Quantitative Real-Time Polymerase Chain Reaction (PCR) Assay for Rapid Monitoring of the Harmful Algal Bloom Species Cochlodinium polykrikoides
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Cochlodinium polykrikoides
2.2. Design of Cochlodinium polykrikoides-Specific Primers
2.3. Development of LSU Standard Materials for Cochlodinium polykrikoides
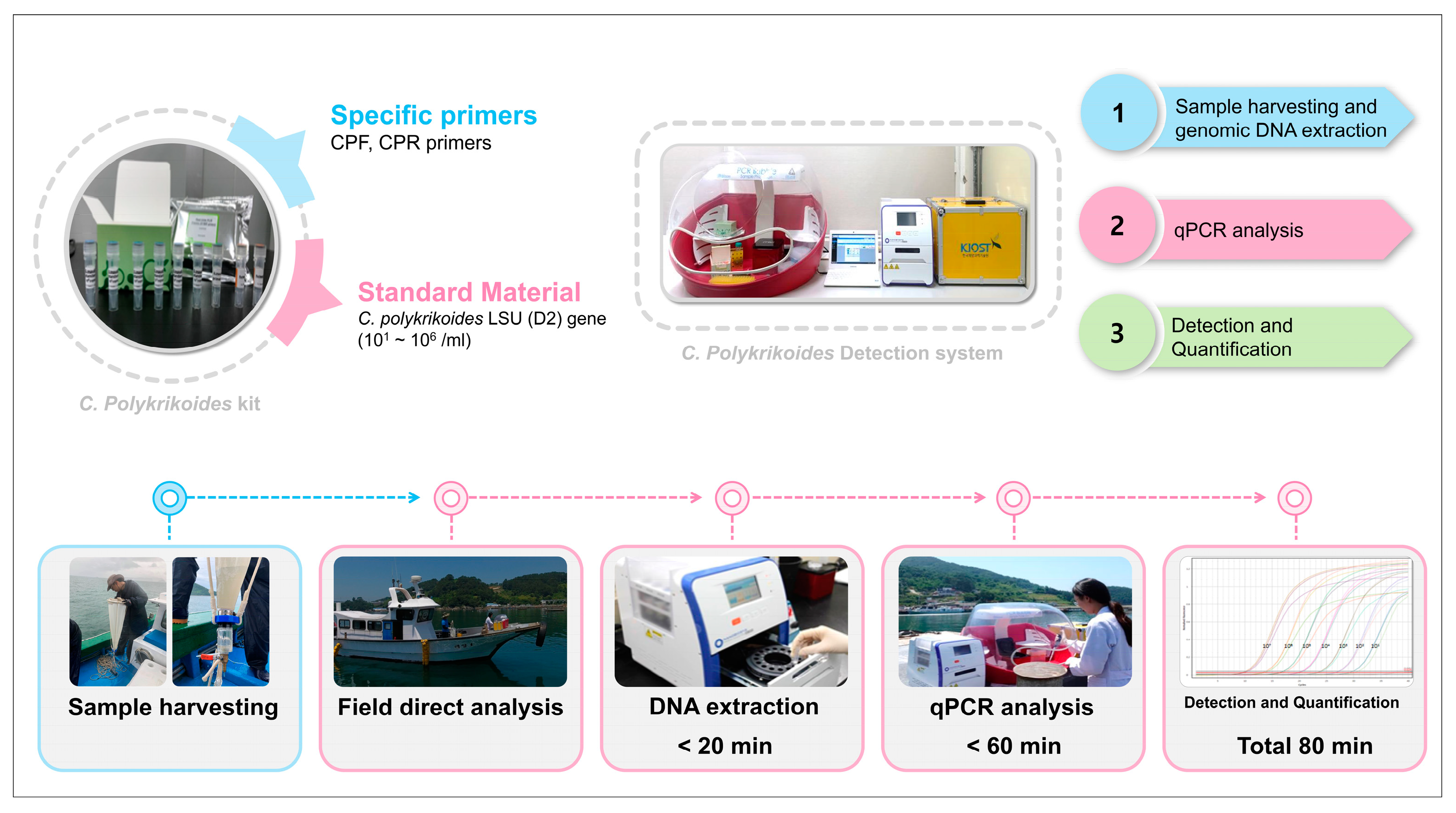
2.4. Development of a Cochlodinium polykrikoides Detection System
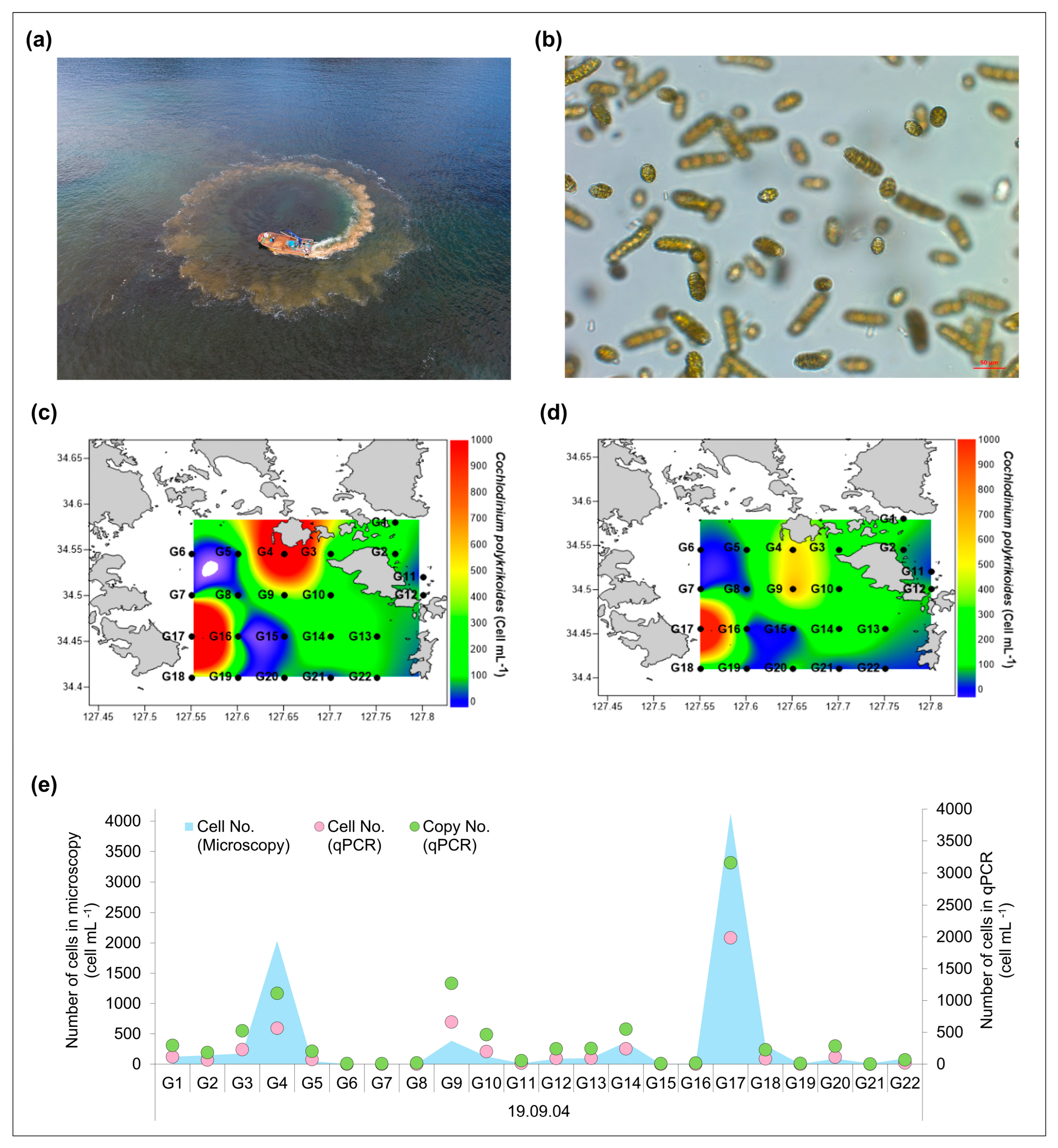
2.5. Field Application of the Cochlodinium polykrikoides Detection System
3. Results and Discussion
3.1. Evaluation of the Cochlodinium polykrikoides-Specific Primers
3.2. Verification Through Field Application
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ternon, E.; Pavaux, A.-S.; Marro, S.; Thomas, O.P.; Lemée, R. Allelopathic interactions between the benthic toxic dinoflagellate Ostreopsis cf. ovata and a co-occurring diatom. Harmful Algae 2018, 75, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Gobler, C.J. Climate change and harmful algal blooms: Insights and perspective. Harmful Algae 2020, 91, 101731. [Google Scholar] [CrossRef] [PubMed]
- Heil, C.A.; Muni-Morgan, A.L. Florida’s harmful algal bloom (HAB) problem: Escalating risks to human, environmental and economic health with climate change. Front Ecol. Evol. 2021, 9, 646080. [Google Scholar] [CrossRef]
- Lee, C.-K.; Park, T.-G.; Park, Y.-T.; Lim, W.-A. Monitoring and trends in harmful algal blooms and red tides in Korean coastal waters, with emphasis on Cochlodinium polykrikoides. Harmful Algae 2013, 30, S3–S14. [Google Scholar] [CrossRef]
- Park, T.G.; Lim, W.A.; Park, Y.T.; Lee, C.K.; Jeong, H.J. Economic impact, management and mitigation of red tides in Korea. Harmful Algae 2013, 30, S131–S143. [Google Scholar] [CrossRef]
- Lee, Y.-C.; Jin, E.; Jung, S.W.; Kim, Y.-M.; Chang, K.S.; Yang, J.-W.; Kim, S.-W.; Kim, Y.-O.; Shin, H.-J. Utilizing the algicidal activity of aminoclay as a practical treatment for toxic red tides. Sci. Rep. 2013, 3, 1292. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-J.; Jeoung, G.; Kim, K.E.; Park, J.S.; Kang, D.; Baek, S.H.; Lee, C.Y.; Kim, H.; Cho, S.; Lee, T.-K.; et al. Co-variance between free-living bacteria and Cochlodinium polykrikoides (Dinophyta) harmful algal blooms, South Korea. Harmful Algae 2023, 122, 102371. [Google Scholar] [CrossRef]
- Kang, D.; Kim, B.K.; Jung, S.W.; Baek, S.H.; Choi, J.-Y.; Cho, H.-Y.; Lee, S.-J.; Kim, H. Development and application of an integrated system for the detection and prediction of harmful algal blooms in Korea. J. Mar. Sci. Eng. 2023, 11, 2207. [Google Scholar] [CrossRef]
- LeGresley, M.; McDermott, G. Counting Chamber Methods for Quantitative Phytoplankton Analysis-Haemocytometer, Palmer-Maloney Cell and Sedgewick-Rafter Cell. In United Nations Educational, Scientific and Cultural Organization (IOC Manuals and Guides); UNESCO: Paris, France, 2010; Volume 55, pp. 25–30. [Google Scholar]
- Antonella, P.; Luca, G. The quantitative real-time PCR applications in the monitoring of marine harmful algal bloom (HAB) species. Environ. Sci. Pollut. Res. 2013, 20, 6851–6862. [Google Scholar] [CrossRef] [PubMed]
- Karlson, B.; Cusack, C.; Bresnan, E. (Eds.) Microscopic and Molecular Methods for Quantitative Phytoplankton Analysis; UNESCO: Paris, France, 2010; p. 110. [Google Scholar]
- Medlin, L. Molecular tools for monitoring harmful algal blooms. Environ. Sci. Pollut. Res. Int. 2013, 20, 6683–6685. [Google Scholar] [CrossRef]
- Lee, H.-G.; Kim, H.M.; Min, J.; Park, C.; Jeong, H.J.; Lee, K.; Kim, K.Y. Quantification of the paralytic shellfish poisoning dinoflagellate Alexandrium species using a digital PCR. Harmful Algae 2020, 92, 101726. [Google Scholar] [CrossRef] [PubMed]
- Feist, S.M.; Lance, R.F. Genetic detection of freshwater harmful algal blooms: A review focused on the use of environmental DNA (eDNA) in Microcystis aeruginosa and Prymnesium parvum. Harmful Algae 2021, 110, 102124. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Y.; Lai, J.; Li, J.; Yu, K. Improvement and application of qPCR assay revealed new insight on early warning of Phaeocystis globosa bloom. Water Res. 2023, 229, 119439. [Google Scholar] [CrossRef]
- Kim, W.; Han, T.; Jung, S.; Kang, D. Analysis on the Optical Absorption Property of Sea Waters Dominated by Alexandrium affine in Coastal Waters off Tongyeong, 2017. Photogramm. Cartogr. 2019, 37, 563–570. [Google Scholar]
- Choi, J.K.; Min, J.E.; Noh, J.H.; Han, T.H.; Yoon, S.; Park, Y.J.; Park, J.H. Harmful algal bloom (HAB) in the East Sea identified by the Geostationary Ocean Color Imager (GOCI). Harmful Algae 2014, 39, 295–302. [Google Scholar] [CrossRef]
- Dizaji, S.Z.; Fariman, G.A.; Zahedi, M.M. Pigment content analysis in two HAB forming dinoflagellate species during the growth period. J. Appl. Phycol. 2021, 33, 807–817. [Google Scholar] [CrossRef]
- Medlin, L.K.; Orozco, J. Molecular techniques for the detection of organisms in aquatic environments, with emphasis on harmful algal bloom species. Sensors 2017, 17, 1184. [Google Scholar] [CrossRef]
- Hernández-López, E.L.; Gasperin, J.; Bernáldez-Sarabia, J.; Licea-Navarro, A.F.; Guerrero, A.; Lizárraga-Partida, M.L. Detection of Alcanivorax spp., Cycloclasticus spp., and Methanomicrobiales in water column and sediment samples in the Gulf of Mexico by qPCR. Environ. Sci. Pollut. Res. Int. 2019, 26, 35131–35139. [Google Scholar] [CrossRef]
- Engesmo, A.; Strand, D.; Gran-Stadniczeñko, S.; Edvardsen, B.; Medlin, L.K.; Eikrem, W. Development of a qPCR assay to detect and quantify ichthyotoxic flagellates along the Norwegian coast, and the first Norwegian record of Fibrocapsa japonica (Raphidophyceae). Harmful Algae 2018, 75, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, R.G.; Bean, T.; Turner, A.D.; Lees, D.N.; Lowther, J.; Lewis, A.; Baker-Austin, C. Development of a TaqMan qPCR assay for detection of Alexandrium spp. and application to harmful algal bloom monitoring. Toxicon X 2019, 2, 100011. [Google Scholar] [CrossRef]
- Jung, S.W.; Joo, H.M.; Park, J.S.; Lee, J.H. Development of a rapid and effective method for preparing delicate dinoflagellates for scanning electron microscopy. J. Appl. Phycol. 2010, 22, 313–317. [Google Scholar] [CrossRef]
- Joanes, D.N.; Gill, C.A. Comparing measures of sample skewness and kurtosis Soc Ser D-Stat 47(1). J. R. Stat. Soc. Ser. D 1998, 47, 183–189. [Google Scholar] [CrossRef]
- Dhar, S.S.; Chakraborty, B.; Chaudhuri, P. Comparison of multivariate distributions using quantile–quantile plots and related tests. Bernoulli 2014, 20, 1484–1506. [Google Scholar] [CrossRef]
- Jung, S.W.; Kang, D.; Kim, H.-J.; Shin, H.H.; Park, J.S.; Park, S.Y.; Lee, T.-K. Mapping distribution of cysts of recent dinoflagellate and Cochlodinium polykrikoides using next-generation sequencing and morphological approaches in South Sea, Korea. Sci. Rep. 2018, 8, 7011. [Google Scholar] [CrossRef] [PubMed]
- Park, T.G.; Kim, J.J.; Kim, W.J.; Won, K.M. Development of real-time RT-PCR for detecting viable Cochlodinium polykrikoides (Dinophyceae) cysts in sediment. Harmful Algae 2016, 60, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Litaker, R.W.; Vandersea, M.W.; Kibler, S.R.; Reece, K.S.; Stokes, N.A.; Steidinger, K.A.; Millie, D.F.; Bendis, B.J.; Pigg, R.J.; Tester, P.A. Identification of Pfiesteria piscicida (Dinophyceae) and Pfiesteria-like organisms using internal transcribed spacer-specific pcr assays 1. J. Phycol. 2003, 39, 754–761. [Google Scholar] [CrossRef]
- Sonnenberg, R.; Nolte, A.W.; Tautz, D. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front. Zool. 2007, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Park, T.G.; Park, Y.T.; Lee, Y. Development of a SYTO9 based real-time PCR probe for detection and quantification of toxic dinoflagellate Karlodinium veneficum (Dinophyceae) in environmental samples. Phycologia 2009, 48, 32–43. [Google Scholar] [CrossRef]
- Handy, S.M.; Demir, E.; Hutchins, D.A.; Portune, K.J.; Whereat, E.B.; Hare, C.E.; Rose, J.M.; Warner, M.; Farestad, M.; Cary, S.C.; et al. Using quantitative real-time PCR to study competition and community dynamics among Delaware Inland Bays harmful algae in field and laboratory studies. Harmful Algae 2008, 7, 599–613. [Google Scholar] [CrossRef]
- Kim, C.-J.; Kim, H.-G.; Kim, C.-H.; Oh, H.-M. Life cycle of the ichthyotoxic dinoflagellate Cochlodinium polykrikoides in Korean coastal waters. Harmful Algae 2007, 6, 104–111. [Google Scholar] [CrossRef]
- Shin, H.H.; Li, Z.; Yoon, Y.H.; Oh, S.J.; Lim, W.A. Formation and germination of temporary cysts of Cochlodinium polykrikoides Margalef (Dinophyceae) and their ecological role in dense blooms. Harmful Algae 2017, 66, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wang, S.; Du, X.; He, Q.; Liu, Y.; Wang, Z.; Feng, K.; Li, Y.; Deng, Y. ddPCR surpasses classical qPCR technology in quantitating bacteria and fungi in the environment. Mol. Ecol. Resour. 2022, 22, 2587–2598. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.-K.; Lin, Z.-R.; Zhang, Q.-C.; Kong, F.-Z.; Cen, J.-Y.; Zeng, Y.-L.; Yu, R.-C. Detection of bloom-forming dinoflagellates Karenia mikimotoi and Prorocentrum donghaiense using qPCR assays. J. Appl. Phycol. 2022, 34, 1483–1496. [Google Scholar] [CrossRef]
- Gałuszka, A.; Migaszewski, Z.M.; Namieśnik, J. Moving your laboratories to the field–Advantages and limitations of the use of field portable instruments in environmental sample analysis. Environ. Res. 2015, 140, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Kim, H.M.; Min, J.; Kim, K.; Park, M.G.; Jeong, H.J.; Kim, K.Y. An advanced tool, droplet digital PCR (ddPCR), for absolute quantification of the red-tide dinoflagellate, Cochlodinium polykrikoides Margalef (Dinophyceae). Algae 2017, 32, 189–197. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Alenazi, T.H.; AlOmaim, W.S.; Wazzan, A.; Alsufayan, A.; Hasanain, R.A.; Aldibasi, O.S.; Althawadi, S.; Altamimi, S.A.; Mutabagani, M.; et al. Portable RT-PCR system: A rapid and scalable diagnostic tool for COVID-19 testing. J. Clin. Microbiol. 2021, 59, 10.1128. [Google Scholar] [CrossRef]
- Spikmans, V. The evolution of environmental forensics: From laboratory to field analysis. WIREs Forensic Sci. 2019, 1, e1334. [Google Scholar] [CrossRef]
- Lepej, S.Z.; Poljak, M. Portable molecular diagnostic instruments in microbiology: Current status. Clin. Microbiol. Infect. 2020, 26, 411–420. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.-J.; Kim, H.-J.; Park, J.S.; Kang, D.; Cho, S.; Kim, H.; Baek, S.H.; Park, J.J.C.; Han, J.; Kim, K.E.; et al. Quantitative Real-Time Polymerase Chain Reaction (PCR) Assay for Rapid Monitoring of the Harmful Algal Bloom Species Cochlodinium polykrikoides. J. Mar. Sci. Eng. 2025, 13, 277. https://doi.org/10.3390/jmse13020277
Kim M-J, Kim H-J, Park JS, Kang D, Cho S, Kim H, Baek SH, Park JJC, Han J, Kim KE, et al. Quantitative Real-Time Polymerase Chain Reaction (PCR) Assay for Rapid Monitoring of the Harmful Algal Bloom Species Cochlodinium polykrikoides. Journal of Marine Science and Engineering. 2025; 13(2):277. https://doi.org/10.3390/jmse13020277
Chicago/Turabian StyleKim, Min-Jeong, Hyun-Jung Kim, Joon Sang Park, Donhyug Kang, Sungho Cho, Hansoo Kim, Seung Ho Baek, Jordan Jun Chul Park, Jeonghoon Han, Kang Eun Kim, and et al. 2025. "Quantitative Real-Time Polymerase Chain Reaction (PCR) Assay for Rapid Monitoring of the Harmful Algal Bloom Species Cochlodinium polykrikoides" Journal of Marine Science and Engineering 13, no. 2: 277. https://doi.org/10.3390/jmse13020277
APA StyleKim, M.-J., Kim, H.-J., Park, J. S., Kang, D., Cho, S., Kim, H., Baek, S. H., Park, J. J. C., Han, J., Kim, K. E., & Jung, S. W. (2025). Quantitative Real-Time Polymerase Chain Reaction (PCR) Assay for Rapid Monitoring of the Harmful Algal Bloom Species Cochlodinium polykrikoides. Journal of Marine Science and Engineering, 13(2), 277. https://doi.org/10.3390/jmse13020277









