Abstract
Microbial biofilms are biological structures composed of surface-attached microbial communities embedded in an extracellular polymeric matrix. In aquatic environments, the microbial colonization of submerged surfaces is a complex process involving several factors, related to both environmental conditions and to the physical-chemical nature of the substrates. Several studies have addressed this issue; however, more research is still needed on microbial biofilms in marine ecosystems. After a brief report on environmental drivers of biofilm formation, this study reviews current knowledge of microbial community attached to artificial substrates, as obtained by experiments performed on several material types deployed in temperate and extreme polar marine ecosystems. Depending on the substrate, different microbial communities were found, sometimes highlighting the occurrence of species-specificity. Future research challenges and concluding remarks are also considered. Emphasis is given to future perspectives in biofilm studies and their potential applications, related to biofouling prevention (such as cell-to-cell communication by quorum sensing or improved knowledge of drivers/signals affecting biological settlement) as well as to the potential use of microbial biofilms as sentinels of environmental changes and new candidates for bioremediation purposes.
1. Introduction
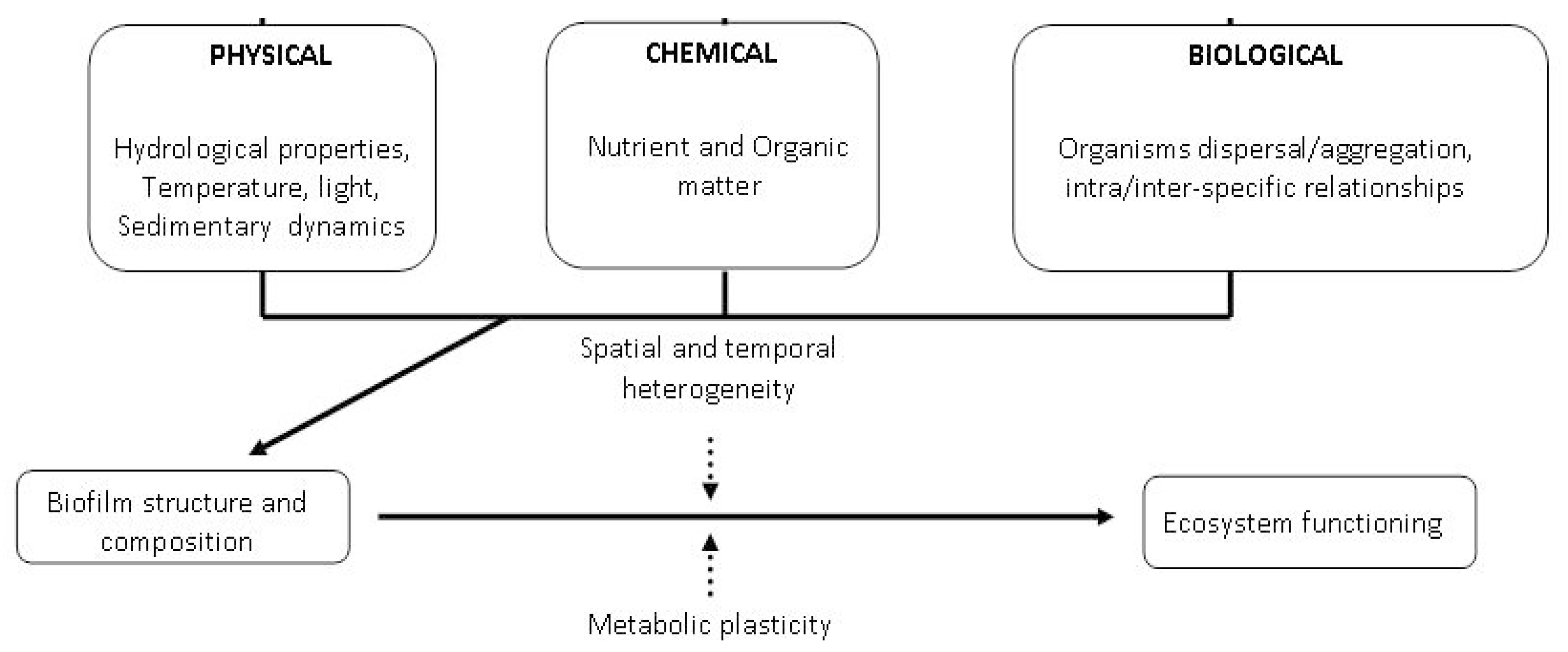
Biofilm formation on substrate surfaces is the first step in biofouling formation [1,2,3,4,5]. Biofouling is a normal process occurring over several kinds of submerged surfaces, including natural aquatic systems such as marine [6,7] and freshwater ecosystems [8], lentic or lotic water bodies, sediments, rocks, or artificial substrates, medical devices or pipelines in industrial or drinking water systems. Biofilms can be considered as a microbial skin that colonizes natural or artificial substrates, adapting a definition reported for epibiotic biofilms growing on marine organisms [9]. These microbial structures include several levels of structural and functional complexity, which are continuously adapted to the environmental conditions of marine ecosystems. The physical structure, community composition and function of microbial biofilms are controlled by a wide range of physical, chemical and biological variables and processes [2,10], including environmental interactions, interactions with topography, nutrient and organic matter cycling, and photosynthesis. In turn, biofilms are able to influence ecosystem functioning (Figure 1).
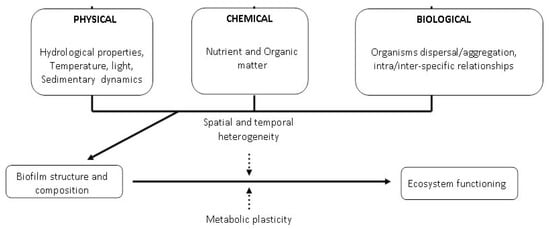
Figure 1.
Physical, chemical and biological drivers affecting marine biofilms.
The spatial and temporal heterogeneity of marine environments in terms of hydrodynamics, topography, nutrients and organic matter availability, biological dispersal and aggregation also at level of micro-niches habitats [11,12] modulates the effects of biofilms on the settlement of larval invertebrates, macroalgal spores, and of all sessile marine organisms in general. In turn, through their complex intra-specific interactions and their structural and functional plasticity, biofilm components are able to adapt to dynamic environments and can affect ecosystem processes and functioning.
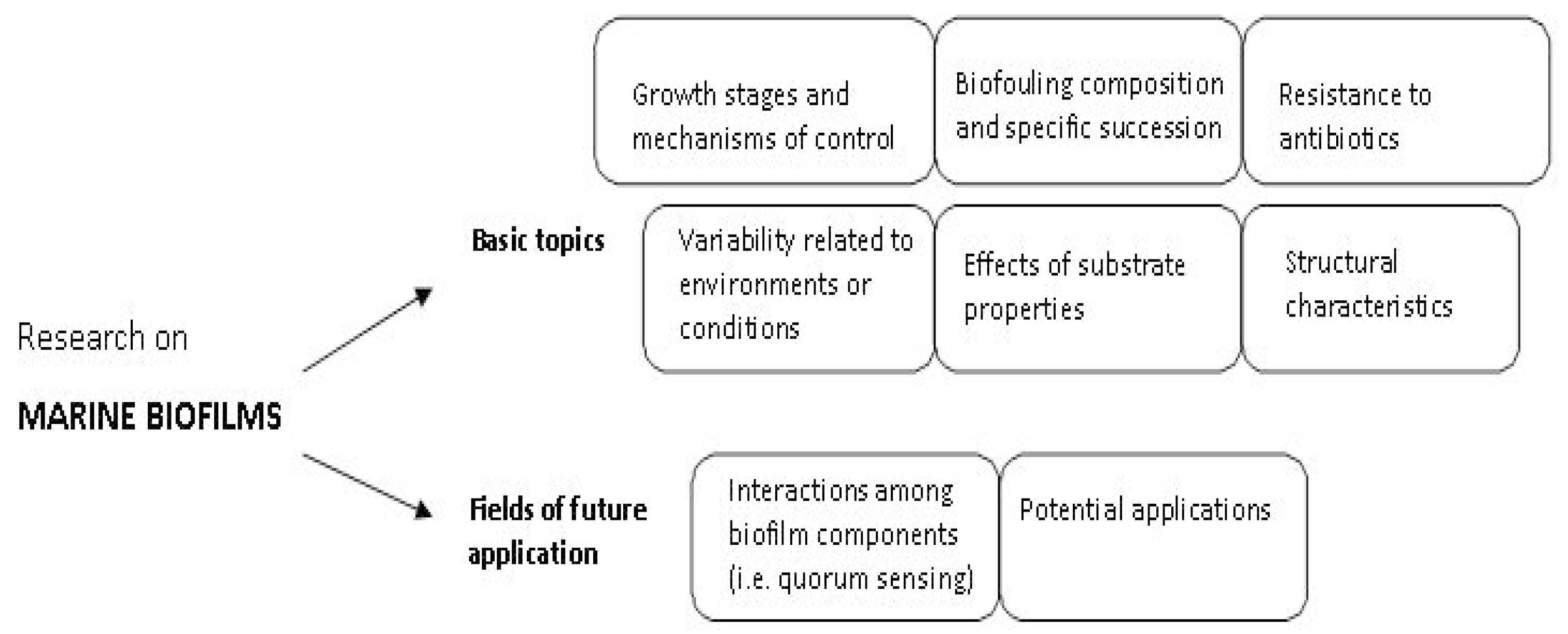
Marine biofilms have been the subject of many studies [2,4,6,7,8,10]; the studies performed in this context have investigated both basic and applicative features, exploring key topics that cover microbial biofilm growth and structure, biofilm–substrate or –environment interactions, resistance of biofilms to antibiotics, microbial cell-to-cell signaling and biotechnological potential of biofilms (i.e., discovery of bioactive molecules/compounds, biofilm application in food, cosmetic, pharmaceutical, bioremediation and industrial fields). A schematic diagram of the main features in marine biofilm research is shown in Figure 2.
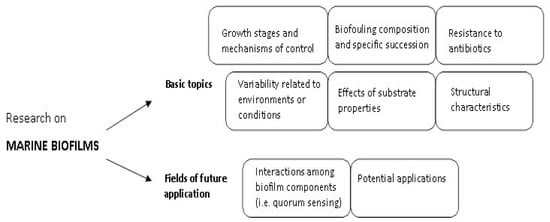
Figure 2.
Basic and emerging topics of research on marine biofilms.
Interest towards microbial biofilms depends not only on their detrimental effects in environmental and medical research (such as in the case of biofilm-related pipeline occlusion, or biofilms acting as a reservoir for antibiotic resistance, respectively) but also on their key role as mediators of substrate colonization and consequent implications in several fields [3,4,10]. Moreover, in parallel with the emerging problem of plastic pollution, a plethora of studies have recently investigated in marine ecosystems the structure, composition and metabolic activities of the microbial communities colonizing the surface of plastic debris, belonging to the size classes of macro- and micro-plastics [13].
According to Donlan [14], microbial biofilms consist of several microorganisms—both prokaryotic (bacteria and archaea) and eukaryotic (algae and fungi)—strictly adhering to a substrate surface. Within microbial biofilms, microcolonies of bacterial cells are wrapped into an exopolysaccharidic matrix (EPS) consisting of “extracellular polymers of biological origin which participate in the formation of microbial aggregates” [15]. Therefore, each microbial biofilm is composed of a complex microbial community, which creates a microenvironment where over 4000 species can co-exist in a complex structure [16,17].
Colonization by microorganisms takes place on all kinds of natural and anthropogenic material present at sea [2]. Attachment of microorganisms to substrate surfaces generally results in the formation of a microbial slime and subsequently of a microbial biofilm. At the initial stage of substrate colonization, pioneer species, recruited from the surrounding medium and generally autotrophic, produce biomolecules that favor bacterial adhesion to the substrate [18]. The ability of bacteria to attach to the substrate depends mainly on their cell size and motility, while bacterial abundance is mostly regulated by the availability of nutrients [19]. In this context, therefore, nutrient cycling by the microbial community plays a key role in supporting biofilm growth. The multiplication of pioneer species subsequently produces a substrate suitable for colonization by secondary heterotrophic species [20,21]; however, after the biofilm reaches its maturation stage, the reciprocal competition among bacterial species for trophic resources leads to the death and/or detachment of the less competitive species [18].
To date, some reviews available in the scientific literature have provided useful insights into different aspects of biofilm formation, structure, biological functions and their biotechnological perspectives [2,4,6,7,8,10,22]. Some research questions, however, need to be further deepened, such as:
- whether and to what extent the community structure of microbial biofilms changes in relation to environmental variables or substrate-related properties;
- whether there are differences in the colonization depending on the chemical nature of the artificial substrates;
- whether there is a selection of biofilm components, with preferential growth of populations that are rare members within the common microflora inhabiting the marine environment.
The rationale of this study relies on the consideration that the composition of the microbial biofilm community can vary with the polymeric nature of substrates supporting microbial adhesion. Since microbial growth is strictly related to environmental variables (i.e., temperature and nutrient availability), these are also expected to be important drivers that may affect microbial colonization.
This review aims at gathering current information on biofilm formation on different artificial substrates in a range of marine ecosystems—from temperate to extreme polar regions—with the objective of evaluating the possible differences in microbial community composition related to the substrate type, as well as to the environmental variables. In particular, the specific points detailed here are: (i) which drivers are potentially affecting microbial colonization; (ii) how microbial colonization varies depending on the type of artificial substrate; (iii) what the role played by microbial biofilms in the recruitment and settlement of invertebrate larvae is. As a case study, a state-of-the art review of microbial colonization in polar regions has also been included.
2. Drivers Potentially Affecting Microbial Colonization
The colonization of a substrate involves both abiotic and biotic conditions [20,23]. Several factors, both environmental and related to the nature of the substrate, may influence microbial colonization at sea. Since the initial studies on the adhesion of some bacterial species [24,25], it has become evident that several physical-chemical variables, such as temperature, salinity, pH, and nutrient concentration, together with the geographic location, seasonal period, light availability, water depth, presence of tides, competition among biofilm components, can affect the formation and composition of biofilm communities in marine environments [2,6,26,27,28,29,30,31,32].
Also, especially during the early stages of biofilm formation, many properties typical of each substrate, such as its wettability, hydrophobicity, surface polarization and tension, may affect the patterns of bacterial adhesion to the surface [2,33]. Substrate roughness has also been shown to be one of the most relevant factors in driving bacterial colonization [34,35]. Indeed, surface irregularities promote biofilm formation and can provide shelter from unfavorable environmental factors [36]. With respect to substrate polarization, microorganisms preferentially colonize hydrophobic, non-polar surfaces, such as Teflon, silicone or other plastics, compared to hydrophilic materials such as glass and metal [37].
Other variables acting at a substrate scale, such as substrate orientation and coating by anti-fouling compounds may affect the structure and composition of bacterial and eukaryotic communities. To this regard, by analyzing through DNA metabarcoding the bacterial and eukaryotic communities developed on settlement plates deployed for three months in a New Zealand port, von Ammon et al. [38] pointed out that surface texture is not relevant in shaping the bacterial communities, and that substrate orientation affects bacterial community structure only slightly. Again, the immersion mode, substrate surfaces and site were found to be the drivers influencing the prokaryotic community composition growing on different artificial surfaces (self-polishing and fouling-release coatings compared to inert plastic) in Toulon and Banyuls Bays (north-western Mediterranean Sea) under different hydrodynamic conditions (dynamic, cyclic and static) [39]. Rhodobacteriaceae and Flavobacteriaceae dominated the biofilm community structure, with distinct genera depending on the surface type or immersion mode. Globally, however, there is still limited knowledge on the relative weight of substrate-related properties in affecting microbial colonization. In this context, there is an urgent need to deepen substrate–microbe interactions, modified by biofilm growth that creates a new interface with different roughness and energy characteristics [40]. The colonization of the substrate surface is generally associated to the selection of specific microbial biofilm communities. Temporal shifts in the composition of the bacterial communities are usually observed; the development of different phenotypes and metabolisms proves that during surface adhesion a selection in bacterial species occurs [41]. The significant differences recorded between biofilm-adherent organisms and planktonic organisms in the surrounding environments [42] provide further evidence of the selection of bacterial species, among those most specialized for biofilm production. On the other hand, over a long time period, exposure to seawater of a surface can modify the physical-chemical characteristics of the substrate itself, which starts to be subject to degradation by microorganisms [43,44].
3. Microbial Colonization of Several Artificial Substrates
It is still a matter of scientific debate whether the nature of the substrate or the initial formation of a conditioning film are more relevant to bacterial adhesion [45]. According to Cooksey and Wigglesworth-Cooksey [46] and Singh et al. [47], the settlement and growth of colonizing organisms are influenced by the physical and chemical properties of the substrate. Similar findings were reported by Donlan [48], who stated that bacterial adhesion to the substrate surface depends on the nature of the substrate on which biofilm develops.
When diverse artificial substrates are compared, different microbial communities are found, especially at early colonization stages; over time, however, such communities become reciprocally more similar, regardless of the settlement substrate [49,50]. Therefore there is evidence that the effect of the substrate could be more relevant in the early stages of biofilm development only; conversely, in advanced stages of biofilm growth, differences in structure and biomass in relation to the substrate type are reduced and the variability in bacterial community structure appears to depend more on changes in light, salinity, turbidity and trophic conditions [41,49,50,51,52].
To date, the influence of the substrate nature on microbial biofilms has been not systematically studied, and current scientific reports on bacterial adhesion to different substrates are rather fragmentary. Reported below is a review of previous investigations performed on microbial biofilms grown on different artificial substrates.
3.1. Stainless
Khandekar and Johns [53] approached the study of marine corrosion using steel plates submerged in a natural temperate seagrass environment. Six days after deployment, they found the formation of an algal film succeeded by a bacterial community and the further development of a new algal community from Day 6 onwards. These authors suggested the involvement in the early stages of biofilm formation of acidic polysaccharides, which were responsible for corrosion of the steel structure. Geesey et al. [15] on 316 L stainless steel unpolished surfaces exposed to aqueous media observed during the initial colonization significant changes in the elemental composition of the surface biofilm, with depletion of Cr and Fe compared to Ni. Analysing by 16S rDNA libraries, fluorescence in situ hybridization (FISH), and denaturing gradient gel electrophoresis (DGGE) the bacterial community structure of biofilms growing on stainless steel and polycarbonate in seawater from the Delaware Bay, Jones et al. [54] observed no differences in the composition of stainless steel and polycarbonate biofilms at the initial stages, while differences were evident after about 1 week of biofilm growth. This suggested a relationship between surface properties and biofilm community structure changes during the biofilm growth. According to FISH and DGGE analyses, the structure of the free-living bacterial community was different from that of the attached bacteria that was dominated by alpha-Proteobacteria. Libraries of 16S rRNA genes revealed that representatives of the Rhodobacterales clade were the most abundant members of the biofilm community.
Comparing the adhesion of five marine bacterial species (Cobetia marina and Aliivibrio fischeri (gamma-Proteobacteria), Sulfitobacter guttiformism and S. mediterraneus (alpha-Proteobacteria) and Salegentibacter flavus (Flavobacteria)) on two glass types (one of which was modified by chemical etching), Mitik-Dineva et al. [55] found a great bacterial attachment and elevated levels of secreted EPS on the chemically etched glass surfaces, unless of the taxonomic affiliation of the selected bacteria. For both types of glass surface, the density of the bacterial attachment was found to correlate with surface wettability, while no correlation was observed between cell surface charge and the bacterial attachment.
An interesting study by Landoulsi et al. [56] highlighted that diatoms dominated the biofilms formed on stainless steel surfaces; these authors reviewed the diatom–stainless steel surface and diatom–bacteria interactions in several aquatic environments, both natural (seawater, estuaries, lentic and lotic freshwaters) and human-related (dam-water, wastewater, tap water). Emphasis on the physico-chemical features of these surfaces, on the composition, structure and biochemical properties of the diatom cell surface, as well as on the metabolic activities that influence the electrochemical response of stainless steel was given. Diatoms were identified to be an important component in the biofouling community that develops on stainless steel, playing a key role in the electrochemical processes that may lead to biocorrosion.
More recently, Li et al. [57] analysed, by targeted 16S rRNA gene (V3-V4 region) sequencing through Illumina MiSeq, the composition of bacterial communities living on steel plates deployed at Sanya and Xiamen, China, together with the influence of ecological factors on these communities. The phylogenetic analysis revealed that bacteria fell into 13 phylotypes (with a similarity level D 97%). Proteobacteria, Firmicutes and Bacteroidetes were the dominant phyla, accounting for 88.84% of the total, while the dominant classes were delta-Proteobacteria, Clostridia and gamma-Proteobacteria, that accounted for 70.90% of the total. Desulfovibrio spp., Desulfobacter spp. and Desulfotomaculum spp. were the dominant genera and accounted for 45.87% of the total. These genera are sulfate-reducing bacteria known to be able to corrode steel. The bacterial community composition was found to be influenced by the immersion time: the bacterial diversity of samples immerged for 6 months was found to be significantly higher than those deployed for 8 years.
3.2. Glass
On the surface of glass slides deployed for up to 3 weeks at different water depths (5,10,20,40,80 and 160 m) in Loch Fyne, Clyde Sea, W Scotland, Head et al. [58] using epifluorescence and bright field microscopy found a seasonal variability in the composition of microbial biofilm. In April, the biofilm community varied significantly with depth, although this effect was ascribable to changes in the diatom community. Diatoms peaked between 10 and 20 m. During August, the numbers of diatoms and bacteria peaked at 5 m and they were positively related; this was not observed in April.
Patil and Chandrashekar Anil [59] used fibreglass and glass coupons submerged for 4 days to investigate over a period of 14 months the influence of time and substrate variability on the structure of biofilm diatom community that are early autotrophic biofouling colonizers in a monsoon-influenced tropical estuary (Dona Paula Bay, located at the mouth of the Zuari estuary on the west coast of India). The planktonic diatom community structure was dominated by centric diatoms, in contrast with the biofilm community, which was dominated by pennate diatoms, among which the genera Navicula, Amphora, Nitzschia, Pleurosigma and Thalassionema were very abundant. Significant differences in density and diversity were also observed in the biofilms formed on the two substrates, although species composition was almost constant. Seasonal variations recorded in the community composition of biofilm diatoms were related to temporal variations in the tychopelagic diatoms as well as physico-chemical and biological changes in both the water and substrate.
In order to evaluate the effects of surface wettability on biofilm formation and subsequent settlement of Hydroides elegans, Huggett et al. [49] treated glass microscope slides with trimethylsilyl and dimethyldichlorosilane to create low surface wettability, with aminopropyltriethoxysilane and 3-chloroproplyltrimethoxysilane to create intermediate wettability, and with trimethylaminopropyltrimethoxysilane to create high surface wettability. Fifteen replicates of each treatment were placed in frames and deployed for 3, 6 and 10 d at a depth of 2 m below the low tide mark at a pier at Ford Island, Pearl Harbor, Hawaii. The biofilm community composition, as determined by a DGGE and FISH combined approach, was similar across all surfaces, regardless of the initial wettability, and all surfaces had distinct temporal shifts in their community structure over a 10-day period. Larvae settled and recruited in higher numbers on slides with a medium-low wettability in May and August, and also on slides with a high wettability in August. Biofilm communities developed similarly on a range of surface wettabilities, and after 10-day deployment all surfaces were equally attractive to larvae of H. elegans, regardless of their initial surface properties.
Through an experiment performed on glass slides deployed in the tropical waters of the Dona Paula Bay for up to 192 h and analysed by flow cytometry, Mitbavkar et al. [60] demonstrated that picophytoplankton is the pioneer autotrophic colonizer in tropical marine biofilms, and that it plays a relevant role in the initial stages of biofilm food web dynamics. A succession of microorganisms were observed, with the presence of three groups of picophytoplankton, two prokaryotes, Prochlorococcus-like organisms, Synechococcus (SYN) and the picoeukaryotes (PEUK). Nanoeukaryotes, which were mostly represented by diatoms, were also monitored. In the total biofilm community, prokaryotes were dominant throughout the study period and the contribution of SYN was highest (50%) in the earlier development stages. The contribution of PEUK and nanoeukaryotes was always below 20%. Picophytoplankton contribution to the total photosynthetic biomass was >60% in the initial period of biofilm formation, both in terms of numbers and biomass, with PEUK as the major contributors. However, after 2 d of incubation, their contribution to the total chlorophyll declined, suggesting that picophytoplankton was succeeded by nanoeukaryotes in terms of biomass. Laboratory experiments revealed that, in the biofilms, heterotrophic bacteria and picophytoplankton appeared within 5 min, but after 5 h they were succeeded by nanoeukaryotes.
3.3. Ceramics
Biofilm formation on unglazed ceramic tiles deployed in the northern Gulf of Eilat, Israel, was followed by Siboni et al. [61]. Already 2 h after deployment, PCR-DGGE of film eluted from the tile surface indicated the presence of a bacterial community. Bacteria were microscopically visible 6 h after deployment, although a developed biofilm was observed 24 h after deployment only. Within the first 4 h following deployment, the total organic carbon content suggested that a conditioning film was built, with the adhesion of organics (e.g., proteins, sugars, and humic substances) coming from the water column. This conditioning film was probably used by the primary adhering bacteria (pioneer species) as a trophic source.
Kang et al. [62] examined the amount of biofouling developed on porous ceramics, which were used to retain red tide organisms by filtration. Analysing six different ceramic porous bodies, it was found that the specimen with a lower porosity and water absorption showed the least amount of biofouling. In addition, by increasing the surface roughness with silica particles, the amount of biofouling (due to barnacles and mussels) was decreased. This observation could be used as an environmental-friendly method for preventing marine biofouling, by controlling the physical properties and the surface roughness of the porous ceramics.
3.4. Plastic Polymers
With their physical properties, plastic debris offer a unique habitat to host and carry over the oceans diverse microbial hitchhikers attached to their surface, creating the so-called "plastisphere", according to the denomination by Zettler et al. [42,63]. A large fraction of plastic waste is composed of polyethylene (PE), followed by polypropylene (PP) and polystyrene (PS) [64]. In recent decades, increasing attention has been paid to microplastics; this term has been assigned to particles of PE, PP, polyamide and polyester smaller than 1 mm in size, which can accumulate in the oceans’ depths, enter the food chain and cause significant damage to the marine ecosystem [65].
The distribution patterns and environmental fate of plastic materials have been reported to depend on water hydrodynamism (i.e., water mass circulation and turbulence) [66,67,68]; also, the variable density plays a relevant role in affecting the sinking of plastic debris and their burial in the sediments. In fact, particle sinking has been found to depend on the polymeric nature of plastics. Generally most of plastics have a density higher than seawater [69]; however, among the variety of existing polymers, PP and PE are low-density plastics that tend to be relatively buoyant, unlike from polyvinyl chloride (PVC), PS, polyester, and polyamide, which, being high-density plastics, are expected to be more prone to sink [68,70,71]. Biofouling formation on plastic surface may increase the density and weight of the microplastic particles, causing their sinking to pelagic or benthic zones [68,71,72,73,74]; conversely, high-density microplastics can be resuspended along the water column by water turbulence [66,67,68].
Submerged plastics are rapidly covered by the “conditioning film” made of inorganic and organic matter, which is then rapidly colonized by bacteria [75]. Cai et al. [76] recently investigated the effects on bacterial adhesion to plastics related to the substrate properties, like surface charge, hydrophobicity, hydrophilicity, roughness, and hardness, concluding that substrate hardness is the key factor driving plastic colonization.
Through a multi-ocean basin biogeographical survey, Amaral-Zettler et al. [77] showed that plastisphere communities developed on different polymer types in the North Pacific and North Atlantic Seas reflected primarily their biogeographic origins, and to a lesser extent the plastic type; therefore, differences in microbial communities among ocean basins were more significant than those among plastic polymers. Similar conclusions were obtained for bacterial communities colonizing plastics along an environmental gradient (from freshwater to marine), which were shaped firstly by the environmental conditions and secondarily by the plastic type [78].
Within plastic-attached microbial biofilms, diatoms have frequently been found, representing the most abundant type of eukaryotes [32,79,80,81]. Diatoms are recognized among the first recruits in marine biofilm formation and likely pioneer species for subsequent heterotrophic microbial colonizers [2,6,82]. They can form specific assemblages of co-associated bacterial epibionts [83]. Among these, Roseobacter, Alteromonas and Pseudoalteromonas are also dominant in the plastic-attached microbial communities. Flavobacteria, belonging to the genera Tenacibaculum and Polaribacter, are major colonizers of diatoms’ detritus [84].
According to a recent review by Jacquin et al. [85], plastisphere communities found in the sub-surface are dominated by photoautotrophic bacteria such as cyanobacteria with the genera Phormidium and Rivularia, while the core microbiome of the seafloor and subsurface plastisphere seems to share some taxa, such as Bacteroidetes (Flavobacteriaceae) and Proteobacteria (Rhodobacteraceae and Alcanivoraceae) [42,86,87,88]. Plastic-attached bacteria were dominated by alpha- and gamma-Proteobacteria, while seawater bacteria were dominated by alpha-Proteobacteria (mainly Pelagibacter sp.) [88].
The current knowledge of plastic-related marine biofilms in terms of community assembly processes could be improved by future research exploring plastic-microbial interactions at an increased spatial resolution. Since the plastisphere in marine environments has been shown to host specific potential microbial degraders [89], biofilm communities on plastic polymers could represent targets for future discovery of novel plastic-degrading microbes and the genes involved in this enzymatic process.
3.4.1. Polyvinyl Chloride
In an experiment performed by Hung et al. [90], polyvinyl chloride (PVC) plates placed into a nylon mesh bag (to prevent the attachment of invertebrate larvae) were used as a substrate to study the recruitment of barnacles Balanus amphitrite in subtropical Hong Kong waters. The obtained results point out significant site-specific variations in larval recruitment, suggesting the ability of barnacle larvae to discriminate among biofilms collected from contrasting environments in the intertidal region, probably mediated by specific signals produced by the biofilms.
Balasubramanian et al. [91] studied the biofilm formation by heterotrophic bacteria on PVC sheets fitted in wooden racks immersed in the Tuticorin waters (India). The samplings were performed over 7 d at 30 min, and 1, 2, 4, 24, 48, 72, 96, 120 and 144 h. Within 30 min, bacteria belonging to the genera Pseudomonas, Enterobacter, Aeromonas, Cytophaga and Flavobacterium were found to be the pioneer microorganisms colonizing the PVC surface. Gram-positive bacteria belonging to Micrococcus and Bacillus sp. were also detected, but only at a later stage (48 h old biofilms). Between 48 and 96 h, both Gram-negative and Gram-positive groups co-existed. After 96 h, the biofilm was constituted only by Gram-positive bacteria.
By flow cytometry, microscopy and high throughput sequencing (HTS) by 454 pyrosequencing, Briand et al. [92] studied the drivers of microbial community composition of the biofilm developing on PVC and four antifouling coatings at two French sites, one eutrophic (Lorient, Atlantic coast) and the other mesotrophic but highly contaminated (Toulon, North-Western Mediterranean Sea). Whereas seasons were not a relevant variable, microbial communities were affected by surface type and environmental variables such as high temperatures, salinity and lead at the Toulon site, and nutrients and dissolved organic carbon at the Lorient site. HTS revealed that bacterial communities were dominated by gamma and alpha-Proteobacteria, together with Bacteroidetes. The percentage of Bacteroidetes overall decreased with the presence of pyrithione as an antifouling coating. Small diatoms (Amphora and Navicula spp.) dominated on all surfaces.
3.4.2. Polyethylene and Polyethylene Terephthalate
Polyethylene terephthalate (PET) bottles were deployed in the North Sea (UK) in order to verify how the bacterial community varied in relation to the season, the geographical location and the type of substrate [75]. After 6 weeks of immersion, the bacterial Phyla colonizing PET included Bacteroidetes, Proteobacteria and Cyanobacteria, together with the eukaryotes Bacillariophyceae and Phaeophyceae, with only a 10% similarity with the community present at sea. Significant variations in the biofilm bacterial/archaeal (16S) communities were detected among stations and seasons, probably reflecting the influence of local physicochemical conditions.
In a further study in the North Sea [32], by 16S rRNA gene sequence analysis, the microbial community structure of biofilms colonizing PET bottles was compared to that of seawater communities (separately for free-living (0.22–3 μm) and particle-associated (>3 μm) fractions) and to glass-colonized biofilm community. The bacterial biofilm community on plastic surfaces showed species-specificity and was significantly influenced by both season and substrate type. A significant difference between the PET-colonizing and seawater free-living bacterial/archaeal communities was observed, but not between PET-attached and particle-associated or glass-attached communities. Similarities detected among plastic-attached, seawater particle-associated or glass-attached microbial communities suggested that PET as a substrate did not play a major role in structuring the plastic-associated biofilm communities. Conversely, significant differences were found between microbial biofilm and seawater free-living microbial communities. In detail, within the PET-attached communities highly abundant OTUs belonged to the phylum Bacteroidetes with the families Flavobacteriaceae, Cryomorphaceae, and Saprospiraceae. Rhodobacteraceae were always identified, with the dominant genera Tenacibaculum (Bacteriodetes, Flavobacteriaceae), Crocinitomix and Owenweeksia. In addition, microbes belonging to Sphingobacteriales (Saprospiraceae, in particular) and Myxococcales, together with members of the Verrucomicrobia phylum (Verrucomicrobia subdivision 1) and of the genus Phormidium were detected. Sphingobacteriales are successful biofilm community members, probably for their ability to produce exopolysaccharides and scavenge biofilm materials for energy and carbon. Myxobacteria excrete a polymeric substance enabling their gliding and swarming, and also complex bioactive secondary metabolites and hydrolytic enzymes, that give them a competitive advantage in limiting resources, such as in a biofilm environment. Sphingobacteriales and Myxococcales were also identified as biofilm components of plastic debris collected from the North Atlantic Sea by Zettler et al. [42].
De Tender et al. [87] identified a core group of 25 single OTUs, belonging to the phyla Proteobacteria, Bacteriodetes and Verrucomicrobia, on PE debris collected from the North Sea, but it remains unproven whether these “core organisms” are specific to an environment or whether they are also found on other types of synthetic polymer.
Using PET bottles in a microcosm experiment lasting 34 days, Misic and Covazzi-Harriague [93] recently investigated the effects on colonization of changes in physical, chemical, and biochemical properties of seawater. Possible influences of variations in temperature and light have also been evaluated. The temperature increase and light limitation have been found to potentially modify the biofilm community, increasing the role of prokaryotic organisms. Particularly, summer conditions could favor the growth of photoautotrophic organisms.
3.4.3. Polystyrene
PS dishes were used in a settlement assay to test the possible interference of bacterial supernatants on the attachment of barnacle cyprids. The attachment of balanids was found to be delayed by biofilms from stationary-phase Deleya marina cultures, while Alteromonas macleodii and Pseudomonas fluorescens did not yield relevant effects [94]. A significant attachment to 96-well PS plates of the marine bacterium Pseudoalteromonas sp. D41 in sterile natural seawater was observed by Leroy et al. [95], who proposed this method for a first screening of the effectiveness of antifouling agents in the early steps of marine biofilm formation.
The succession changes observed in multispecies biofilms growing on PS Petri dishes deployed in a subtidal zone (Port Shelter, Hong Kong) for 20 days were evaluated by Chung et al. [50] with molecular methods (PhyloChip microarray and DGGE). The extracts of biofilms of different ages induced larval settlements in relation to the biofilm age, but for different substrates of the same age the larval settlement was not significantly different.
PS Petri dishes were also chosen as a colonization substrate by Chiu et al. [26], in order to examine the composition and biomass of mixed algal and bacterial biofilms in two seasonal periods (summer and winter) in relation to differences in temperature and salinity. Biofilms were produced in a laboratory over a 20-d time period using natural seawater; a greater biofilm biomass (ranging from 10 to 46 μg dry weight cm−2) was obtained in summer than in winter and at a salinity of 34 compared to 20. In summer, both bacterial and diatom community compositions differed significantly depending on salinity, while temperature exerted a major influence on community composition in winter.
3.4.4. Polyurethane
Polyurethane (PU) is another plastic material that has been tested as an artificial substrate in studies on microbial colonization. Using a modified glass slide method, followed by living observation and silver impregnation method, Xu et al. [96] studied the colonization of PU foam enveloped slide by periphytic ciliates in marine ecosystems and identified a total of 27 ciliate species. Although the ciliate colonizations had similar species compositions, they showed considerable differences in both structural and functional parameters between the PU foam slide system and the conventional slide system. The species diversity, evenness and the colonization rate were distinctly higher, but the time for reaching 90% equilibrium species number was shorter in the PU foam slide system than on the naked slides. The results suggest that the PU foam slide system is more effective than the conventional slide method for periphytic ciliate colonization, with high species diversity, evenness, and colonization rate.
Using PU sponges as adsorbents for lithium recovery from seawater, Jeong et al. [97] studied the microbial community attached to three types of polymers differing for shape and size (2 µm sphere-shaped, 2 mm circular and 2 mm rod-shaped adsorbents), which were immersed in seawater for 30 days. Primary colonization was performed by gamma-Proteobacteria, with the genera Vibrio, Alteromonas, and Pseudoalteromonas. Confocal microscopy and scanning electron microscopy (SEM) images showed that bacterial distribution on surfaces strongly depended on the shape of the adsorbent; also, bacterial composition was pore-size dependent, with only Alteromonas on the 2 µm sphere-shaped adsorbent, while Vibrio was found on the other shapes and was succeeded by Alteromonas at advanced biofilm stages.
3.4.5. Acrylic
Poly (methyl methacrylate), also known as acrylic glass or plexiglass, is a transparent thermoplastic material often used in sheet form as a lightweight or shatter-resistant alternative to glass. Lee et al. [98] isolated from acrylic coupons submerged in Goje Islands (Korea) a total of 115 bacteria, 70 of which were identified according to their 16S rDNA sequences. Within them, alpha-Proteobacteria were predominant, followed by gamma-Proteobacteria, low GC Gram-positive bacteria, high GC Gram positive bacteria and Cytophaga/Flexibacter/Bacteroides group bacteria. The potential application of the bacterial isolates as standard strains for testing new antifouling agents or as enhancers of the settlement of invertebrate larvae was suggested. After 6 days, the microbial biofilm appeared, which was covered by serpulids and balanids at later stages. After 6 months, the surface was covered by adult bryozoa, mussels and algal coverage.
Dobretsov et al. [4] studied the effect of substrate colour on the settlement of micro- and macro-fouling communities on acrylic plastic slides and tiles, respectively. The studies were conducted over a time period of 5, 10 and 20 d. The densities of bacteria on the black and white substrates were similar, with the exception of day 10, when the black substrates had a higher abundance than white ones. 454 pyrosequencing of 16S rRNA genes of bacteria from white and black substrates revealed that alpha-Proteobacteria and Firmicutes were the dominant groups. Similarity percentages (SIMPER) analysis demonstrated that bacterial phylotypes (uncultured gamma-Proteobacteria, Actibacter, Gaetbulicola, Thalassobius and Silicibacter) and diatoms (Navicula directa, Navicula sp. and Nitzschia sp.) contributed to the dissimilarities between communities developed on white and black substrates. On day 20, the highest amount of chlorophyll a was recorded in biofilms developed on black substrates. SIMPER analysis showed that Folliculina sp., Ulva sp. and B. amphitrite were the major macrofouling species that contributed to the dissimilarities between the communities formed on white and black substrates. Higher densities of these species were observed especially on black tiles.
Plexiglass plates were used by Mejdandžić et al. [99] to study the succession and settling of benthic microalgae during biofilm formation in the northeast Adriatic Sea. The quantitative and qualitative composition of diatoms and bacteria on plexiglass plates immersed for 30 d was investigated using a combination of various methods (epifluorescence and electron microscopy). Among the pioneer species, planktonic diatoms belonging to the species Dactyliosolen fragilissimus, Proboscia alata, Thalassionema nitzschioides, and Leptocylindrus danicus were detected. The dominant species was Pseudo-nitzschia pseudodelicatissima, which reached its greatest abundance (10.2 × 104 cells L−1) after a month of exposure and whose distribution correlated positively with temperature. After the first week of exposure, benthic diatoms of the genera Licmophora, Cocconeis and Achnantes substituted the planktonic diatoms, becoming the most abundant colonizers. Insights into the probable mutual influence of bacteria and diatoms on the growth and development of the biofilm were also obtained.
3.5. Mixed Substrates
Several different artificial substrates were deployed at a deep-sea hydrothermal site (Snake Pit) along the Mid Atlantic Ridge in order to investigate the influence of the nature of the substrate on microbial colonization rates, population structure and succession over a period of 12 days [100]. The results demonstrate the rapid in situ colonization of artificial substrates by hydrothermal vent microbial populations, irrespective of the substrate type, although the greatest microbial biomass already present in large amounts after 4 d of exposure was observed to accumulate on 316L stainless steel and titanium substrates. Within the biofilm microbial community, two main bacterial morphotypes dominated, namely rod-shaped bacteria and large filamentous forms. Sulfate-reducing and filamentous sulfur-oxidizing bacteria were detected by polar lipid fatty acid and fatty acids analysis, respectively.
A study was conducted in the Bay of Bengal over a period of twelve months on four different substrates (polycarbonate, low density polyethylene LDPE and high-density polyethylene-HDPE and PP), found in large quantities in marine waters [101]. Bacterial adhesion was found to correlate positively with the energy of the material surface (calculated as the sum of the dispersion and polar forces, always greater than 20 mN m−1) only in the first three months of the experiment, while it was negatively correlated with its roughness. The largest amount of biomass accumulated on polycarbonate, which has polar and hydrophilic molecules, while the lowest density was found on LDPE.
Previous studies documented that different hard substrates (including ceramic, glass, plastic, aluminum, and coral skeleton) host similar communities, both in marine [102] and freshwater environments [103]; the differences detected comparing microbial communities colonizing soft (i.e., leaf litter) and hard (i.e., plastic, aluminium, tile, glass) substrates were explained by the different microbial abilities to degrade such substrates [103].
Microbial biofilms developed on PS, Teflon and antifouling paints, submerged for 2 weeks in two sites of the French Mediterranean coast and characterized by molecular (PCR-DGGE) and microscopic methods, showed variations in both microbial community abundance and structure depending on the location and substrate type [79]. Lower fouling densities were observed at Porquerolles Island compared to Toulon harbour. Two diatom species, Licmophora gracilis and Cylindrotheca closterium, dominated the pioneer microalgal communities at both sites, irrespective of the substrates.
In the Bay of Brest (France), differences in the composition of bacterial biofilm community colonizing different polymer particles (PE, PP, and PS) were also found [104], with the orders Pseudomonadales, Oceanospirillales and the Propionispira genus prevailing on PE samples, while PP was colonized by the alpha-Proteobacteria class and PS by the family Rhodospirillaceae and the genus Nitrosomonas. Bacterial strains belonging to Vibrio aestuarinus and V. splendidus were also detected.
Using aquaria filled with natural circulating seawater, Dussud et al. [105] detected over 6 weeks the marine microorganisms associated with the successive phases of colonization, growing, and maturation of the biofilms developing on non-biodegradable [i.e., polyolefins such low-density PE, PE additivated with pro-oxidant (OXO)] compared to biodegradable polymers [i.e., artificially aged OXO (AA-OXO) and a polyester, poly(3-hydroxybutyrate-co-3-hydroxyvalerate) (PHBV)]. All these substrates showed variable surface properties in terms of hydrophobicity and roughness. Depending on polymer type, different trends in microbial biofilm abundance, activity and taxonomic composition were observed. Higher colonization by active and specific bacteria was found on the biodegradable (AA-OXO and PHBV) polymers compared to non-biodegradable ones (PE and OXO). Moreover, bacterial population displayed a succession during the three colonization phases, with hydrocarbonoclastic bacteria being highly abundant on all plastic types.
Kirstein et al. [106] conducted a long-term (15 months) incubation experiment with nine different synthetic polymer films and glass substrates, using a natural seawater flow-through system. 16S and 18SrRNA gene tag sequencing was used to identify colonizing microorganisms and SEM to visualize the microbial biofilms. Biofilm communities attached to synthetic polymers were different from the glass-associated ones; apparently there was no specific synthetic polymer community, but a more general marine biofilm core community was shared among all the polymers, even if some taxa discriminatory of a given substrate were identified.
Oberbeckmann et al. [78], investigating microbial colonization of wood, HDPE and PS over 14 days along a gradient ranging from marine (coastal Baltic Sea) to freshwater (wastewater treatment plant) conditions, found that microbial communities did not differ significantly among the polymers.
Ogonowski et al. [107] incubated cellulose, glass, PE, PP and PS for two weeks in pre-filtered seawater from the Baltic Sea and found significant differences between plastic and non-plastic substrates, with higher abundance of Burkholderiales within the plastic-associated bacterial flora, while Actinobacteria and Cytophaga prevailed within the bacterial flora attached to non-plastic substrates.
Recently, Cai et al. [76], studying bacterial adhesion onto four types of plastic most frequently encountered in aquatic environments such as PVC, PE, PET and PP, found that the amount of bacteria that adhered to PE and PVC surfaces was much greater than that on PP and PET.
Muthukrishnan et al. [108] investigated microbial biofouling at two locations in the Sea of Oman (Arabian Gulf) by comparing the microbial communities developed on PET and PE with those on steel and wood. Substrate and location-specific effects were found, with the bacterial genera Microcystis and Hydrogenophaga and the diatoms Licmophora and Mastogloia specifically attached to plastics, while Desulfovibrio and Pseudomonas spp. on steel and Corynebacterium spp. on wood. The total biomass was lower on plastics than on steel and wood.
Pinto et al. [109] recently analyzed the composition of microbial biofilms growing on LDPE and HDPE, PVC and PP. The bacterial communities developing on the plastics clustered in two groups; one group was detected on PVC, while the other group on all the other plastics and on glass, used as an inert control. Specific bacterial taxa were found on specific surfaces in all stages of biofilm development. Differences in bacterial community composition among the different plastics and light exposures were observed after one week rather than at later incubation stages. The common families detected on all plastic types were Flavobacteriaceae, Rhodobacteraceae, Planctomycetaceae and Phyllobacteriaceae. Another part of the bacterial community was plastic-type specific and was more abundant after one week of incubation than at later stages.
4. A Case Study: Microbial Colonization in Polar Regions
In addition to the substrate type and surface properties, microbial biofilm composition is affected by regional factors. In polar regions, only a few studies have focused on microbial colonization. At first, the research performed in these extreme environments focused on the study of biofilm formation on stainless steel surfaces related to biocorrosion of this material. The effects of biofilm settlement on several stainless steel samples, immersed and recovered after one year, were tested in Ny Alesund (Svalbard Islands, Norway, Arctic) [110,111]. The formation of a microbial slime on stainless steel caused a change in oxygen reduction that favored the metal corrosion.
In Antarctica, Maki et al. [112], studying biofilm formation on titanium and copper/nickel foils shortly (15 min to 4 h) exposed to seawater, showed that a higher bacterial abundance was associated with the titanium substrate compared with copper/nickel, and that processes controlling bacterial adhesion to substrates took place within 15 min after the surfaces were deployed in the environment.
In the framework of biofilm research in polar ecosystems, a particularly interesting area concerns the bioprospecting of microorganisms isolated from these cold regions, in view of their unique metabolic characteristics (see a review by Urbanek et al. [43]). As bacteria respond quickly to changing environmental patterns, microorganisms inhabiting polar regions may have unique applications in the degradation of plastics. In fact, the peculiar environmental conditions of polar oceans, in terms of low temperatures (from −1.8 to −6 ℃) and extreme seasonal variations in irradiation and in light hours, could result in a site-specific microbial biofilm community, whose composition might be strongly influenced by the interactions with the environment.
Using heated settlement panels immersed for 18 months at 15 m depth near Rothera Research Station (Adelaide Island, Antarctic Peninsula), Clark et al. [113] found that biofilm bacterial communities did not differ significantly in their community structure with temperature, while other marine encrusting communities (such as metazoans) lacked long-term acclimation to either +1 ℃ or +2 ℃ above ambient temperatures. This suggested that bacterial vulnerability to warming was different to that of metazoans and that ecosystem responses to future climate change could be more complex than previously expected.
Funded by the Italian Antarctic Research Program (PNRA16_00105 Microbial colonization of benthic environments in Antarctica: responses of abundance, diversity and microbial activity), an in situ experiment with short-term and long-term observations over a two-year period on microbial colonization as a primary step for invertebrate settlements is currently being performed in the Ross Sea (Antarctica), in correspondence with the Italian Mario Zucchelli research Station (MZS). Two areas (Tethys Bay and Road Bay) of the Ross Sea, affected by a salinity gradient due to a glacier proximity and by anthropogenic pressure related to the treated wastewater effluents of the Italian station, respectively, were selected as the study areas. Artificial substrates (PVC panels and PE coupons) were deployed in November 2017 through ice-drilled holes at two different depths (−5 and −20 m). The main results obtained from seawater analysis [114] show higher proteolytic activity rates in Road Bay at the site impacted by the wastewaters with respect to the control site; in Tethys Bay, high microbial activity rates are found in correspondence with the site close to the glacier, probably in relation to a higher availability of detritus released from ice melting. Regarding biofilm, a higher abundance of heterotrophic bacteria is found on PVC compared to PE; pigmented bacteria, ascribable to Flavobacterium sp., are abundant.
5. Significance of Microbial Biofilms in Larval Recruitment and Settlement
The formation of microbial biofilm starts with the production of a conditioning film [115] that favors further colonization by other sessile macroscopic organisms, such as porifera, polychetes (serpulids), cnidarians (hydrozoa), crustaceans (barnacles), molluscs (mussels), macroalgae, and epiphytic invertebrates.
Substrate colonization by biofouling organisms is a dynamic process that occurs via sequential steps, from surface conditioning to attachment and growth of biofilm (by micro-foulers) and to successive settlement and final colonization by larvae and spore of macro-organisms (macro-foulers). Four main stages can be identified in biofouling process: (1) immediately after immersion, surface conditioning, with formation of a primary film by absorption of organic/inorganic macromolecules (seconds to minutes); (2) initial attachment and growth of bacterial cells and eukaryotes (micro-foulers) to the substrate surface (minutes to hours); (3) settlement of larvae and spores of macro-organisms (macro-foulers, days to weeks); (4) colonization of the substrates with development of a complex community of multicellular species (weeks to years) [6,97]. In the biofouling formation process, the critical step seems to be the first colonization with the initial bacterial attachment, because it can either induce or inhibit the succession to the next fouling stage [116]. Biofilm formation generally takes a few minutes, while hours are required for its growth [117].
Larval settlement of marine polychaetes (H. elegans), bryozoans (Bugula neritina) and barnacles (Balanus spp.) was found to be related to larval morphology and swimming capability, as well as to flow velocity in laboratory and field experiments using plastic (PVC) tubes as experimental models to mimic intake pipe environments [118]. When the effect of different substrates [PVC1 (Tygon), PVC2 (Nalgene), PVC3 (Nalge-Nunc), PE, PU, Teflon and glass tubes] on larval settlement of these macrofouling organisms was investigated [119], complex interactions among substratum characteristics, flow rates, and larval settlement behavior were observed to affect macrofouling formation. Teflon tubes were characterized by the highest settlement of bryozoans and polychaetes and the lowest one of barnacles, while glass tubes by the highest settlement of barnacles and bryozoans. PE and PU tubes showed the lowest settlement of polychaetes.
Since microbial biofilms have been suggested to exert both inductive or inhibitive effects on larval settlement for several benthic fouling species [120,121], increasing studies have explored how the bacterial community structure of microbial biofilms can mediate larval settlement [27,30,90,122,123], also giving emphasis to the chemical cues that modulate the settlement and succession of macrofouling organisms [10].
During the colonization process, particular significance is given to the mechanism of cell-to-cell communication ("quorum sensing", QS), a process occurring both in Gram-negative and-positive bacteria, which is activated as an adaptive response of a microbial population to high cell density. By QS, bacteria are able to monitor their population in a cell-density dependent manner through the synthesis and exchange of intracellular signal molecules (autoinducers, mainly represented by the acylhomoserine lactones-AHLs) [124]. Within marine environments, many bacteria belonging to the genera Pseudoalteromonas, Thalassomonas, Vibrio and Pseudomonas have been described to produce AHLs [125]. The QS process allows the expression of genes directly under the control of cell density and can regulate the production of membrane exopolysaccharides and also virulence determinants in pathogenic bacteria [126].
6. Future Research Directions
The data reported in this review demonstrate that marine biofilms host complex and dynamic microbial communities and that research in this field is far from being exhausted. The gathered information on microbial colonization of aquatic environments can provide useful baseline findings on the different susceptibilities to biofouling of a variety of artificial materials and on the environmental variables that control biofilm formation. Considering the high economic costs caused by marine biofilms to many sectors including maritime transport, aquaculture, oil and gas industries, desalination plants and other activities [2,127], the relevance of marine biofilms in multiple research fields appears evident.
Integration among several disciplines, such as biology, chemistry, genetics and environmental sciences, could allow future progress in marine biofilm research. From a strictly ecological perspective, a multidisciplinary research approach is required to integrate marine biofilm into the larger context of microbial ecology and more generally of aquatic sciences. The effects of climate changes, such as increased water temperature, decreased salinity and pH, could modify the composition of microbial biofilms and their bioactive molecules, which in turn could affect larval settlement and survival. As marine biofilms have been recognized to be sensitive to ocean warming and acidification related to climate change [128], the consequences of large-scale climate changes on microbial biofilm formation and biodiversity from an ecological and biochemical point of view need to be explored.
Future studies on marine biofilms addressing the exploration of their biodiversity [129] and their ecological role in ecosystem functioning should be encouraged, also considering the role of microbes as catalysts of biogeochemical nutrient cycling [130,131]. Marine biofilms could also be used as indicators of water quality, in relation to the well-known ability of microbes as sentinels of environmental changes in several temperate and polar marine environments [132,133,134] as well as in tropical coral reef ecosystems [135].
Regarding the role of marine microbial biofilms as pioneer colonizers in the biofouling process, actual settlement cues associated with biofilm communities remain unknown and represent another interesting subject for further investigations [50]. In this context, an important research issue is represented by QS, which is directly involved in the biofilm development. A better understanding of biofilm dynamics and chemical signals from biofilms could provide an environmentally friendly approach for new antifouling technologies, such as those based on the use of enzymes and QS inhibitors [3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131,132,133,134,135,136]. Similarly, knowledge of antibiotic susceptibility profiles and tolerance to biocides of bacterial biofilm components could open new perspectives for biofilm control.
Some of the key points in marine biofilm research that require further deepening are summarized in Table 1.

Table 1.
Main features regarding key points for future studies on marine biofilms.
7. Conclusions
An increasing number of reports on the plastisphere are giving an idea of the widespread occurrence of microbial biofilms in marine ecosystems, but the factors that affect the composition of microbial communities attached to artificial substrates such as plastic debris are still a matter of ongoing debate. Further studies on this topic, related to the main physical and chemical properties of the substrates, will provide a clearer picture of the response to the colonizable surfaces of these complex biological matrices. Specific features such as cell-to-cell communication, interactions among species or substrate species-specificity, the biogeochemical role of selected components of microbial biofilms, bacterial inhibition and/or induction towards larval and macro-organism settlements, the response of microbial biofilm to acidification or temperature warming are only some examples of the still unexplored variety of research themes that could improve current understanding of these microbial structures.
Funding
This research was funded by PROGRAMMA NAZIONALE DI RICERCHE IN ANTARTIDE (PNRA), National Antarctic Research Program with the grant number PNRA16_00105 (Microbial colonization of benthic environments in Antarctica: responses of microbial abundance, diversity and activities and larval settlement to natural or anthropogenic disturbances and search for secondary metabolites; ANT-Biofilm project).
Acknowledgments
The Author is grateful to Maurizio Azzaro (CNR-ISP, Messina, Italy) and Ombretta Dell’Acqua (DISTAV, University of Genoa, Italy) for their help with the experiments performed in Ross Sea. Acknowledgments are also addressed to the other components of ANT-Biofilm Core group: M. Chiantore, S. Fazi, A. Lo Giudice, M. Papale, C. Caroppo, F. Azzaro, R. La Ferla, G. Maimone, P. Laganà, F. Marinelli, F. Berini, E. Binda. Special thanks are due to Gerard Pichon (Barbier Group, Sainte-Sigolène, France), who provided the polyethylene coupons used in the Antarctic experiments.
Conflicts of Interest
The Author declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016, 80, 91–138. [Google Scholar] [CrossRef]
- Qian, P.-Y.; Lau, S.C.K.; Dahms, H.-U.; Dobretsov, S.; Harder, T. Marine biofilms as mediators of colonization by marine macroorganisms: Implications for antifouling and aquaculture. Mar. Biotechnol. 2007, 9, 399–410. [Google Scholar] [CrossRef]
- Dobretsov, S.; Abed, R.M.; Teplitski, M. Mini-review: Inhibition of biofouling by marine microorganisms. Biofouling 2013, 29, 423–441. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef]
- Salta, M.; Wharton, J.A.; Blache, Y.; Stokes, K.R.; Briand, J.F. Marine biofilms on artificial surfaces: Structure and dynamics. Environ. Microbiol. 2013, 15, 2879–2893. [Google Scholar] [CrossRef] [PubMed]
- Anderson, O.R. Marine and estuarine natural microbial biofilms: Ecological and biogeochemical dimensions. AIMS Microbiol. 2018, 2, 304–331. [Google Scholar] [CrossRef]
- Battin, T.J.; Besemer, K.; Bengtsson, M.M.; Romani, A.M.; Packmann, A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016, 14, 251–263. [Google Scholar] [CrossRef]
- Wahl, M.; Goecke, F.; Labes, A.; Dobretsov, S.; Weinberger, F. The second skin: Ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 2012, 3, 292. [Google Scholar] [CrossRef]
- Antunes, J.; Leão, P.; Vasconcelos, V. Marine biofilms: Diversity of communities and of chemical cues. Environ. Microbiol. Rep. 2019, 11, 287–305. [Google Scholar] [CrossRef]
- Dobretsov, S.; Abed, R.M.; Voolstra, C.R. The effect of surface colour on the formation of marine micro and macrofouling communities. Biofouling 2013, 29, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-F.; Guo, X.-P.; Chen, Y.-R.; Ding, D.-W.; Yang, J.-L. Comparative analysis of biofilm community on different coloured substrata in relation to mussel settlement. J. Mar. Biol. Assoc. UK 2016, 97, 81–89. [Google Scholar] [CrossRef]
- Arthur, C.; Baker, J.; Bamford, H. (Eds.) NOAA Technical Memorandum NOS-OR&R-30. In Proceedings of the International Research Workshop on the Occurrence, Effects, and Fate of Microplastic Marine Debris, Tacoma, WA, USA, 9–11 September 2008; NOAA: Silver Spring, MD, USA, 2009. [Google Scholar]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Geesey, G.G.; Gillis, R.J.; Avci, R.; Daly, D.; Hamilton, M.; Shope, P.; Harkin, G. The influence of surface features on bacterial colonization and subsequent substratum chemical changes of 316L stainless steel. Corros. Sci. 1996, 38, 73–95. [Google Scholar] [CrossRef]
- Tolker-Nielsen, T.; Molin, S. Spatial organization of microbial biofilm communities. Microb. Ecol. 2000, 40, 75–84. [Google Scholar] [CrossRef]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef]
- Besemer, K.; Singer, G.; Limberger, R.; Chlup, A.K.; Hochedlinger, G.; Hödl, I.; Baranyi, C.; Battin, T.J. Biophysical controls on community succession in stream biofilms. Appl. Environ. Microbiol. 2007, 73, 4966–4974. [Google Scholar] [CrossRef]
- Cowan, M.M.; Warren, T.M.; Fletcher, M. Mixed-species colonization of solid surfaces in laboratory biofilms. Biofouling 1991, 3, 23–34. [Google Scholar] [CrossRef]
- Dang, H.; Lovell, C.R. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 2000, 66, 467–475. [Google Scholar] [CrossRef]
- Lappin-Scott, H.M.; Costerton, J.W. Bacterial biofilms and surface fouling. Biofouling 1989, 1, 323–342. [Google Scholar] [CrossRef]
- Di Donato, P.; Poli, A.; Taurisano, V.; Abbamondi, G.R.; Nicolaus, B.; Tommonaro, G. Recent Advances in the Study of Marine Microbial Biofilm: From the Involvement of Quorum Sensing in Its Production up to Biotechnological Application of the Polysaccharide Fractions. J. Mar. Sci. Eng. 2016, 4, 34. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M. Attachment of Pseudomonas fluorescens to glass and influence of electrolytes on bacterium-substratum separation distance. J. Bacteriol. 1988, 170, 2027–2030. [Google Scholar] [CrossRef] [PubMed]
- Characklis, W.G.; McFeters, G.A.; Marshall, K.C. Physiological ecology in biofilm systems. In Biofilms; Characklis, W.G., Marshall, K.C., Eds.; John Wiley & Sons: New York, NY, USA, 1990; pp. 341–394. [Google Scholar]
- Chiu, J.M.Y.; Thiyagarajan, V.; Tsoi, M.M.Y.; Qian, P.Y. Qualitative and quantitative changes in marine biofilms as a function of temperature and salinity in summer and winter. Biofilms 2005, 2, 183–195. [Google Scholar] [CrossRef]
- Lau, S.C.K.; Thiyagarajan, V.; Cheung, S.C.K.; Qian, P.Y. Roles of bacterial community composition in biofilms as a mediator for larval settlement of three marine invertebrates. Aquat. Microb. Ecol. 2005, 38, 41–45. [Google Scholar] [CrossRef]
- Webster, N.S.; Smith, L.D.; Heyward, A.J.; Watts, J.E.M.; Webb, R.I.; Blackall, L.L.; Negri, A.P. Metamorphosis of a scleractinian coral in response to microbial biofilms. Appl. Environ. Microbiol. 2004, 70, 1213–1221. [Google Scholar] [CrossRef]
- Chiu, J.; Zhang, R.; Wang, H.; Thiyagarajan, V.; Qian, P. Nutrient effects on intertidal community: From bacteria to invertebrates. Mar. Ecol. Prog. Ser. 2008, 358, 41–50. [Google Scholar] [CrossRef][Green Version]
- Dobretsov, S.; Qian, P.-Y. Facilitation and inhibition of larval attachment of the bryozoan Bugula neritina in association with mono-species and multi-species biofilms. J. Exp. Mar. Biol. Ecol. 2006, 333, 263–274. [Google Scholar] [CrossRef]
- Babin, M.; Roesler, C.S.; Cullen, J.J. Real-time observation systems for ecosystem dynamics and harmful algal blooms: Theory, instrumentation and modelling. In Biofouling and Underwater Measurements; Lehaitre, M., Compère, C., Eds.; Oceanographic Methodology Series; UNESCO: Paris, France, 2008; pp. 463–493. [Google Scholar]
- Oberbeckmann, S.; Osborn, A.M.; Duhaime, M.B. Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PLoS ONE 2016, 11, e0159289. [Google Scholar] [CrossRef]
- Scheuerman, T.R.; Camper, A.K.; Hamilton, M.A. Effects of substratum topography on bacterial adhesion. J. Colloid Interf. Sci. 1998, 208, 23–33. [Google Scholar] [CrossRef]
- Crawford, R.J.; Webb, H.K.; Truong, V.K.; Hasan, J.; Ivanova, E.P. Surface topographical factors influencing bacterial attachment. Adv. Colloid Interface Sci. 2012, 179, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.J.; Lee, N.; Jo, W.; Jung, W.K.; Lim, J. Effects of substrates on biofilm formation observed by atomic force microscopy. Ultramicroscopy 2009, 109, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Mitik-Dineva, N.; Wang, J.; Mocanasu, R.C.; Stoddart, P.R.; Crawford, R.J.; Ivanova, E.P. Impact of nano-topography on bacterial attachment. Biotechnol. J. 2008, 3, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, M.; Loeb, G.I. Influence of substratum characteristics on the attachment of a marine Pseudomonad to solid surfaces. Appl. Environ. Microbiol. 1979, 37, 67–72. [Google Scholar] [CrossRef]
- von Ammon, U.; Wood, S.A.; Laroche, O.; Zaiko, A.; Tait, L.; Lavery, S.; Inglis, G.; Pochon, X. The impact of artificial surfaces on marine bacterial and eukaryotic biofouling assemblages: A high-throughput sequencing analysis. Mar. Environ. Res. 2018, 133, 57–66. [Google Scholar] [CrossRef]
- Catão, E.C.P.; Pollet, T.; Misson, B.; Garnier, C.; Ghiglione, J.-F.; Barry-Martinet, R.; Maintenay, M.; Bressy, C.; Briand, J.-F. Shear stress as a major driver of marine biofilm communities in the NW Mediterranean Sea. Front. Microbiol. 2019, 10, 1768. [Google Scholar] [CrossRef]
- Stewart, P.S.; Hamilton, M.A.; Goldstein, B.R.; Schneider, B.T. Modeling biocide action against biofilms. Biotechnol. Bioeng. 1996, 49, 445–455. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Tian, R.; Bougouffa, S.; Yang, B.; Cao, H.L.; Zhang, G.; Wong, Y.H.; Xu, W.; Batang, Z.; et al. Species sorting during biofilm assembly by artificial substrates deployed in a cold seep system. Sci. Rep. 2015, 4, 6647. [Google Scholar] [CrossRef]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “Plastisphere”: Microbial Communities on Plastic Marine Debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef]
- Urbanek, A.K.; Rymowicz, W.; Mirończuk, A.M. Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl. Microbiol. Biotechnol. 2018, 102, 7669–7678. [Google Scholar] [CrossRef]
- Sudhakar, M.; Priyadarshini, C.; Doble, M.; Murthy, S.; Venkatesan, R. Marine bacteria mediated degradation of nylon 66 and 6. Int. Biodeterior. Biodegrad. 2007, 60, 144–151. [Google Scholar] [CrossRef]
- Terlizzi, A.; Faimali, M. Fouling on artificial substrata. In Biofouling; Dürr, S., Thomason, J.C., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2010; pp. 170–179. [Google Scholar]
- Cooksey, K.E.; Wigglesworth-Cooksey, B. Adhesion of bacteria and diatoms to surfaces in the sea: A review. Aquat. Microb. Ecol. 1995, 9, 87–96. [Google Scholar] [CrossRef]
- Singh, R.; Paul, D.; Jain, R.K. Biofilms: Implications in bioremediation. Trends Microbiol. 2006, 14, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms and device-associated infections. Emerg. Infect. Dis. 2001, 7, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Huggett, M.J.; Nedved, B.T.; Hadfield, M.G. Effects of initial surface wettability on biofilm formation and subsequent settlement of Hydroides elegans. Biofouling 2009, 25, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Lee, O.; Huang, Y.; Mok, S.Y.; Kolter, R.; Qian, P.Y. Bacterial community succession and chemical profiles of subtidal biofilms in relation to larval settlement of the polychaete Hydroides elegans. ISME J. 2010, 4, 817–828. [Google Scholar] [CrossRef]
- Witt, V.; Wild, C.; Anthony, K.R.; Diaz-Pulido, G.; Uthicke, S. Effects of ocean acidification on microbial community composition of, and oxygen fluxes through, biofilms from the Great Barrier Reef. Environ. Microbiol. 2011, 13, 2976–2989. [Google Scholar] [CrossRef]
- Bellou, N.; Papathanassiou, E.; Dobretsov, S.; Lykousis, V.; Colijn, F. The effect of substratum type, orientation and depth on the development of bacterial deep-sea biofilm communities grown on artificial substrata deployed in the Eastern Mediterranean. Biofouling 2012, 28, 199–213. [Google Scholar] [CrossRef]
- Khandekar, N.; Johns, R.B. Marine corrosion studies-II. A biomarker study tracing the early formation of biofilms on steel plates in a marine environment. Org. Geochem. 1990, 15, 531–538. [Google Scholar] [CrossRef]
- Jones, P.R.; Cottrell, M.T.; Kirchman, D.L.; Dexter, S.C. Bacterial community structure of biofilms on artificial surfaces in an estuary. Microb. Ecol. 2007, 53, 153–162. [Google Scholar] [CrossRef]
- Mitik-Dineva, N.; Wang, J.; Truong, V.K.; Stoddart, P.R.; Malherbe, F.; Crawford, R.J.; Ivanova, E.P. Differences in colonisation of five marine bacteria on two types of glass surfaces. Biofouling 2009, 25, 621–631. [Google Scholar] [CrossRef]
- Landoulsi, J.; Cooksey, K.E.; Dupres, V. Review—Interactions between diatoms and stainless steel: Focus on biofouling and biocorrosion. Biofouling 2011, 27, 1105–1124. [Google Scholar] [CrossRef]
- Li, X.; Duan, J.; Xiao, H.; Li, Y.; Liu, H.; Guan, F.; Zhai, X. Analysis of Bacterial Community Composition of Corroded Steel Immersed in Sanya and Xiamen Seawaters in China via Method of Illumina MiSeq Sequencing. Front. Microbiol. 2017, 8, 1737. [Google Scholar] [CrossRef]
- Head, R.; Davenport, J.; Thomason, J.C. The effect of depth on the accrual of marine biofilms on glass substrata deployed in the Clyde Sea, Scotland. Biofouling 2004, 20, 177–180. [Google Scholar] [CrossRef]
- Patil, J.S.; Chandrashekar Anil, A. Biofilm diatom community structure: Influence of temporal and substratum variability. Biofouling 2005, 21, 189–206. [Google Scholar] [CrossRef]
- Mitbavkar, S.; Raghu, C.; Rajaneesh, K.M.; Pavan, D. Picophytoplankton community from tropical marine biofilms. J. Exp. Mar. Biol. Ecol. 2012, 426, 88–96. [Google Scholar] [CrossRef]
- Siboni, N.; Lidor, M.; Kramarsky-Winter, E.; Kushmaro, A. Conditioning film and initial biofilm formation on ceramics tiles in the marine environment. FEMS Microbiol. Lett. 2007, 274, 24–29. [Google Scholar] [CrossRef]
- Kang, J.; Kang, S.; Kim, Y.-T. Study on the control of marine biofouling developed on the surface of porous ceramics. J. Korean Cryst. Growth Cryst. Technol. 2015, 25, 218–224. [Google Scholar] [CrossRef]
- Harrison, H.P.; Sapp, M.; Schratzberger, M.; Osborn, A.M. Interactions between microorganisms and marine microplastics: A call for research. Mar. Technol. Soc. J. 2011, 45, 12–20. [Google Scholar] [CrossRef]
- Auta, H.S.; Emenike, C.U.; Fauziah, S.H. Distribution and importance of microplastics in the marine environment: A review of the sources, fate, effects, and potential solutions. Environ. Int. 2017, 102, 165–176. [Google Scholar] [CrossRef]
- Thompson, R.C.; Olsen, Y.; Mitchell, R.P.; Davis, A.; Rowland, S.J.; John, A.W.; McGonigle, D.; Russell, A.E. Lost at sea: Where is all the plastic? Science 2004, 304, 838. [Google Scholar] [CrossRef]
- Moore, C.J.; Moore, S.L.; Weisberg, S.B.; Lattin, G.L.; Zellers, A.F. A comparison of neustonic plastic and zooplankton abundance in southern California’s coastal waters. Mar. Pollut. Bull. 2002, 44, 1035–1038. [Google Scholar] [CrossRef]
- Lattin, G.L.; Moore, C.J.; Zellers, A.F.; Moore, S.L.; Weisberg, S.B. A comparison of neustonic plastic and zooplankton at different depths near the southern California shore. Mar. Pollut. Bull. 2004, 49, 291–294. [Google Scholar] [CrossRef]
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as contaminants in the marine environment: A review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef]
- Ballent, A.; Pando, S.; Purser, A.; Juliano, M.F.; Thomsen, L. Modelled transport of benthic marine microplastic pollution in the Nazaré Canyon. Biogeosciences 2013, 10, 7957–7970. [Google Scholar] [CrossRef]
- Browne, M.A.; Galloway, T.; Thompson, R. Microplastic—An emerging contaminant of potential concern? Integr. Environ. Assess. Manag. 2007, 3, 559–566. [Google Scholar] [CrossRef]
- Andrady, A.L. Microplastics in the marine environment. Mar. Pollut. Bull. 2011, 62, 1596–1605. [Google Scholar] [CrossRef]
- Lobelle, D.; Cunliffe, M. Early microbial biofilm formation on marine plastic debris. Mar. Pollut. Bull. 2011, 62, 197–200. [Google Scholar] [CrossRef]
- Corcoran, P.L. Benthic plastic debris in marine and fresh water environments. Environ. Sci. Process. Impacts 2015, 8, 1363–1369. [Google Scholar] [CrossRef]
- Corcoran, P.L.; Biesinger, M.C.; Grifi, M. Plastics and beaches: A degrading relationship. Mar. Pollut. Bull. 2009, 58, 80–84. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Loeder, M.G.; Gerdts, G.; Osborn, A.M. Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. FEMS Microbiol. Ecol. 2014, 90, 478–492. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Wu, D.; Xia, J.; Shi, H.; Kim, H. Influence of physicochemical surface properties on the adhesion of bacteria onto four types of plastics. Sci. Total Environ. 2019, 671, 1101–1107. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; Zettler, E.R.; Slikas, B.; Boyd, G.D.; Melvin, D.W.; Morrall, C.E.; Proskurowski, G.; Mincer, T.J. Biogeography of the Plastisphere: Implications for Policy. Front. Ecol. Environ. 2015, 13, 541–546. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2018, 8, 2709. [Google Scholar] [CrossRef] [PubMed]
- Briand, J.-F.; Djeridi, I.; Jamet, D.; Coupé, S.; Bressy, C.; Molmeret, M.; Le Berre, B.; Rimet, F.; Bouchez, A.; Blache, Y. Pioneer marine biofilms on artificial surfaces including antifouling coatings immersed in two contrasting French Mediterranean coast sites. Biofouling 2012, 28, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Carson, H.S.; Nerheim, M.S.; Carroll, K.A.; Eriksen, M. The plastic-associated microorganisms of the North Pacific Gyre. Mar. Pollut. Bull. 2013, 75, 126–132. [Google Scholar] [CrossRef]
- Reisser, J.; Shaw, J.; Hallegraeff, G.; Proietti, M.; Barnes, D.K.; Thums, M.; Wilcox, C.; Hardesty, B.D.; Pattiaratchi, C. Millimeter-sized marine plastics: A new pelagic habitat for microorganisms and invertebrates. PLoS ONE 2014, 9, e100289. [Google Scholar] [CrossRef] [PubMed]
- Khandeparker, L.; D’Costa, P.M.; Anil, A.C.; Sawant, S.S. Interactions of bacteria with diatoms: Influence on natural marine biofilms. Mar. Ecol. 2014, 35, 233–248. [Google Scholar] [CrossRef]
- Amin, S.A.; Parker, M.S.; Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol. Mol. Biol. Rev. 2012, 76, 667–684. [Google Scholar] [CrossRef] [PubMed]
- Abell, G.C.; Bowman, J.P. Colonization and community dynamics of class Flavobacteria on diatom detritus in experimental mesocosms based on southern ocean seawater. FEMS Microbiol. Ecol. 2005, 53, 379–391. [Google Scholar] [CrossRef]
- Jacquin, J.; Cheng, J.; Odobel, C.; Pandin, C.; Conan, P.; Pujo-Pay, M.; Barbe, V.; Meistertzheim, A.L.; Ghiglione, J.F. Microbial Ecotoxicology of Marine Plastic Debris: A Review on Colonization and Biodegradation by the “Plastisphere”. Front. Microbiol. 2019, 10, 865. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.A.; Clemente, T.M.; Viviani, D.A.; Fong, A.A.; Thomas, K.A.; Kemp, P.; Karl, D.M.; White, A.E.; DeLong, E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems 2016, 1, e00024-16. [Google Scholar] [CrossRef] [PubMed]
- De Tender, C.; Devriese, L.I.; Haegeman, A.; Maes, S.; Vangeyte, J.; Cattrijsse, A.; Dawyndt, P.; Ruttink, T. Temporal dynamics of bacterial and fungal colonization on plastic debris in the North Sea. Environ. Sci. Technol. 2017, 51, 7350–7360. [Google Scholar] [CrossRef] [PubMed]
- Dussud, C.; Meistertzheim, A.L.; Conan, P.; Pujo-Pay, M.; George, M.; Fabre, P.; Coudane, J.; Higgs, P.; Elineau, A.; Pedrotti, M.L.; et al. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ. Pollut. 2018, 236, 807–816. [Google Scholar] [CrossRef]
- Delacuvellerie, A.; Cyriaque, V.; Gobert, S.; Benali, S.; Wattiez, R. The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 2019, 380, 120899. [Google Scholar] [CrossRef]
- Hung, O.S.; Thiyagarajan, V.; Zhang, R.; Wu, R.S.S.; Qian, P.Y. Attachment of Balanus amphitrite larvae to biofilms originating from contrasting environments. Mar. Ecol. Prog. Ser. 2007, 333, 229–242. [Google Scholar] [CrossRef][Green Version]
- Balasubramanian, V.; Palanichamy, S.; Subramanian, G.; Rajaram, R. Development of polyvinyl chloride biofilms for succession of selected marine bacterial populations. J. Environ. Biol. 2012, 33, 57–60. [Google Scholar]
- Briand, J.-F.; Barani, A.; Garnieri, C.; Réhel, K.; Urvois, F.; LePoupon, C.; Bouchez, A.; Debroas, D.; Bressy, C. Spatio-temporal variations of marine biofilm communities colonizing artificial substrata including antifouling coatings in contrasted French coastal environments. Microb. Ecol. 2017, 74, 585–598. [Google Scholar] [CrossRef]
- Misic, C.; Harriague, A.C. Development of marine biofilm on plastic: Ecological features in different seasons, temperatures, and light regimes. Hydrobiologia 2019, 835, 129–145. [Google Scholar] [CrossRef]
- O’Connor, N.J.; Richardson, D.L. Attachment of barnacle (Balanus amphitrite Darwin) larvae: Responses to bacterial films and extracellular materials. J. Exp. Mar. Biol. Ecol. 1998, 226, 115–129. [Google Scholar] [CrossRef]
- Leroy, C.; Delbarre-Ladrat, C.; Ghillebaert, F.; Rochet, M.J.; Compère, C.; Combes, D. A marine bacterial adhesion microplate test using the DAPI fluorescent dye: A new method to screen antifouling agents. Lett. Appl. Microbiol. 2007, 44, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Min, G.-S.; Choi, J.-K.; Jung, J.-H.; Park, M.-H. An approach to analyses of periphytic ciliate colonization for monitoring water quality using a modified artificial substrate in Korean coastal waters. Mar. Pollut. Bull. 2009, 58, 1278–1285. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Kim, J.-A.; Kim, H.; Chung, K.-S.; Yoon, H.-O. Identification of preponderant marine bacteria and their biofouling characteristics on adsorbents of different sizes and shapes in seawater. J. Mar. Sci. Technol. 2018, 26, 458–464. [Google Scholar] [CrossRef]
- Lee, Y.K.; Kwon, K.K.; Cho, K.H.; Park, J.H.; Lee, H.K. Isolation and identification of Bacteria from marine biofilms. Key Eng. Mater. 2005, 277, 612–617. [Google Scholar] [CrossRef]
- Mejdandžić, M.; Ivanković, T.; Pfannkuchen, M.; Godrijan, J.M.; Pfannkuchen, D.M.; Hrenović, J.; Ljubešić, Z. Colonization of diatoms and bacteria on artificial substrates in the northeastern coastal Adriatic Sea. Acta Bot. Croat. 2015, 74, 407–422. [Google Scholar] [CrossRef]
- Guezennec, J.; Ortega-Morales, O.; Raguenes, G.; Geesey, G. Bacterial colonization of artificial substrate in the vicinity of deep-sea hydrothermal vents. FEMS Microbiol. Ecol. 1998, 26, 89–99. [Google Scholar] [CrossRef]
- Artham, T.; Sudhakar, M.; Venkatesan, R.; Madhavan Nair, C.; Murty, K.V.G.K.; Doble, M. Biofouling and stability of synthetic polymers in sea water. Int. Biodeterior. Biodegrad. 2009, 63, 884–890. [Google Scholar] [CrossRef]
- Witt, V.; Wild, C.; Uthicke, S. Effect of substrate type on bacterial community composition in biofilms from the Great Barrier Reef. FEMS Microbiol. Lett. 2011, 323, 188–195. [Google Scholar] [CrossRef]
- Hoellein, T.; Rojas, M.; Pink, A.; Gasior, J.; Kelly, J. Anthropogenic litter in urban freshwater ecosystems: Distribution and microbial interactions. PLoS ONE 2014, 9, e98485. [Google Scholar] [CrossRef]
- Frère, L.; Paul-Pont, I.; Rinnert, E.; Petton, S.; Jaffré, J.; Bihannic, I.; Soudant, P.; Lambert, C.; Huvet, A. Influence of environmental and anthropogenic factors on the composition, concentration and spatial distribution of microplastics: A case study of the Bay of Brest (Brittany, France). Environ. Pollut. 2017, 225, 211–222. [Google Scholar] [CrossRef]
- Dussud, C.; Hudec, C.; George, M.; Fabre, P.; Higgs, P.; Bruzaud, S.; Delort, A.-M.; Eyheraguibel, B.; Meistertzheim, A.-L.; Jacquin, J.; et al. Colonization of Non-biodegradable and Biodegradable Plastics by Marine Microorganisms. Front. Microbiol. 2018, 9, 1571. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, I.V.; Wichels, A.; Krohne, G.; Gerdts, G. Mature biofilm communities on synthetic polymers in seawater—Specific or general? Mar. Environ. Res. 2018, 142, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Ogonowski, M.; Motiei, A.; Ininbergs, K.; Hell, E.; Gerdes, Z.; Udekwu, K.I.; Bacsik, Z.; Gorokhova, E. Evidence for selective bacterial community structuring on microplastics. Environ. Microbiol. 2018, 20, 2796–2808. [Google Scholar] [CrossRef] [PubMed]
- Muthukrishnan, T.; Al Khaburi, M.; Abed, R.M.M. Fouling Microbial Communities on Plastics Compared with Wood and Steel: Are They Substrate- or Location-Specific? Microb. Ecol. 2019, 78, 361–374. [Google Scholar] [CrossRef]
- Pinto, M.; Langer, T.M.; Hüffer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165. [Google Scholar] [CrossRef]
- Scotto, V.; Alabiso, G.; Marcenaro, G. An example of microbiologically influenced corrosion: The behaviour of stainless steels in natural seawater. Bioelectrochem. Bioenerg. 1986, 16, 347–355. [Google Scholar] [CrossRef]
- Alabiso, G.; Montini, U.; Milillo, M. One year corrosion test of four stainless steel grades at Ny-Alesund (Svalbard Islands): June 2004–June 2005. In CNR-IAMC Technical Report; National Research Council: Taranto, Italy, 2005. [Google Scholar]
- Maki, J.S.; Little, B.J.; Wagner, P.; Mitchell, R. Biofilm formation on metal surfaces in Antarctic waters. Biofouling 1990, 2, 27–38. [Google Scholar] [CrossRef]
- Clark, M.S.; Villota Nieva, L.; Hoffman, J.I.; Davies, A.J.; Trivedi, U.J.; Turner, F.; Ashton, G.V.; Peck, L.S. Lack of long-term acclimation in Antarctic encrusting species suggests vulnerability to warming. Nat. Commun. 2019, 10, 3383. [Google Scholar] [CrossRef]
- Caruso, G.; Azzaro, M.; Dell’Acqua, O.; Lo Giudice, A.; Fazi, S.; Caroppo, C.; Azzaro, F.; La Ferla, R.; Maimone, G.; Laganà, P.; et al. Microbial response to anthropic and natural forcings in two coastal Antarctic sites (Ross Sea). Biol. Mar. Medit. 2019, in press. [Google Scholar]
- Loeb, G.I.; Neihof, R.A. Marine conditioning films. Adv. Chem. 1975, 145, 319–335. [Google Scholar] [CrossRef]
- Bowman, J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs 2007, 5, 220–241. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Qian, P.-Y.; Rittschof, D.; Sreedhar, B.; Chia, F.S. Macrofouling in unidirectional flow: Miniature pipes as experimental models for studying the effects of hydrodynamics on invertebrate larval settlement. Mar. Ecol. Prog. Ser. 1999, 191, 141–151. [Google Scholar] [CrossRef]
- Qian, P.-Y.; Rittschof, D.; Sreedhar, B. Macrofouling in unidirectional flow: Miniature pipes as experimental models for studying interaction of flow and surface characteristics on the attachment of barnacle, bryozoan and polychaete larvae. Mar. Ecol. Prog. Ser. 2000, 207, 109–121. [Google Scholar] [CrossRef][Green Version]
- Maki, J.S.; Rittschof, D.; Costlow, J.D.; Mitchell, R. Inhibition of attachment of larval barnacles, Balanus amphitrite, by bacterial surface films. Mar. Biol. 1988, 97, 199–206. [Google Scholar] [CrossRef]
- Wieczorek, S.; Todd, C. Inhibition and facilitation of bryozoan and ascidian settlement by natural multi-species biofilms: Effects of film age and the roles of active and passive larval attachment. Mar. Biol. 1997, 128, 463–473. [Google Scholar] [CrossRef]
- Qian, P.-Y.; Thiyagarajan, V.; Lau, S.C.K.; Cheung, S.C.K. Relationship between bacterial community profile in biofilm and attachment of the acorn barnacle Balanus amphitrite. Aquat. Microb. Ecol. 2003, 33, 225–237. [Google Scholar] [CrossRef]
- Chiu, J.M.Y.; Thiyagarajan, V.; Pechenik, J.A.; Hung, O.S.; Qian, P.Y. Influence of bacteria and diatoms in biofilms on metamorphosis of the marine slipper limpet Crepidula onyx. Mar. Biol. 2007, 151, 1417–1431. [Google Scholar] [CrossRef]
- Huang, J.; Shi, Y.; Zeng, G.; Gu, Y.; Chen, G.; Shi, L.; Hu, Y.; Tang, B.; Zhou, J. Acyl-homoserine lactone- based quorum sensing and quorum quenching hold promise to determine the performance of biological wastewater treatments: An overview. Chemosphere 2016, 157, 137–151. [Google Scholar] [CrossRef]
- Dobretsov, S.; Dahms, H.-U.; YiLi, H.; Wahl, M.; Qian, P.-Y. The effect of quorum-sensing blockers on the formation of marine microbial communities and larval attachment. FEMS Microbiol. Ecol. 2007, 60, 177–188. [Google Scholar] [CrossRef]
- Landini, P.; Antoniani, D.; Burgess, J.G.; Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 2010, 86, 813–823. [Google Scholar] [CrossRef]
- de Carvalho, C.C.C.R. Marine biofilms: A successful microbial strategy with economic implications. Front. Mar. Sci. 2018, 5, 126. [Google Scholar] [CrossRef]
- Dobretsov, S.; Coutinho, R.; Rittschof, D.; Salta, M.; Ragazzola, F.; Hellio, C. The oceans are changing: Impact of ocean warming and acidification on biofouling communities. Biofouling 2019, 35, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ding, W.; Li, Y.-X.; Tam, C.; Bougouffa, S.; Wang, R.; Pei, B.; Chiang, H.; Leung, P.; Lu, Y.; et al. Marine biofilms constitute a bank of hidden microbial diversity and functional potential. Nat. Commun. 2019, 10, 517. [Google Scholar] [CrossRef] [PubMed]
- Battin, T.; Kaplan, L.; Denis Newbold, J.; Hansen, C.M.E. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 2003, 426, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Bond, P.L. Microbial community proteomics: Elucidating the catalysts and metabolic mechanisms that drive the Earth’s biogeochemical cycles. Curr. Opin. Microbiol. 2009, 12, 310–317. [Google Scholar] [CrossRef]
- Moss, J.A.; Nocker, A.; Lepo, J.E.; Snyder, R.A. Stability and change in estuarine biofilm bacterial community diversity. Appl. Environ. Microbiol. 2006, 72, 5679–5688. [Google Scholar] [CrossRef]
- Webster, N.S. and Negri, A.P. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 2006, 8, 1177–1190. [Google Scholar] [CrossRef]
- Dang, H.; Li, T.; Chen, M.; Huang, G. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 2008, 74, 52–60. [Google Scholar] [CrossRef]
- Kriwy, P.; Uthicke, S. Microbial diversity in marine biofilms along a water quality gradient on the Great Barrier Reef. Syst. Appl. Microbiol. 2011, 34, 116–126. [Google Scholar] [CrossRef]
- Huang, Y.; Zeng, Y.; Yu, Z.; Zhang, J.; Feng, H.; Lin, X. In silico and experimental methods revealed highly diverse bacteria with quorum sensing and aromatics biodegradation systems—A potential broad application on bioremediation. Bioresour. Technol. 2013, 148, 311–316. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).