |
|---|---|---|---|---|---|
| SVM | Accuracy | 0.79 | 0.79 | 0.62 | 0.62 |
| Recall | 0.63 | 0.63 | 0.27 | 0.26 | |
| Precision | 0.64 | 0.65 | 0.32 | 0.32 | |
| F1 | 0.64 | 0.64 | 0.29 | 0.29 | |
| RVM | Accuracy | 0.62 | 0.76 | 0.55 | 0.59 |
| Recall | 0.73 | 0.72 | 0.58 | 0.47 | |
| Precision | 0.42 | 0.58 | 0.35 | 0.35 | |
| F1 | 0.53 | 0.64 | 0.43 | 0.40 | |
| NB | Accuracy | 0.52 | 0.54 | 0.47 | 0.47 |
| Recall | 0.85 | 0.78 | 0.72 | 0.73 | |
| Precision | 0.37 | 0.37 | 0.33 | 0.32 | |
| F1 | 0.52 | 0.50 | 0.45 | 0.45 | |
| ANN | Accuracy | 0.74 | 0.71 | 0.61 | 0.60 |
| Recall | 0.57 | 0.56 | 0.33 | 0.40 | |
| Precision | 0.55 | 0.51 | 0.34 | 0.34 | |
| F1 | 0.56 | 0.53 | 0.34 | 0.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.F.; Glibert, P.M.; Lyubchich, V. Machine Learning Classification Algorithms for Predicting Karenia brevis Blooms on the West Florida Shelf. J. Mar. Sci. Eng. 2021, 9, 999. https://doi.org/10.3390/jmse9090999
Li MF, Glibert PM, Lyubchich V. Machine Learning Classification Algorithms for Predicting Karenia brevis Blooms on the West Florida Shelf. Journal of Marine Science and Engineering. 2021; 9(9):999. https://doi.org/10.3390/jmse9090999
Chicago/Turabian StyleLi, Marvin F., Patricia M. Glibert, and Vyacheslav Lyubchich. 2021. "Machine Learning Classification Algorithms for Predicting Karenia brevis Blooms on the West Florida Shelf" Journal of Marine Science and Engineering 9, no. 9: 999. https://doi.org/10.3390/jmse9090999
APA StyleLi, M. F., Glibert, P. M., & Lyubchich, V. (2021). Machine Learning Classification Algorithms for Predicting Karenia brevis Blooms on the West Florida Shelf. Journal of Marine Science and Engineering, 9(9), 999. https://doi.org/10.3390/jmse9090999







