Abstract
Bioremediation is one of the promising environment-friendly approaches to eliminate oil contamination. However, heavy oil is known to degrade slowly due to its hydrophobicity. Therefore, microorganisms capable of producing biosurfactants are gaining substantial interest because of their potential to alter hydrocarbon properties and thereby speed up the degradation process. In this study, six bacterial consortia were obtained from the oil-spilled beach areas in Miyagi, Japan, and all of which exhibited high potential in degrading heavy oil measured by gas chromatography with flame ionization detector (GC-FID). The polymerase chain reaction—denaturing gradient gel electrophoresis (PCR-DGGE) and next-generation sequencing (NGS) revealed that the diverse microbial community in each consortium changed with subculture and became stable with a few effective microorganisms after 15 generations. The total petroleum hydrocarbons (TPH) degradation ability of the consortia obtained from a former gas station (C1: 81%) and oil refinery company (C6: 79%) was higher than that of the consortia obtained from wastewater treatment plant (WWTP) (C3: 67%, and C5: 73%), indicating that bacteria present in C1 and C6 were historically exposed to petroleum hydrocarbons. Moreover, it was intriguing that the consortium C4, also obtained from WWTP, exhibited high TPH degradation ability (77%). The NGS results revealed that two bacteria, Achromobacter sp. and Ochrobactrum sp., occupied more than 99% of the consortium C4, while no Pseudomonas sp. was found in C4, though this bacterium was observed in other consortia and is also known to be a potential candidate for TPH degradation as reported by previous studies. In addition, the consortium C4 showed high biosurfactant-producing ability among the studied consortia. To date, no study has reported the TPH degradation by the combination of Achromobacter sp. and Ochrobactrum sp.; therefore, the consortium C4 provided an excellent opportunity to study the interaction of and biosurfactant production by these two bacteria during TPH degradation.
1. Introduction
Soil and water pollution caused by petroleum and its derivatives are among the most severe global environmental problems because petroleum hydrocarbons are highly persistent in the environment, toxic, and pose serious health risks to humans [1,2]. In marine areas, the oil spill pollution occurs not only because of anthropogenic activities but also because of natural disasters, such as earthquakes and tsunamis. Currently, petroleum hydrocarbons are recognized as the third most common pollutants in some sea areas [3,4]. Although several physical, chemical, and biological methods are available to remove petroleum hydrocarbons from the contaminated environments, biological processes, also known as bioremediation processes, are considered to be the most exciting, cost-effective, and environmentally friendly approaches for the removal of petroleum hydrocarbons as compared to chemical or physical processes [5,6,7]. However, the mechanism of the biological processes exerts a slow degradation rate, limiting their widespread implementation [8].
Petroleum hydrocarbons like heavy oil are degraded slowly, owing to their extremely hydrophobic properties [9]. As a result, several previous studies suggested using the synthetic chemical surfactants in order to increase the apparent solubility of particular hydrocarbon and to speed the contaminant phase transfer [9,10]. Nevertheless, it is also known that chemical surfactants are nonbiodegradable and, thus, have relatively high potential to cause toxicity to living organisms, hydrocarbon-degrading bacteria, and the surrounding environment. They can also inhibit biodegradation via toxic interaction and sequestration of hydrocarbon into surfactant micelles and can cause a decrease in oxygen uptake rate [11].
Thus, one of the helpful approaches in enhancing the effectiveness of the process is the use of biosurfactant-producing microbes. Several biosurfactants have been identically recognized as the synthetic chemical surfactants that can alter the interfacial properties [12]. Biosurfactants increase the surface area of petroleum hydrocarbons and maintain the uptake inside the bacterial cell surface [13]. In correlation with chemical surfactants, biosurfactants offer several environmentally friendly advantages such as biodegradable, less toxic, and nonhazardous [14]. The most engaging characteristic of biosurfactants is the balance structure at different temperatures, pH, and salinity [11]. Biosurfactants can also be generated from waste materials and thereby produce fewer contaminants and reduce the production cost, making biosurfactants fascinating to further research [15,16].
There have been many studies on the bioremediation of petroleum hydrocarbons by potential biosurfactant-producing microbes. However, most of the studies on biosurfactants-mediated biodegradation focused on the use of a single culture and rarely the mixed culture [17]. Furthermore, in situ biosurfactant production by co-culture or the biosurfactant-producing and hydrocarbon-degrading bacteria is relatively less studied as compared to the research done with the addition of biosurfactant alone [1]. In this study, we investigated the heavy oil degradation ability of six biosurfactant-producing bacterial consortia enriched from previously oil-spilled beach areas in Miyagi, Japan, and compared the microbial community structures of the studied consortia to reveal their potentiality in degradation while producing biosurfactants.
2. Materials and Methods
2.1. Sample Collection and Chemicals
Six soil samples were collected from Miyagi, Japan, in September 2017. Samples 1 and 2 were obtained from a former gas station near the beach of Shiogama, Miyagi, Japan (38°17′14.8″ N, 141°03′42.0″ E). Samples 3, 4, and 5 were collected from three different points near the wastewater treatment plant (WWTP), which is also located on the shoreline beach of Shiogama (38°25′24.1716″ N, 141°18′25.3404″ E). These sampling sites of Shiogama beach areas were severely impacted by oil spills that occurred due to a tsunami caused by the Great East Japan Earthquake, 2011. Sample 6 was collected from an oil refinery company in Miyagi, Japan. Samples 1, 2, and 6 had a soil texture, samples 3 and 4 were sandier, while sample 5 was an asphalt rock. Samples 1 and 3 also had more moisture than the others. Heavy oil (type A) was obtained from a private oil company in Miyagi, Japan, for this study. All the chemicals and solvents were purchased from Wako Pure Chemical Industries, Ltd., Osaka, Japan.
2.2. Enrichment of Bacterial Consortia
The bacterial consortia were enriched inside 100-mL glass vials with 20 mL of mineral salt medium (MSM), which contained 1.8 g K2HPO4, 4.0 g NH4Cl, 0.2 g MgSO4·7H2O, 0.1 g NaCl, and 0.01 g FeSO4·7H2O in 1 L of distilled water [18]. During enrichment, the heavy oil was added directly to MSM without dissolving in an organic solvent at 2.5% volume per volume (v/v) as the sole carbon and energy source. The pH of the medium was adjusted to 7.0. The glass vials were incubated at 30 °C with agitation at 140 rpm. Subculture to the next generation was regularly done with 10% inoculum from the previous generation and incubation for 15 days. The concentration of total petroleum hydrocarbon (TPH) and the microbial community was investigated regularly.
2.3. TPH Degradation Analysis
The TPH extraction from the culture vials was performed by the method reported by Dominguez et al. with a slight modification [19]. The TPH was extracted from 10 mL of liquid culture by an equal volume of hexane. The vials were washed and vortexed twice for 1 min, exposed to ultra-sonication for 2 min, and centrifuged at 6500× g for 2 min (TOMY TX-160). After that, the solvent phase was analyzed by manually injecting 1 (μL) into a GC-4000 gas chromatograph (GL Science Inc., Eindhoven, The Netherlands) equipped with a flame ionization detector (FID) and a specific column InertCap 17 MS column. The column temperature was held at 50 °C for 5 min, then increased to a maximum temperature of 250 °C at the rate of 10 °C/min and held for 16 min. Both the injector and detector temperatures were maintained at 300 °C. Helium was used as the carrier gas. The percentage of TPH degradation in the culture was calculated using the following formula:
The hydrocarbon loss in the blank sample during the sampling period was 5–15%.
2.4. DNA Extraction
Bacterial cells were harvested from 1 mL of liquid culture by centrifugation at 14,000 rpm for 2 min (TOMY TX-160) in 1.5 mL sterile plastic tubes. DNA extraction was carried out by UltraClean®Microbial DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) according to the manufacturer’s instructions. DNA extraction from soil samples was carried out by PowerSoil®DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA). All the operations were done following the protocol provided by the kit instruction manual. The concentration of extracted DNA solution was measured by nanophotometer®C40 (IMPLEN, Munich, Germany), and the quality was checked by agarose gel electrophoresis. The DNA solution was then stored at 4 °C until further use.
2.5. Microbial Community Analyses
2.5.1. Polymerase Chain Reaction—Denaturing Gradient Gel Electrophoresis (PCR-DGGE)
The following primers were used for PCR amplification: 341F-GC (5′ CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GCC TAC GGG AGG CAG CAG-3′), which comprises a GC clamp attached to the 5′ end of the primer, and 518R (5′-ATT ACC GCG GCT GCT GG-3′) as forward and reverse primers, respectively [20]. The PCR amplification was carried out in the ABI 9700 Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) with preheating for 30 s at 94 °C, and each cycle consisting of 30 s at 94 °C, 30 s touchdown temperature from 61 to 56 °C with a temperature decreasing rate of 0.5 °C/cycle, 60 s at 72 °C, and a final extension at 72 °C for 7 min. After PCR, 15 µL of products were loaded onto polyacrylamide gels containing a gradient of 30–70% denaturants. For the 30% gel, the composition of the gel was as follows: 5 mL of 40% acrylamide, 0.4 mL of 50× Tris-acetate-EDTA (TAE) buffer, 2.4 mL of formamide (deionized), and 2.52 g of urea, finally up to 20 mL by distilled H2O (dH2O). For the 70% gel, the composition of the gel was as follows: 5 mL of 40% acrylamide, 0.4 mL of 50× TAE buffer, 5.6 mL of formamide (deionized), and 5.88 g of urea, finally up to 20 mL by dH2O.
DGGE was achieved using the DCodeTM Universal Mutation Detection System (BIO RAD Laboratories, Hercules, CA, USA). Then DGGE gel electrophoresis was performed at 70 V, 60 milliamperes (mA), and 60 °C for 16 h. The gels were stained with ethidium bromide and photographed under UV illumination. A sterile scalpel was used to cut the target bands from the gel and purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The products then were amplified for cycle sequence reaction under the conditions: 96 °C for 1 min, 25 cycles of denaturation at 96 °C for 10 s, annealing at 50 °C for 10 s, elongation at 72 °C for 2 min, and a final extension at 72 °C for 5 min. The amplicons were purified with Centri-Sep and sequenced using BigDyeTM Terminator v3.1 chemistry in 3130 Genetic Analyzers (Applied Biosystems Inc., Waltham, MA, USA), and finally, the sequences were compared in the GenBank database using Basic Local Alignment Search Tool (BLAST) to identify the closest relatives.
2.5.2. Next-Generation Sequencing (NGS)
One nanogram of extracted DNA was used as a template to amplify the V4 region of 16S ribosomal DNA gene of bacteria and Archaea by primer 515F_MIX (5′-ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT-NNNNN-GTG CCA GCM GCC GCG GTA A-3′) and 806R_MIX (5′-GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T-NNNNN-GGA CTA CHV GGG TWT CTA AT-3′). The PCR conditions were as follows: 94 °C for 1 min, followed by 25 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, elongation at 72 °C for 30 s, and a final extension at 72 °C for 5 min [21]. The PCR amplicons were then sent to the Bioengineering Lab. Co. Ltd. (Seibutsu Giken Inc., Atsugi, Japan), where the amplicons were sequenced by the Miseq system (Illumina, Inc. CA, USA) using 2 × 300 base pair (bp), paired-end reads. Only the sequences containing completed primer sequences were selected for further analysis by platform Qiime2. After removing primer sequences, the selected sequences were applied to database Greengene (Ver.13_8), which represents 97% of the operational taxonomic unit (OTUs) similarity of all bacteria and archaea.
2.6. Real-Time qPCR Analysis
Real-time PCR assays were performed using a CFX ConnectTM Real-Time PCR Analysis System (Bio-Rad, Hercules, CA, USA) with 96-well plates to quantify the 16S ribosomal RNA (rRNA) gene. The 15-µL reaction mixture contained 2 µL template DNA (<100 ng), 7.5 µL TB GreenTM Premix Ex TaqTM (Tli RNaseH Plus) (Takara Bio Inc., Kusatsu, Japan), 0.3 µL (10 micromolar (µM) each of primer 341F and 518R (Eurofins Genomics Co., Ltd., Yokohama, Japan), and 4.9 µL milli-Q. The cycle conditions were: 30 s at 95 °C for enzyme activation and 40 cycles of 5 s at 95 °C, 30 s at 57 °C, and 30 s at 72 °C. After real-time PCR, a melt curve was plotted during heating from 55 to 95 °C at a transition rate of 0.5 °C/s. The standard curve was obtained from the amplification of serial dilution of 109 copies of DNA/mL, which was inserted into a plasmid of chemically competent TOP10 E. coli cells (Thermo Fisher Scientific, Waltham, MA, USA). The standard was prepared from 1 × 102 − 1 × 107 copy number/mL with Milli-Q as the negative control. All the amplicons were measured in duplicate and the correlation coefficient was around 0.99.
2.7. Biosurfactants’ Measurements
2.7.1. Qualitative Assay
Parafilm M Test
Parafilm M test was conducted by transferring 10 uL of the cell-free culture on the hydrophobic surface of the parafilm. The shape of the drop was observed after 1 min and the diameter of the droplets was evaluated [22]. MSM was used as a negative control.
Emulsification Index E24
The emulsifying potential of biosurfactants was measured by emulsion index E24 [23,24]. For this study, hexadecane and the heavy oil samples were added to the cell-free culture. The mixture was vortexed at high speed for 2 min. After 24 h, E24 was calculated as the ratio of the height of the emulsion layer and the total height of the liquid.
Oil-Spreading Assay
Oil-spreading assay, also called oil displacement activity assay, was performed by adding 10 µL of heavy oil to the surface of distilled water in a Petri dish to form a thin oil layer. Later, 10 µL of cell-free culture was lightly placed on the center of the oil layer. The clear zone was observed if the biosurfactant was present and, subsequently, the diameter of the clear zone was measured [23].
2.7.2. Surface Tension Measurement
The capillary rise method with Dyne Gauge DG-1 from Surfgauge Instrument was used to conduct surface tension measuring test. The measurement was performed in triplicate.
2.7.3. Statistic Analysis
The level of significance was obtained using the student’s t-test. The correlation was tested using Pearson’s correlation coefficient. Principal component analysis (PCA) biplot using BioVinci software by BioTuring was also performed.
3. Results
3.1. Comparison of Microbial Community Analysis
3.1.1. Microbial Community Analysis by PCR-DGGE
The PCR-DGGE was performed to investigate the microbial community shift from the initial generation and after ten times subculture. As identified from 200 bp nucleotides, the excised bands in Figure 1 had close similarities to (A–C) Pseudomonas sp., (D) Acinetobacter sp., (E) Rhodanobacter sp., Dyella sp., Luteibacter sp., (F) Pseudomonas sp., and (G) Achromobacter sp. The obtained DNA bands from DGGE depicted that the microbial community in all consortia changed after the 10th subculture to adapt and acclimate with the composition of the heavy oil. Along with the alteration of the microbial community structures, the degradation ability of each consortium also changed.
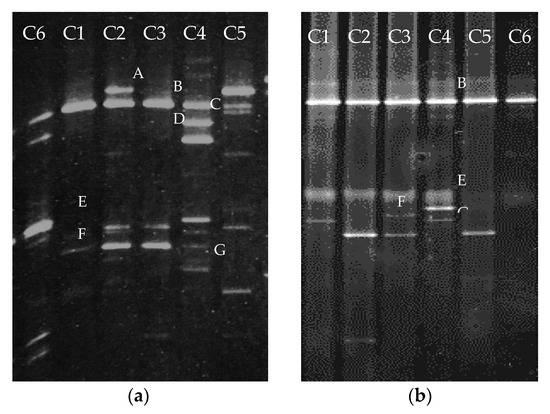
Figure 1.
Comparison of PCR-DGGE profile of six different bacterial consortia in (a) initial generation and (b) after 10th generation.
3.1.2. Microbial Community Analysis by NGS
In comparison, high-throughput analysis with NGS was done to obtain deeper sequencing and detect novel or rare variants. Figure 2a showed that the bacteria community in the consortium C1 was dominated by Pseudomonadaceae in the beginning (46.64%) and also at the end of the 15th generation (87.46%). From 11 OTUs of Pseudomonadaceae, the dominant was identified as Pseudomonas nitroreducens, P. citronellolis, and P. aeruginosa. The other identified bacteria (with percentage > 3%) in the original sample were Pseudonocardia halophobica, Achromobacter sp., Caulobacteraceae sp., Rhodanobacter sp., Ochrobactrum sp., Nocardiaceae, Rhodococcus sp., Spingomonas sp., and Chelatococus asccharovorans. With time, Pseudomonas sp., Stenotrophomonas sp., and Achromobacter sp. became the predominant species after the 15th subculture.

Figure 2.
NGS profile of the consortia (a) Consortium C1 and (b) Consortium C2 enriched from a former gas station dominated by Pseudomonadeceae, Xanthomonadaceae, Alcaligenaceae, and Moxarelaceae after the 15th generation.
On the other hand, the consortium C2 was not only dominated by Pseudomonas sp. and Achromobacter sp., but also by Dyella jiangningensis. From 84 OTUs, C2 became stable with 29 OTUs. However, the consortium C2 presented a more diverse bacterial community than the consortium C1. The consortia C3, C4, and C5, obtained from soil nearby WWTP, also evinced particular bacterial composition, as shown in Figure 3a–c. According to Figure 3, Pseudomonadaceae, Xanthomonadacea, and Moxarelaceae dominated the consortia C3, C4, and C5 in the beginning. After the 15th subculture, C3 and C5 showed almost the same patterns, while the C3 was dominated with 50.54% of Pseudomonas sp. and 28.47% of Rhodanobacter sp., and the C5 was practically dominated by Pseudomonas sp. up to 79.79%. Achromobacter sp. from Alcaligenes family and Ochrobactrum sp. from Brucellaceae family only gave a small portion of around 4% to both the 15th generations of C3 and C5.
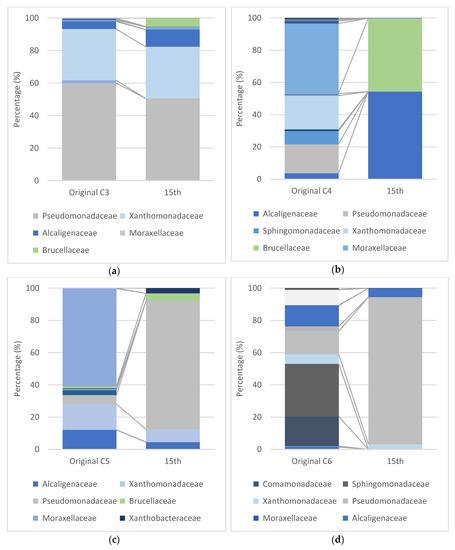
Figure 3.
NGS profile of the consortia (a) C3, (b) C4, and (c) C5 enriched from WWTP near the beach; and (d) C6 from an oil refinery company. The predominant family was Pseudomonadeceae, Xanthomonadaceae, Alcaligenaceae, Moxarelaceae, and Brucellaceae.
Surprisingly, the shift in microbial community structure during the biodegradation of heavy oil by the consortium C4 was found to be different when compared with that of C3 and C5. In the beginning, Gammaproteobacteria (Pseudomonadaceae, Xanthomonadacea, and Moxarelaceae) was the predominant class, up to 82.16%, leading Acinetobacteria, Alphaproteobacteria, and Betaproteobacteria, which were just about 1.66%, 10.16%, and 6.66%, respectively. Nevertheless, after the 15th subculture, the number of Alphaproteobacteria (Brucellaceae) and Betaproteobacteria (Alcaligenes) increased significantly, up to 45.51% and 54.33%, and left no Gammaproteaobacteria in the consortium, contrasting with the other consortia which were dominated by Pseudomonadaceae from Gammaproteobacteria. The Alphaproteobacteria (Brucellaceae) obtained from OTU can be cultured independently and identified by 16s rRNA as Ochrobactrum sp., while culturable Betaproteobacteria (Alcaligenaceae) identified as Achromobacter sp.
The consortium C6 in Figure 3d exhibited similar patterns with the consortium C1, wherein the amount of family Pseudomonadaceae, Alcaligenacea, and Xanthomonadaceae dominated the cultures after the 15th generation. Intriguingly, Sphingomonadaceae and Comamonadaceae in the original sample were eliminated in stable consortia with the heavy oil as carbon source.
3.2. TPH Degradation and Biosurfactant Production
The degradation of the TPH was shown in Figure 4, which indicated that the hydrocarbon could be degraded up to 75–80%. According to the t-test with a 90% confidence interval, the percentage of TPH degradation between C1, C2, C4, and C6 was not significantly different, as shown in Figure 5. It was also revealed from Figure 5 that the consortium C1 exhibited better ability in degrading TPH up to 81%, as compared to consortia C2, C3, C4, C5, and C6, which showed the TPH degradability of up to 73%, 67%, 77%, 73%, and 79%, respectively. The consortia C1, C2, C4, and C6, showed a statistically significant difference in their TPH degradability in comparison with the consortia C3 and C5 (p < 0.1). The rate of degradation can be seen in detail in Figure S1, where almost all of the consortia showed no significant difference in the biodegradation rate. However, the consortium C1 removed the hydrocarbon at a rate of 5.38%/day, which was faster than the degradation rates achieved by other consortia C2 (5.02%/day), C3 (4.47%/day), C4 (5.12%/day), C5 (4.47%/day), and C6 (5.25%/day).
Figure 4.
The representative GC-FID profile for (a) initial sample and (b) sample after 15 days of incubation.
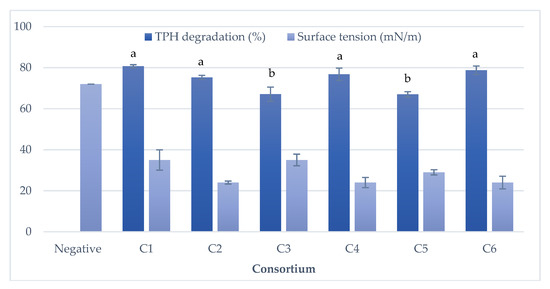
Figure 5.
Percentage of TPH degradation and surface tension reduction by the 15th generation of six consortia (After 15 days of incubation at 30 °C and 140 rpm).
The consortium C1, C4, and C6 showed a better ability to reduce surface tension from 72 millinewtons per meter (mN/m) to 24 mN/m. This can also be demonstrated from the alteration of the appearance of liquid culture, which became more turbid than the liquid cultures of other consortia in the beginning of subculture (until the 10th generation). At the end of the 15th generation, only C4 was found with constant turbidity.
The biosurfactant-producing ability of different consortia by reducing surface tension was presented in Figure 5 and Table 1. As compared to the other consortia, C2 showed a higher ability to produce biosurfactants according to the turbidity of liquid culture from the beginning until the fifth generation, as can be seen in Figure S2.

Table 1.
Qualitative analysis of biosurfactants among the consortia.
4. Discussion
In this study, different approaches were used for a better understanding of the microbial community shifting as well as to distinguish the effectiveness of using PCR-DGGE and high-throughput sequencing with NGS. Recently, with the advancements in molecular technology, it has become easier to understand the profile of microbial communities in the environment [25]. After years, researchers found the explanation and perception, according to PCR-DGGE bands, that may lead to some problems. The bands on a DGGE have a possibility not to represent true bacterial amplicons but rather the form of chimeric [26]. Another problem is that DNA in the DGGE band sometimes could not be sequenced after recovery from the gels. On the other hand, even the cutout DNA from the DGGE gel shows a single band, it may result in several different references in GenBank. Other researchers also reported that one isolate produced multiband, which did not correlate [27]. Due to its limitation, NGS was provided to study the consortium, learn the interaction inside it, and discover novel or rare variants with deep sequencing.
For developing efficient bioremediation technology, characterization of the diversity of the indigenous microbial communities, their metabolic potential, and the response of community composition is of paramount importance [28]. In some research, single bacteria have been reported to have broad-spectrum in degrading hydrocarbon, both monoaromatic and polyaromatic, but almost no bacterium has been identified that can degrade the entire petroleum hydrocarbon fraction [29]. Even from the hydrocarbon-utilizing bacteria that are continually exposed to oil contamination, no single bacterium has been reported yet that is capable of degrading all the oil components [30].
In constructing microbial consortium, there are two different approaches, called top-down and bottom-up ways. The top-down approach depends on the development of consortium from several samples and lets the adapted strain survive, and the other is eliminated. In the other way, the bottom-up approach depends on the immensely stable and efficient consortium, which has different metabolic pathways [31]. In this study, both approaches were applied through subculturing the consortia regularly. By using heavy oil as the only carbon source, diverse bacteria from broad taxonomic groups with distinct capabilities were enriched. However, some of the bacteria were eliminated because the heavy oil contained more aliphatic carbon chain than mono/polycyclic aromatic hydrocarbon (PAH).
The enrichment step in this study was not to enhance and increase the number of bacteria but to obtain stable consortium, which can degrade the heavy oil. Because of the composition of the heavy oil, some of the indigenous species that were abundant in the original microbial community might be eliminated. For example, the potentially predominant Spingomonodaceae, which is known to have the ability to degrade a wide range of PAH compounds [32], was excluded from the consortium C6 and also Pseudomonadaceae from C4. A faster biodegradation rate was demonstrated by C1 and C6, compared to the other consortia. The consortium C6 was expected to give the highest TPH degradability with a faster degradation rate because it came from an oil refinery company, followed by C1 and C2, as obtained from a former gas station.
The Pseudomonadaceae was present in all the original consortium samples. Among the OTUs, the most abundant family of Pseudomonadaceae was OTU_001, which was identified as Pseudomonas aeruginosa. The P. aeruginosa was already known as a player in biodegradation of many substrates such as glycerol [33], diesel oil [34], water column-contaminated oil [35], polluted coastal area [36,37], PAH [38], and more.
The Gammaproteobacteria (in this study, Pseudomonadaceae, Xanthomonadacea, and Moxarelaceae) appeared in the original sample. However, after the 15th subculture, they still grew in all consortia, except in C4. Those Gammaproteobacteria were regularly discovered in the oil-impacted marine environments and were found competent in degrading oil [39]. They were also explored for their ability to degrade low molecular weight alkanes, aromatics, BTEX (benzene, toluene, ethylbenzene and xylenes), proline, catechol, cyclohexanone, and nitroalkanes. However, no Gammaproteobacteria was found in the consortium C4, where the impressive microbial shifting was observed.
The results of NGS showed that Achromobacter sp. and Ochrobactrum sp. were predominantly abundant (99%) in the consortium C4. Achromobacter sp. can be found both in soil and water (fresh and marine). The halotolerant Achromobacter sp. isolated from the marine environment was reported to have the ability to degrade both aliphatic hydrocarbons and PAHs while producing biosurfactants by utilizing citric acid as the carbon source. Even the pathways for alkane degradation had not been entirely resolved in its systematical genome analysis [40,41,42].
The Ochrobactrum intermedium, which was acknowledged as an opportunistic pathogen in humans, rarely isolated from an oil field, lately was being studied to give a better understanding of the mechanism on how it adapts to the environment [43]. It was also reported to be isolated from waste lubricants and could metabolize linear n-alkanes C12–C28, phenanthrene, anthracene [44], and benzo[a]pyrene [45]. Ochrobactrum sp. had also been isolated from marine sediments in China and was reported to have the ability to degrade benzo[a]pyrene, phenanthrene, pyrene, and fluoranthene [45]. Both of the functional gene alkB genes and P450 genes were detected in the genome of O. intermedium strain 2745-2, which made the isolate capable of utilizing tetradecane and hexadecane as the main compound of crude oil [43].
In some research, Achromobacter sp., Pseudomonas sp., and Ochrobactrum sp. were found as the dominant genera in oil spilled-contaminated sites from Japan and Vietnam and were reported to have the ability to eliminate 50% and 25% of the n-alkanes and PAHs, respectively [46]. Marine microorganisms generated exclusive metabolites such as biosurfactants. Nonetheless, such surface-active compounds have not been explored because of the adversity correlated with the isolation and growth of marine microorganisms under laboratory conditions [47]. Some researchers have applied biosurfactants of indigenous marine microorganisms for environmental applications such as bioremediation process and microbial-enhanced oil recovery (MEOR) [48]. Due to their widespread demand in industrial and environmental applications, a number of economical biosurfactants have already been registered as patents. Some of the patent-registered biosurfactants were cultivated from Pseudomonas sp., Bacillus sp., Candida sp., and sophorolipid producer [13]. Moreover, other well-known biosurfactant producers are Mycobacterium sp., Nocardia sp., Corynebacterium sp., Rhodococcus sp., Arthrobacter sp., Acinetobacter sp., Thiobacillus sp., Alcanivorax sp., Serratia sp., Spingomonas sp., and Burkholderia sp. [13,14,48], but very rarely the Achromobacter sp. and Ochrobactrum sp.
The efficiency of biosurfactants can be measured by a property called critical micelle concentration (CMC). A low CMC, which means only fewer biosurfactants are required to decrease the surface tension, signifies the biosurfactants as more efficient ones [49]. Based on the molecular weight, biosurfactants are divided into high-molecular-mass (HMM) and low-molecular-mass (LMM) surfactants; the latter one is more efficient in lowering the surface and interfacial tensions. On the other hand, the HMM biosurfactants are more efficient at stabilizing oil-in-water emulsion [13]. The presence of biosurfactants in enhancing hydrocarbon biodegradation occurs by three various pathways, including mobilization, solubilization, and emulsification. If the concentration of LMM biosurfactants is below the CMC, the mechanism of oil removal is mobilization. On the contrary, if the concentration of LMM biosurfactant is above the CMC, solubilization occurs. Meanwhile, the emulsification exists if the biosurfactants have HMM property [50].
From Table 1, all consortia showed a distinct ability to produce biosurfactants as conforming to some indirect assay for screening the interfacial activity of the biosurfactants. Principal component analysis (PCA) biplot was used to analyze five variables from Figure 5 (% TPH degradation and surface tension) and Table 1 (parafilm test, oil-spreading assay, and E24). From the PCA, five principal components, specific direction, and the angle between each variable were analyzed, as shown in Figure 6, and the detailed statistical summary of PCA biplot was presented in Table S1. Those variables were transformed into two. The most significant equation refers to variance, where 77.50% was interpreted from the original data. PC1 was dominated by % TPH degradation, surface tension, parafilm test, and oil-spreading assay with a 52.71% proportion of variance. Meanwhile, E24 only contributed 24.79% to PC1 and was dominant to PC2.

Figure 6.
Principal component analysis (PCA) biplot depicting the relationship between the percentage of TPH degradation by six different consortia, surface tension reduction, and some qualitative and indirect assay for biosurfactant measurements. Data visualization analyzed with software BioVinci by BioTuring. Note: Solid circles represented the six consortia. Arrows indicate the direction variables associated with the consortia.
Therefore, according to PC1 and PC2 in Figure 6, the degradation of TPH revealed a strong positive correlation with the parafilm test screening, while a moderate positive correlation was observed with the oil-spreading assay. On the other hand, the consortia C1, C2, C4, and C6 showed close cluster distribution and no significant difference in their ability to reduce the TPH (p < 0.1), explaining that the stability of drops was dependent on the surface tension.
Nevertheless, the biosurfactant screening with the E24 assay presented approximately no correlation with the TPH degradation or surface tension of the consortia. The E24 assay is a simple screening method suitable for the first screening of biosurfactant-producing microbes. However, surface activity and emulsification capacity do not always correlate [23]. The results of three indirect qualitative assays explained that the biosurfactant properties of each consortium were different. All the consortia had a distinct mechanism in TPH biodegradation. Furthermore, Figure 6 also presented a robust significant negative relationship between TPH degradation and surface tension reduction, r (16) = 0.79, p < 0.01, where the lower alteration of surface tension caused the higher percentage of TPH degradation.
The circumstances inside consortia conceivably explained the hypothesis that the presence of surfactant molecules has a high possibility to promote alteration, not only to hydrocarbon characteristics but also in the microbial community, which correlated to different degradation patterns [14]. Also, the existence of biosurfactants in the consortium C4 demands more verification about how the biosurfactants affect the other microorganisms because only two stable, dominant bacteria grew in the consortium C4. Santos et al. [14] also reported that the biosurfactant rhamnolipids has the potential to exert toxicity to natural vegetation but also reduce the toxicity of specific compounds by altering the solubilization. Additionally, rhamnolipids are not only biodegradable but also can be co-degraded or utilized as a carbon and energy source by numerous mono-cultures.
There were several studies on the potential consortia enriched from various sites to remediate various contaminants. Comprehensively, after frequently subculturing the six different consortia obtained from different sampling sites, the study found the consortium C4 as the most interesting one, which was not known to be historically exposed to petroleum hydrocarbons. The consortium C4 was obtained from the WWTP, which was struck by a tsunami caused by Great East Japan Earthquake, 2011, and accidentally exposed by an oil spill from factories, including a 5000-barrel/day-capacity oil refinery around Shiogama beach, Miyagi, Japan [51,52]. As compared to the consortia C1 and C6 that were supposed to get exposed to the petroleum hydrocarbons due to the presence of gas station and oil refinery company, the consortium C4 demonstrated a fascinating finding. This ecological resilience showed that the indigenous consortia, from the previously infected area, still had the ability to utilize petroleum hydrocarbons. However, it can also be stated that the consortium with no previous exposure to petroleum hydrocarbons can also degrade heavy oil.
The combination of Achromobacter sp. and Ochrobactrum sp. in mixed culture has been rarely studied. Only one report has been found in the literature that demonstrated the ability of those mixed bacteria to degrade organophosphonates [53]. To the best of our knowledge, it is the first report that revealed the coexistence of these two bacteria, Achromobacter sp. and Ochrobactrum sp., while playing a vital role in degrading heavy oil and its derivates as well as producing the biosurfactant. Moreover, further research is suggested to identify the type of biosurfactants they produce and the underlying mechanism responsible for eliminating other bacteria from the consortium.
5. Conclusions
The majority of the previous studies on both oil-degrading and biosurfactants-producing bacteria were carried out by mono-culture and occasionally by a microbial consortium. In our study, six stable, heavy oil-degrading bacterial consortia were constructed from three different sampling sites. The consortium C1 showed better ability in degrading TPH (81%) while producing biosurfactants. Interestingly, all of the consortia consisted of Pseudomonadaceae, except for C4. After the 15th generation, the consortium C4 was found to undergo significant microbial shifting, which resulted in two predominant bacteria, Achromobacter sp. and Ochrobactrum sp., that efficiently removed about 77% of the TPH and lowered the surface tension to 24 mN/m. A few studies on the hydrocarbon-degrading and biosurfactants-producing ability of these two bacteria in mixed culture is available in the existing literature. Therefore, our study provided new scientific information in this regard. However, further study will be continuing to understand the property of biosurfactants produced from C4, the hydrocarbon used preferentially, and the factors influencing the biosurfactants production.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-1312/8/8/577/s1, Figure S1: Percentage of TPH degradation, 16s rRNA gene copy/mL of culture, and surface tension reduction of consortia (a) C1, (b) C2, (c) C3, (d) C4, (e) C5, and (f) C6; of the 15th generation, Figure S2: The turbidity of culture among consortia C1, C2, C3, C4, C5, and C6 at the (a) 5th, (b) 10th, and (c) 15th generation, Table S1: Statistical summary of principal component analysis (PCA) biplot of Figure 6.
Author Contributions
Conceptualization, C.I. and S.P.; methodology, S.P.; software, S.P.; validation, S.P.; formal analysis, S.P.; investigation, S.P.; resources, S.P.; data curation, S.P.; writing—original draft preparation, S.P.; writing—review and editing, S.P., C.I., and M.-F.C.; visualization, S.P.; supervision, C.I. and M.-F.C.; project administration, C.I.; funding acquisition, S.P., C.I., and M.-F.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Indonesia Endowment Fund for Education/ Lembaga Pengelola Dana Pendidikan (LPDP) Indonesia and Laboratory of Geoenvironmental Remediation, Tohoku University, Japan.
Acknowledgments
The authors thank anonymous reviewers for constructive comments. The authors also appreciate the support from Tohoku University Center for Gender Equality Promotion (TUMUG) Support Project (Project to Promote Gender Equality and Female Researchers).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mnif, I.; Sahnoun, R.; Ellouz-Chaabouni, S.; Ghribi, D. Application of bacterial biosurfactants for enhanced removal and biodegradation of diesel oil in soil using a newly isolated consortium. Process Saf. Environ. Prot. 2017, 109, 72–81. [Google Scholar] [CrossRef]
- Ismail, W.; Al-Rowaihi, I.S.; Al-Humam, A.A.; Hamza, R.Y.; Nayal, A.M.; Bououdina, M. Characterization of a lipopeptide biosurfactant produced by a crude-oil-emulsifying Bacillus sp. I-15. Int. Biodeterior. Biodegrad. 2013, 84, 168–178. [Google Scholar] [CrossRef]
- Nishino, T.; Takagi, Y. Numerical analysis of tsunami-triggered oil spill fires from petrochemical industrial complexes in Osaka Bay, Japan, for thermal radiation hazard assessment. Int. J. Disaster Risk Reduct. 2020, 42. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Guo, J.; Lv, Y.; Li, Y. Biodegradation of marine oil spill residues using aboriginal bacterial consortium based on Penglai 19-3 oil spill accident. Ecotoxicol. Environ. Saf. 2018, 159, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.K. Bioremediation: A green approach for restoration of polluted ecosystems. Environ. Sustain. 2018, 1, 305–307. [Google Scholar] [CrossRef]
- Azubuike, C.C.; Chikere, C.B.; Okpokwasili, G.C. Bioremediation techniques–classification based on site of application: Principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 2016, 32. [Google Scholar] [CrossRef]
- Cui, J.-Q.; He, Q.-S.; Liu, M.-H.; Chen, H.; Sun, M.-B.; Wen, J.-P. Comparative study on different remediation strategies applied in petroleum-contaminated soils. Int. J. Environ. Res. Public Health 2020, 17, 1606. [Google Scholar] [CrossRef]
- Patowary, K.; Patowary, R.; Kalita, M.C.; Deka, S. Development of an efficient bacterial consortium for the potential remediation of hydrocarbons from contaminated sites. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Abdel-Moghny, T.; Mohamed, R.S.A.; El-Sayed, E.; Aly, S.M.; Snousy, M.G. Removing of hydrocarbon contaminated soil via air flushing enhanced by surfactant. Appl. Petrochem. Res. 2012, 2, 51–59. [Google Scholar] [CrossRef]
- Christofi, N.; Ivshina, I.B. Microbial surfactants and their use in field studies of soil remediation. J. Appl. Microbiol. 2020, 93, 915–929. [Google Scholar] [CrossRef]
- Mohanty, S.; Jasmine, J.; Mukherji, S. Practical considerations and challenges involved in surfactant enhanced bioremediation of oil. BioMed Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Park, T.; Jeon, M.K.; Yoon, S.; Lee, K.S.; Kwon, T.H. Modification of interfacial tension and wettability in oil–brine–quartz system by in situ bacterial biosurfactant production at reservoir conditions: Implications for microbial enhanced oil recovery. Energy Fuels 2019, 33, 4909–4920. [Google Scholar] [CrossRef]
- Pacwa-Płociniczak, M.; Płaza, G.A.; Piotrowska-Seget, Z.; Cameotra, S.S. Environmental applications of biosurfactants: Recent advances. Int. J. Mol. Sci. 2011, 12, 633–654. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.K.F.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional biomolecules of the 21st century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.G.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Applications of biosurfactants in the petroleum industry and the remediation of oil spills. Int. J. Mol. Sci. 2014, 15, 12523–12542. [Google Scholar]
- Makkar, R.S.; Cameotra, S.S.; Banat, I.M. Advances in utilization of renewable substrates for biosurfactant production. AMB Express 2011, 1. [Google Scholar] [CrossRef]
- Carmen, R.; Alessandro, C.; Luigi, M.; Emilio, D.; Carlos, R.; Antonio, C.; Angelina, G. Efficiency in hydrocarbon degradation and biosurfactant production by Joostella sp. A8 when grown in pure culture and consortia. J. Environ. Sci. 2018, 67, 115–126. [Google Scholar]
- Zajic, J.E.; Supplisson, B. Emulsification and degradation of ‘Bunker C’ fuel oil by microorganisms. Biotechnol. Bioeng. 1972, 14, 331–343. [Google Scholar] [CrossRef]
- Dominguez, J.J.A.; Inoue, C.; Chien, M.F. Hydroponic approach to assess rhizodegradation by sudangrass (Sorghum x drummondii) reveals pH- and plant age-dependent variability in bacterial degradation of polycyclic aromatic hydrocarbons (PAHs). J. Hazard. Mater. 2020, 387. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Caporaso, J.G. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Youssef, N.H.; Duncan, K.E.; Nagle, D.P.; Savage, K.N.; Knapp, R.M.; McInerney, M.J. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods 2004, 56, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Walter, V.; Syldatk, C.; Hausmann, R. Screening concepts for the isolation of biosurfactant producing microorganisms. In Biosurfactants. Advances in Experimental Medicine and Biology; Sen, R., Ed.; Springer: New York, NY, USA, 2010; Volume 672, pp. 1–13. [Google Scholar]
- Peele, K.A.; Kodali, V.P. Emulsifying activity of a biosurfactant produced by a marine bacterium. 3 Biotech 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Grace, L.; Chang, T.C.; Whang, L.M.; Kao, C.H.; Pan, P.T.; Cheng, S.S. Bioremediation of petroleum hydrocarbon contaminated soil: Effects of strategies and microbial community shift. Int. Biodeterior. Biodegrad. 2011, 65, 1119–1127. [Google Scholar] [CrossRef]
- Paulson, D.S. Applied Biomedical Microbiology: A Biofilms Approach; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Sekiguchi, H.; Tomioka, N.; Nakahara, T.; Uchiyama, H. A single band does not always represent single bacterial strains in denaturing gradient gel electrophoresis analysis. Biotechnol. Lett. 2001, 23, 1205–1208. [Google Scholar] [CrossRef]
- Jayeeta, S.; Sufia, K.; Abhishek, G.; Avishek, D.; Balaram, M.; Ajoy, R.; Paramita, B.; Adinpunya, M.; Pinaki, S. Biostimulation of indigenous microbial community for bioremediation of petroleum refinery sludge. Front. Microbiol. 2016, 7, 1407. [Google Scholar] [CrossRef]
- Xu, X. Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef]
- Venosa, A.D.; Zhu, X. Biodegradation of crude oil contaminating marine shorelines and freshwater wetlands. Spill Sci. Technol. Bull. 2003, 8, 163–178. [Google Scholar] [CrossRef]
- Ibrar, H.; Zhang, H. Construction of a hydrocarbon-degrading consortium and characterization of two new lipopeptides biosurfactants. Sci. Total Environ. 2020, 714, 136400. [Google Scholar] [CrossRef]
- Kertesz, M.A.; Kawasaki, A. Hydrocarbon-degrading sphingomonads: Sphingomonas, sphingobium, novosphingobium, and sphingopyxis. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1693–1705. [Google Scholar]
- Zhang, G.; Wu, Y.; Qian, X.; Meng, Q. Biodegradation of crude oil by Pseudomonas aeruginosa in the presence of rhamnolipids. J. Zhejiang Univ. Sci. B 2005, 6, 725–730. [Google Scholar] [CrossRef]
- Ueno, A.; Hasanuzzaman, M.; Yumoto, I.; Okuyama, H. Verification of degradation of n-alkanes in diesel oil by pseudomonas aeruginosa strain watg in soil microcosms. Curr. Microbiol. 2006, 52, 182–185. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muriel-Millán, L.F. Functional and genomic characterization of a pseudomonas aeruginosa strain isolated from the southwestern gulf of Mexico reveals an enhanced adaptation for long-chain alkane degradation. Front. Mar. Sci. 2019, 6, 572. [Google Scholar] [CrossRef]
- Chaerun, S.K.; Tazaki, K.; Asada, R.; Kogure, K. Bioremediation of coastal areas 5 years after the Nakhodka oil spill in the Sea of Japan: Isolation and characterization of hydrocarbon-degrading bacteria. Environ. Int. 2004, 30, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.H.; Ahsan, M.; Yoshizawa, S.; Hosoya, S.; Yokota, A.; Kogure, K. Multilocus sequence typing and phylogenetic analyses of Pseudomonas aeruginosa Isolates from the ocean. Appl. Environ. Microbiol. 2008, 74, 6194–6205. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): A review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef]
- John, M.; Stephen, T.; Julian, F.; Nagissa, M.; Dominique, J.; Jiang, L.; Scott, O.; Eric, A.; Adolfo, F.; Piero, G.; et al. Oil hydrocarbon degradation by caspian sea microbial communities. Front. Microbiol. 2019, 10, 995. [Google Scholar] [CrossRef]
- Yue-Hui, H.; Cong-Cong, Y.; Qian-Zhi, Z.; Xiao-Ying, W.; Jian-Ping, Y.; Juan, P.; Hailin, D.; Jiang-Hai, W. Genome sequencing reveals the potential of Achromobacter sp. hz01 for bioremediation. Front. Microbiol. 2017, 8, 1507. [Google Scholar] [CrossRef]
- Dave, B.P.; Ghevariya, C.M.; Bhatt, J.K.; Dudhagara, D.R.; Rajpara, R.K. Enhanced biodegradation of total polycyclic aromatic hydrocarbons (TPAHs) by marine halotolerant Achromobacter xylosoxidans using Triton X-100 and β-cyclodextrin--a microcosm approach. Mar. Pollut. Bull. 2014, 79, 123–129. [Google Scholar] [CrossRef]
- Deng, M.-C. Characterization of a novel biosurfactant produced by marine hydrocarbon-degrading bacterium Achromobacter sp. HZ01. J. Appl. Microbiol. 2016, 120, 889–899. [Google Scholar] [CrossRef]
- CHAI, L. Isolation and characterization of a crude oil degrading bacteria from formation water: Comparative genomic analysis of environmental Ochrobactrum intermedium isolate versus clinical strains. J. Zhejiang Univ. Sci. B 2015, 16, 865–874. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Biswas, D.; Sana, S.; Datta, S. Biodegradation of waste lubricants by a newly isolated Ochrobactrum sp. 3 Biotech 2015, 5, 807–817. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Isolation of marine benzo pyrene-degrading Ochrobactrum sp. BAP5 and proteins characterization. J. Environ. Sci. 2009, 21, 1446–1451. [Google Scholar] [CrossRef]
- Doan, C.D.P.; Sano, A.; Tamaki, H.; Pham, H.N.D.; Duong, X.H.; Terashima, Y. Identification and biodegradation characteristics of oil-degrading bacteria from subtropical Iriomote Island, Japan, and tropical Con Dao Island, Vietnam. Tropics 2016, 25, 147–159. [Google Scholar] [CrossRef]
- Gudiña, E.J.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactants produced by marine microorganisms with therapeutic applications. Mar. Drugs 2016, 14, 38. [Google Scholar] [CrossRef] [PubMed]
- Floris, R.; Rizzo, C.; Giudice, A.L. Biosurfactants from marine microorganisms. Metab. New Insights Biol. Med. 2018. [Google Scholar] [CrossRef]
- Souza, E.C.; Vessoni-Penna, T.C.; de Souza, R.P. Biosurfactant-enhanced hydrocarbon bioremediation: An overview. Int. Biodeterior. Biodegrad. 2014, 89, 88–94. [Google Scholar] [CrossRef]
- Urum, K.; Pekdemir, T. Evaluation of biosurfactants for crude oil contaminated soil washing. Chemosphere 2004, 57, 1139–1150. [Google Scholar] [CrossRef]
- Bacosa, H.P.; Inoue, C. Polycyclic aromatic hydrocarbons (PAHs) biodegradation potential and diversity of microbial consortia enriched from tsunami sediments in Miyagi, Japan. J. Hazard. Mater. 2015, 283, 689–697. [Google Scholar] [CrossRef]
- Bird, W.A.; Grossman, E. Chemical aftermath: Contamination and cleanup following the tohoku earthquake and tsunami. Environ. Health Perspect. 2011, 119, 290–301. [Google Scholar] [CrossRef]
- Ermakova, I.T. Organophosphonates utilization by soil strains of Ochrobactrum anthropi and Achromobacter sp. Arch. Microbiol. 2017, 199, 665–675. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).