Abstract
This study investigates the effect of intertidal zonation on sediment characteristics, organic matter content, and microbial community distribution in Gomso Bay, Republic of Korea—an ecologically significant estuarine system and part of the UNESCO-designated Getbol, Korean Tidal Flats. It was hypothesized that physicochemical properties and microbial communities differ significantly among the inner, middle, and outer tidal zones due to variations in tidal exposure, sediment texture, and organic matter accumulation. Sediment samples were collected from nine sites across these zones and analyzed for total organic carbon (TOC), acid volatile sulfide (AVS), and trace metals (As, Cd, Cr, Cu, Hg, Li, Ni, Pb, Zn), along with sediment texture. Microbial community structure was examined using 16S rRNA gene amplicon sequencing to evaluate the influence of zonation on microbial diversity and composition. Results revealed distinct spatial variations along the intertidal gradient. Inner tidal zones with finer sediments showed higher TOC, AVS, and metal concentrations, whereas outer zones with coarser sediments exhibited lower values. Microbial composition also varied, with aerobic microorganisms dominating the high tidal flats and anaerobic taxa prevailing in the low tidal flats. Heterotrophic and autotrophic bacteria were more abundant in the outer zone, while chemolithotrophs predominated in the inner zone. Redundancy and Pearson’s correlation analyses further indicated significant relationships between sediment texture, TOC, AVS, heavy metals, and microbial diversity. Overall, the findings confirm that tidal zonation drives distinct sedimentary and microbial patterns, highlighting the ecological complexity of intertidal ecosystems.
1. Introduction
The intertidal ecosystem is of the greatest significance due to its function as a transitional zone between the marine, atmospheric, and terrestrial environments. The intertidal zone is often classified into two separate ecosystems, namely the upper intertidal zone and the lower intertidal zone, based on the duration of inundation. According to Ortega-Morales et al. [1], the flooding of the upper intertidal zone is limited to periods of high tides. The lower intertidal zone is mostly submerged, only emerging during periods of low tide. The reactivity of organic matter is seen to be higher in higher intertidal sediments compared to lower intertidal sediments, attributed to the increased oxygen input [2,3]. Moreover, the injection of freshwater has the potential to alter the characteristics of the sediment. Hwang et al. [4] reported the presence of vertical fluctuations in salinity and nutrient content within the intertidal zone. The changes that have been observed may be related to the interchange of pore water and overlying water within the uppermost 10 cm of surface sediment. The development of a gradient surface sediment is a consequence of dynamic environmental circumstances, resulting in an inherently unstable microorganism habitat [5]. As a result, the microorganisms residing in these zones have significant impacts [6,7].
Microbial communities have a widespread presence throughout marine intertidal habitats. These communities proliferate inside and above sedimentary surfaces and play an essential role in various biogeochemical processes, such as the nitrogen and carbon cycles, as well as the transformation and removal of contaminants [7,8]. Furthermore, the habitats of these organisms are significantly impacted by the chemical composition of sediments and environmental natural processes. Previous studies have shown variations in microbial activities across different sediment depths and within the upper and lower intertidal zones [9]. According to previous research using metabarcoding analysis targeting the 16S rRNA gene, it has been shown that the microbial diversity within tidal flat sediments is influenced by many factors, including the presence of diverse heavy metals, varying levels of nutrients, and the various zonation patterns in the intertidal areas [10,11,12].
Gomso Bay is located on the mid-west coast about 15 km south of the Saemangeum Dyke. Gomso Bay consists of 80% tidal flats and 20% subtidal zone and is divided into an outer bay and an inner bay by the Jujin fluvial channel. It has a typical estuary characterized by a well-developed sand-mud tidal flat with a funnel shape, but receives fresh runoff only from a disproportionately small stream, the Jujin [13]. The majority of the estuary is comprised of large intertidal flats, accounting for almost 80% of its total area. The tidal patterns in the estuary intertidal zone exhibit a semidiurnal [14]. During high tide, tidal flats undergo submersion in water, whereas during low tide, these tidal flats are exposed to atmospheric conditions. Consequently, it can be seen that the physicochemical characteristics of intertidal surface sediment in the estuary appear to be influenced by exposure time [15]. Landsat imagery has revealed the continuous formation of cheniers near the bay’s low-water line, which are gradually pushed shoreline by winter storms and summer typhoons. In the subtidal areas, sediment transport activity is prominent during the summer, especially in the central tidal channel [13]. Notably, suspended mud has been observed to move in two opposite directions simultaneously around the midpoint of the channel-towards both the channel entrance and the bay head-indicating the complex hydrodynamic nature of the system. Due to its ecological significance, particularly as a vital habitat for migratory birds and diverse marine life, Gomso Bay forms part of the Gochang Tidal Flat, one of four core areas designated under the “Getbol, Korean Tidal Flats” UNESCO World Natural Heritage site in 2021. This designation underscores the global importance of the bay’s unique tidal flat ecosystem and its role in supporting biodiversity and coastal environmental stability.
While previous studies have examined either sediment characteristics or microbial communities in tidal flats, comprehensive research integrating sediment texture, chemical composition, and microbial community structure across distinct tidal zones remains limited, particularly in East Asian tidal flat systems. This gap in knowledge is significant as understanding these relationships is essential for predicting ecosystem responses to environmental changes and for effective coastal management. The above-mentioned facts suggest that microbial communities in marine environments are subject to the effects of both physical and chemical factors. Because Gomso Bay is located close to an estuary and has an intertidal flat, variations in the chemical composition of the surface sediment and the structure of the microbial community are expected. In order to get a better understanding of the impact of intertidal zonation on the structure of microbial populations, we performed a study of the surface sediments collected from Gomso Bay. Water content, loss on ignition (LOI), acid volatile sulfide (AVS), heavy metals, and sediment texture were discussed. Using 16S rRNA gene amplicon sequencing, the structure of the microbial community was determined, and the diversity of bacteria present in the sediments was explored. By identifying the relationships between sediment characteristics and microbial community structures across different tidal zones, this study aims to provide baseline data for future monitoring of this UNESCO World Heritage site and to contribute to the broader understanding of microbial ecology in intertidal environments.
2. Materials and Methods
2.1. Study Area and Sediment Sampling
The research focused on the intertidal region of Gomso Bay (Figure 1). The selection of examination sites was based on the geographical characteristics of the coastal zone and tidal surface. Specifically, the sites were categorized into three zones: inner, middle, and outer. Each zone was further divided into three subzones based on the tidal levels: high tide flat (IH, MH, OH), mid-tide flat (IM, MM, OM), and low tidal flat (IL, ML, OL). Surface sediment samples for physicochemical and microbiological examination were obtained in late summer (September) 2023 using a plastic scoop from the outermost layer (~2 cm). The samples were then transported to the laboratory in sealed plastic bags to evaluate the physicochemical characteristics of the sediments. The microbial analysis samples were collected separately in sterile DNase and RNase-free high-density polyethylene (HDPE) tubes (SPL, Pocheon, Republic of Korea). The samples were maintained at a temperature of −20 °C and transported to Macrogen Inc. in Seoul, Republic of Korea, for performing 16S rRNA gene amplicon sequencing.
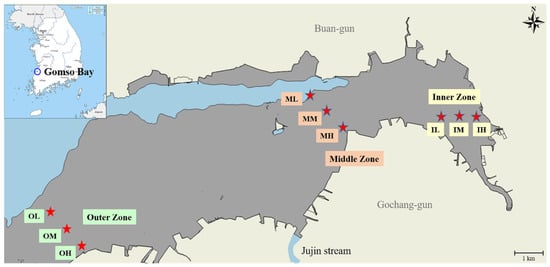
Figure 1.
Map shows Gomso Bay’s intertidal zone on the western coast of Republic of Korea. Tidal zones are indicated by grey area and places of surface sediment sampling are indicated by red stars.
2.2. Physicochemical Analysis
The sediment samples were analyzed for grain size using the standard grain-size approach described by Ingram [16]. To remove the salts in the sediment, wet sediment samples were soaked in purified water and agitated. After decanting purified water, hydrogen peroxide (10%) was used to remove organic matter from the sediment. The sediments were separated into coarse and fine portions using a 63 μm nylon sieve. Particles smaller than 63 μm were analyzed using a laser diffraction particle size analyzer (Mastersizer 3000; Malvern Panalytical, Malvern, UK) and particles larger than 63 μm were analyzed using the standard dry sieving method.
Total organic carbon (TOC) content in sediments was measured using an elemental analyzer (EA, Elementar, Germany) after removing inorganic carbon using 1 M hydrochloric acid (HCL, Sigma Aldrich). The acid volatile sulfide (AVS) level of the sediments was determined using a detection tube (Gastec, 201 L and 201 H). Loss on ignition (LOI) was determined by comparing the weight difference in the oven-dried (100 °C, 24 h) sediment samples before and after combustion at 600 °C for 4 h to measure organic matter contents [17]. To determine the sediment-water content, sediment samples were dried at 100 °C for 4 h in an oven and calculated using the following equation:
The sediment metals (Cu, Li, Zn, Ni, Cr, Cd, Pb, As, Hg) composition analysis was performed as per the reported procedure [18]. After reaction and evaporation, the residue was dissolved in 1% HNO3. Metals were measured using inductively coupled plasma spectrometry (ICP-MS; Parkin Elmer, NexION 2000 ICP, Waltham, MA, USA).
2.3. Metabarcoding Analysis
Genomic DNA was extracted from the sediment samples utilizing the DNeasy PowerMax Soil Kit (Qiagen, MD, USA) following the manufacturer’s provided instructions. The primers Bakt_341F (5′-CCTACGGGNGGCWGCAG-3′) and Bakt_805R (5′-GACTACHVGGGTATCTAATCC-3′) were utilized for amplification. The 16S rRNA gene amplicon libraries containing the targeted region V3-V4 were generated utilizing the Herculase II Fusion DNA polymerase Nextera XT Index Kit V2, in accordance with the instructions provided by the manufacturer. The pair-end sequencing was performed on the Illumina Miseq (Illumina, San Diego, CA, USA) platform at Macrogen. The raw data that were obtained (301 bp) were assembled into high-quality scores (avg. score > 20). Cutadapt was used to eliminate the primer and adapter sequences [19]. Divisive Amplicon Denoising Algorithm 2 (DADA2) v1.16.1 was used to analyze raw reads produced by Illumina Miseq in order to acquire amplicon sequence variants (ASVs) [20]. Unless otherwise noted, the default software settings were utilized throughout the inquiry. BLAST was used to determine the taxonomic resolution of bacterial communities at the species level against the NCBI’s 16S microbial database [21]. The Shannon index was used to measure diversity and Chao1 was used to estimate richness in the statistical data analysis.
2.4. Statistical Analysis
Redundancy analysis (RDA) was performed in R (v4.4.2) using the vegan package to explore relationships between grain size variables and environmental factors. All variables were standardized prior to analysis. The resulting RDA scores were exported and visualized in Python (v3.13.1) using matplotlib (v3.10.1), using custom color schemes, marker shapes, and annotations. Pearson’s correlation analysis was performed in R (version 4.4.2) using the psych package to examine the relationships between environmental parameters (TOC, AVS, LOI, water content, and total metal conc.) and alpha diversity indices (Shannon, Chao1, Gini–Simpson, and Good’s coverage). The analysis was conducted separately for each intertidal zone (Inner, Middle, and Outer) to assess group-specific patterns. Correlation coefficients (r) and associated p-values were calculated using the Pearson method. Statistically significant correlations were identified based on a threshold of p < 0.05. The results were visualized using heatmaps with dual triangular matrices.
3. Results
3.1. Sediment Properties
Figure 2 illustrates the proportional distribution of gravel, sand, silt, and clay within the intertidal sediment of Gomso Bay. The present study results show the inner zone of the Bay has silt (55.5–89.7%) as a major composition, followed by sand (3.7–35.4%) and then clay (6.5–9.1%). On the other hand, the outer zone of Bay comprises sand (77.3–100%) as a major composition, followed by silt (0.0–20.3%) and clay (0.0–2.4%), whereas the middle zone of the Bay shows mixed types of sediment composition in includes silt (25.5–73.2%), sand (18.8–71.9%), and clay (2.6–10.5%). Gravel was observed (0.1%) at the IH sediment only. The sedimentary compositions of Gomso Bay intertidal sediments exhibited significant spatial variations as a consequence of grain size analysis, which may be attributed to the unique regional features of the coastal environment, such as topography and tidal variations.
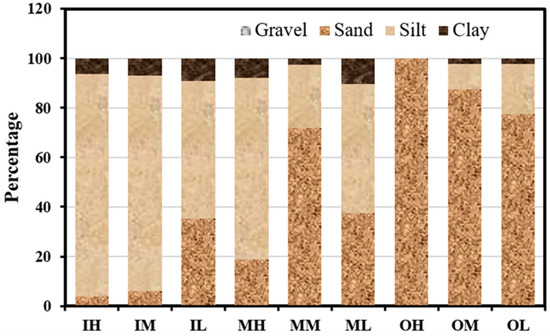
Figure 2.
The distribution of sediment texture in Gomso Bay, Republic of Korea.
The results indicate that the water content (Figure 3a), LOI (Figure 3b), and TOC (Figure 3c) vary within the range of 23.36% (OH) to 36.18% (IM), 0.82% (OH) to 3.33% (IM), and 0.06% (OL) to 0.54% (IM), respectively. The water content, LOI, and TOC demonstrated a decreasing trend in the sediments from the inner zone to the middle zone and further to the outside zone. The AVS analysis results are shown in Figure 3d. AVS conc. was highest in IM (0.0045%) and lowest in IL (0.0010%). However, the AVS was not detected in the IH, OH, and OL and does not reflect any pattern in the tidal flat zones.
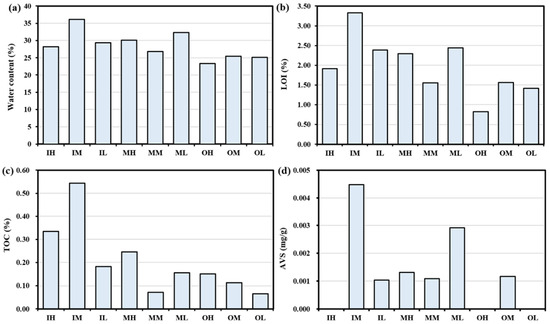
Figure 3.
Changes in the sediment (a) water content, (b) loss on ignition (LOI), (c) total organic carbon (TOC), and (d) acid volatile sulfide (AVS).
3.2. Spatial Distribution of Metals in Sediment
The concentrations of heavy metals detected in the intertidal sediments of Gomso Bay are shown in Table 1. The distribution of heavy metals in three distinct zones of the bay exhibits a notable pattern of decreasing concentration from the inner to the middle to the outer zone. The following is a range of varying amounts (mg/kg) of heavy metals that were detected: Cu: 4.436–12.720, Li: 9.943–44.132, Zn: 16.644–60.500, Ni: 3.674–18.704, Cr: 9.543–56.145, Cd: 0.015–0.061, Pb: 19.724–24.266, As: 3.442–7.230, and Hg: 0.001–0.013 mg/kg. The results indicate that the average cumulative amount of heavy metals is 156.48 ± 48.92 mg/kg, with a range of 68.42 to 221.70 mg/kg. With a concentration of 221.70 mg/kg, the OL had the highest concentration, while the ML had the lowest, at 68.42 mg/kg. The average concentration of the individual metals, appeared as follows: Zn (40.97 ± 15.92 mg/kg) > Cr (38.00 ± 13.35 mg/kg) > Li (29.09 ± 10.94 mg/kg) > Pb (21.86 ± 1.55 mg/kg) > Ni (13.55 ± 4.66 mg/kg) > Cu (7.19 ± 3.95 mg/kg) > As (5.76 ± 1.17 mg/kg) > Cd (0.04 ± 0.01 mg/kg) > Hg (0.006 ± 0.004 mg/kg).

Table 1.
Concentration (mg/kg) of heavy metals in sediments of Gomso Bay, Republic of Korea.
Redundancy analysis (RDA) was conducted to evaluate the relationship between particle size composition and environmental variables across intertidal sediment samples (Figure 4a). The first two RDA axes explained 34.7% and 24.5% of the total variance, respectively. Inner zone sediments were distributed in the positive direction of RDA1, whereas all outer zone samples were positioned in the positive region of RDA2, indicating a clear separation along the vertical axis. As for particle size variables, Sand loaded strongly positively on RDA2, while Clay and Silt loaded negatively on both axes. Environmental variables such as TOC, AVS, LOI, and water content showed strong negative correlations with clay and silt. In contrast, heavy metals such as Cu, Hg, Ni, and Zn were projected along the positive RDA1 axis, indicating that these contaminants were more related to sand fractions.
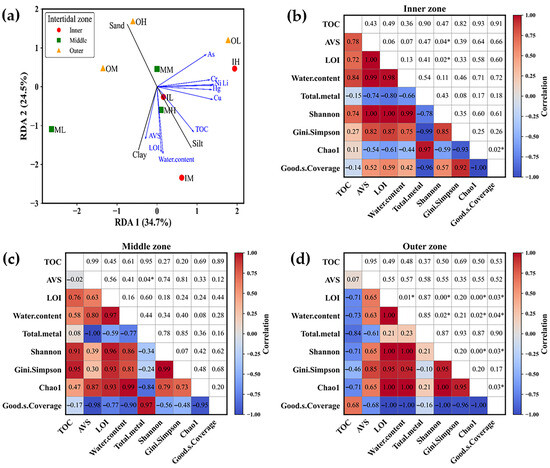
Figure 4.
(a) Redundancy analysis (RDA) biplot showing relationships between particle size components (black vectors) and environmental variables (blue vectors) across intertidal sediments. (b–d) Pearson correlation matrices between environmental parameters and alpha diversity indices for the (b) inner, (c) middle, and (d) outer zones. Lower triangles show correlation coefficients, upper triangles show p-values, and asterisks (*) indicate significant correlations (p < 0.05).
3.3. Overall Intertidal Sediment Microbial Community Composition
A total of nine samples underwent sequencing in order to generate libraries of the 16S rRNA gene. The collected samples resulted in a total of 542,299 reads that passed the quality filter. Additionally, 11,031 ASVs were identified, as shown in Table 2. Based on Good’s coverage estimate, the library sizes demonstrated adequate coverage for 98.91–99.97% of the bacterial communities. In accordance with the diversity and richness indices (Chao1, Shannon, and Simpson), the average diversity indices of sediment microbial communities in samples IM, ML, and OM (compared within zones) were found to be higher when there was a higher concentration of sediment-water contents, organic matter contents (LOI), and AVS. In contrast, the average diversity indices in samples IH, MM, and OH were lower when there was a low concentration of sediment-water contents, LOI, and AVS (Figure 3). This observation suggests that tidal zones characterized by greater water content, as well as elevated levels of LOI and AVS, may be associated with increased levels of species diversity.

Table 2.
Bacterial community richness and diversity in the intertidal surface sediments of Gomso Bay, Republic of Korea.
Pearson correlation analysis revealed distinct relationships between environmental variables and microbial diversity across intertidal zones (Figure 4b–d). In the Inner zone (Figure 4b), strong positive correlations were observed among AVS, LOI, and water content (r > 0.98), while total metal content showed negative correlations with these parameters (r < −0.66). Diversity indices such as Shannon and Gini–Simpson were highly correlated (r = 0.85) but negatively associated with total metals (r < −0.78). Good’s coverage was inversely related to diversity indices, especially Chao1 (r = −0.93). Significant correlations were found between LOI and Shannon, and between Chao1 and Good’s coverage (p < 0.05), suggesting that organic enrichment enhances microbial diversity, whereas metal accumulation suppresses it. In the Middle zone (Figure 4c), TOC, LOI, and water content were positively correlated (r > 0.58), whereas total metals were negatively correlated with AVS, LOI, and water content (r < −0.77). Diversity indices were highly interrelated (r > 0.73) and negatively correlated with total metals and Good’s coverage (r < −0.84, p < 0.05). In the Outer zone (Figure 4d), strong positive correlations were observed among LOI, water content, AVS, total metals, and diversity indices (r > 0.85), several of which were statistically significant (p < 0.05). TOC showed negative correlations (r < −0.71), and Good’s coverage was inversely related to all major parameters (r < −0.96). These results collectively demonstrate that organic matter enrichment supports microbial diversity, while high metal concentrations reduce it, with varying intensity across intertidal zones.
The analysis of bacterial diversity in nine sediment samples at the phylum level reveals the presence of 28 distinct phyla (Figure 5), with Pseudomonadota (synonym Proteobacteria) [22]) being identified as the predominant phylum. The sequential relative abundance of the top six phyla (Table 3) is as follows: Pseudomonadota (26.24–35.07%), Thermodesulfobacteriota (3.01–22.53%), Bacterioidota (7.95–16.05%), Cyanobacteriota (2.37–14.82%), Actinomycetota (5.94–12.39%), and Chloroflexoda (1.07–9.30%). The classes of Pseudomonadota exhibited varying levels of relative abundance, with Gammaproteobacteria (16.13–26.93%) being the most prevalent, followed by Alphaproteobacteria (3.80–11.59%), and then Betaproteobacteria (0.34–5.38%).
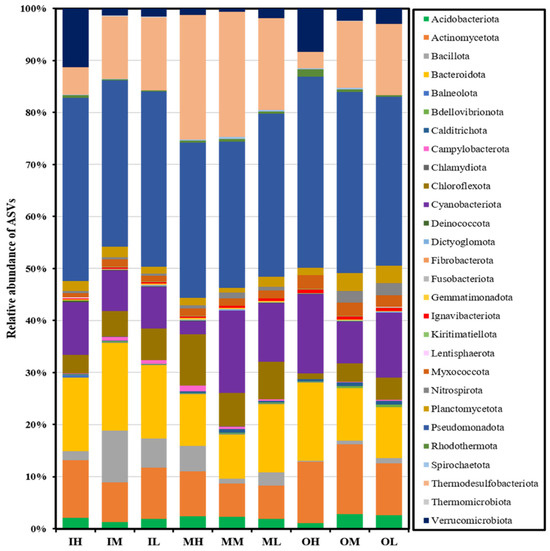
Figure 5.
Relative abundance of bacterial community structure at the phylum level based on the sequencing of 16S rRNA gene amplicon sequence variants.

Table 3.
The relative abundance of top phyla and the classes of Phylum Pseudomonadota (Proteobacteria) in the intertidal sediments of Gomso Bay, Republic of Korea.
The diversity of bacterial communities in sediments, as determined by ASV assignments, exhibited a gradient from highest to lowest when comparing the inner zone, middle zone, and outside zone at the family level (220–236, 187–222, 130–193), at the genera level (407–451, 334–410, 256–345), and at the genera level (527–605, 405–515, 321–425), as shown in Table 4. The middle tidal flat sediments of the inner zone (IM) and outer zone (OM) exhibited the maximum abundance of bacterial genera and species. Specifically, the IM had 451 genera and 605 species, while the OM had 345 genera and 425 species. Additionally, the low tidal flat sediment of the middle zone (ML) included 410 genera and 515 species. The high tidal flat sediments of the inner zone (IH) and outer zone (OH), as well as the mid-tidal zone of the middle zone (MM), exhibited the lowest abundance of bacterial genera and species. Specifically, the IH sediments had 407 genera and 529 species, while the OH sediments had 256 genera and 321 species. The MM sediments included 334 genera and 405 species.

Table 4.
Bacterial community structure in the intertidal sediments of Gomso Bay, Republic of Korea.
The relative changes in the abundance of bacterial species among the sediment samples are shown in a heatmap (Figure 6). The prevalent species seen in all samples are Actinomarinicola tropica (0.98–7.38%), Dulcicalothrix necridiiformans (1.36–13.28%), Thioprofundum lithotrophicum (3.12–8.70%), and Woeseia oceani (2.84–7.10%). The relative abundance of a few Anaerolineae species, including Ornatilinea apprima and Thermomarinilinea lacunifontana, decreases from the outer to the middle and then to the inner zones. This is due to the lower exposure of the outer zone surface sediment to atmospheric conditions. Similarly, decreasing relative abundance was observed in Desulfobacteria species, such as Desulfonatronobacter acidivorans, Desulfocastanea catecholica, Desulfogranum aggregans, Desulfolithobacter dissulfuricans, Geothermobacter hydrogeniphilus, and Syntrophobacter fumaroxidans, from the outer zone to the middle zone and then to the inner zone. In addition, the species relative abundance is much greater in the sediment samples collected from the high tidal flat areas (IH, MH, OH) of the inner, middle, and outer zones of the Bay, compared to the mid-tidal zones (IM, MM, OM) and finally the low tidal zones (IL, ML, OL). This indicates the changes in species abundance are associated with the tidal effect caused by sediment exposure to atmospheric conditions.
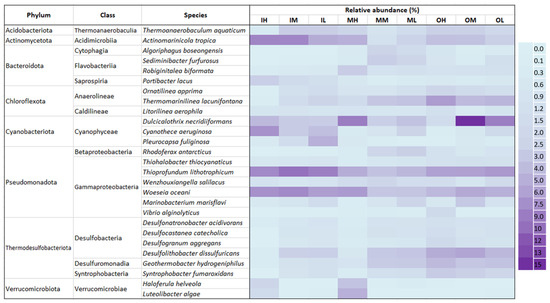
Figure 6.
The heatmap image indicates the relative abundance of bacterial species in the surface sediment samples. Only the species that make up more than 1% of the composition in at least one sample are included in the heat map.
4. Discussion
The sediment dynamics in coastal zones may be distinguished by the way they are affected by multiple factors such as rivers, tides, wind, and waves. Tidal flats that are dominated by tidal processes often exhibit the deposition of fine sediments, while coastlines that are dominated by wave action tend to have surfaces composed mostly of sand or rock. The findings of our study (Figure 2) reveal that the surface sediment in the outer (OH, OM, OL) and inner (IH, IM, IL) zones exhibits a greater proportion of sand and silt, respectively. This pattern was also reflected in the RDA plot (Figure 4), where inner zone sediments were closely aligned with vectors for silt and clay, while outer zone sediments were separated along the positive RDA2 axis near sand.
The water content in the sediments shows an increasing trend from the outside to the middle to the inner zone, as seen in Figure 3a. This is possibly due to the increasing amount of silt and clay in the surface sediments (Figure 2). The sediment water content variation is due to grain size, composition, and permeability [23]. In particular, sediments with a very fine-grained composition have the lowest permeability, whereas sediments consisting of clay and silt show relatively greater permeability due to their larger grain size and increased porosity [24]. Based on the results shown in Figure 3b,c, there is a noticeable decline in the LOI and TOC values from the sediment samples of inner (IH, IM, IL) to middle zones (MH, MM, ML), followed by a further decrease in the outer zone (OH, OM, OL) of Gomso Bay. This could be because of higher organic matter deposition in the inner zone’s sediment than in the outer zone, as well as because of human activities, local algal blooms, and halophytes surrounding the inner zone [25,26]. Furthermore, it is expected that the TOC content in the muddy cores will be higher because the inner zone’s fine-grained sediments have been shown to contain higher levels of organic material than the outer zone’s coarse-grained sediments [27]. Additionally, the outer zone sediment’s prolonged inundation and increased anoxic conditions may slow down organic matter decomposition [2]. Also, the degradation of organic matter is dependent on microbial processes that take place in the sediments [28]. Hwang et al. [29] observed that the concentrations of organic matter and AVS in coastal sediments showed fluctuation according to the particle size of the sediment. Notably, higher concentrations were found in finer sediment in comparison to coarser sediment. Similar observations were noticed in the present study. This study’s results showed the concentration of AVS (Figure 3d) was much higher in the fine sediment of the inner zone (except IH, not detected) in comparison to the mixed sediment of the middle zone and the coarser sediment of the outer zone (except OH and OL, not detected). These individual trends were further supported by redundancy analysis (RDA), which revealed that organic matter (TOC, LOI) and AVS were positively associated with fine sediment fractions, particularly silt and clay (Figure 4). This multivariate pattern confirms the influence of sediment type on the accumulation and retention of organic matter and sulfur in the inner zone. Furthermore, Pearson correlation analysis demonstrated strong positive correlations among AVS, LOI, and water content (r > 0.98) and significant negative correlations between total metal content and organic matter parameters (r < −0.66, p < 0.05), suggesting that fine sediments promote organic and sulfur accumulation while reducing metal mobility.
The contamination of Korean coastal sediments with metals has received considerable attention due to environmental issues along the coast and concerns regarding the safety of seafood products. Hwang et al. [29] investigated 74 intertidal sectors along Korea’s western and southern coasts and discovered that there was a significant spatial variation in the average metal concentrations in the intertidal sediments; the metal concentrations (mean) were in the following order: Zn (67 mg/kg) > Pb (23.1 mg/kg) > Cu (12.8 mg/kg) > As (7.4 mg/kg) > Cd (0.09 mg/kg) > Hg (0.014 mg/kg). The results from our analysis suggest that the average concentration of metals (Table 1) in the intertidal sediment of Gomso Bay is lower than the mean concentration previously reported. Furthermore, the descending order of metal concentrations also aligns with the previous findings [29,30]. The distribution of metals throughout the intertidal zonation and tidal flats did not exhibit a clear trend. This observation is in line with the RDA results (Figure 4a), where metal vectors (e.g., Cu, Zn, Cr) showed weak associations with sediment type variables, suggesting that particle size was not a primary driver of metal distribution in the study area. However, Pearson correlation analysis indicated negative associations between total metal content and microbial diversity indices (r < −0.78), implying that even moderate metal levels may exert selective pressure on microbial communities in finer sediments.
The analysis of 16S rRNA gene amplicon sequencing revealed a significant level of bacterial diversity in the surface sediment samples collected from Gomso Bay. This was evident from the identification of 11,031 ASVs (Table 2) that were classified into 28 different phyla (Figure 5). The observed high diversity might be due to the effect of tides and the introduction of organic matter from terrestrial or estuarine sources, as well as the variability in sulfide content within the surface sediment [31]. The fine sediment of IM exhibited the highest microbial diversity (Table 4), extending from the Class to the species level. This observation might perhaps be attributed to the elevated levels of TOC and AVS present in this sediment (Figure 3). In contrast, the coarse sediment of OH had the lowest microbial diversity. These relationships were supported by both RDA and Pearson analyses, which showed that microbial diversity indices (Shannon, Gini–Simpson, Chao1) were strongly associated with organic-rich and fine-textured sediments (r > 0.85, p < 0.05), whereas metal content exhibited negative correlations with diversity (r < −0.78). This indicates that organic enrichment enhances microbial diversity while higher metal loads suppress it, particularly in the inner and middle tidal zones.
The bacteria belonging to the phylum Pseudomonadota have been identified as the major heterotrophic, chemolithotrophic, facultative, and obligatory anaerobic organisms in marine sediments and are recognized for their significant contributions to biogeochemical processes [17,31,32]. The presence of a large number of heterotrophic microbes is associated with the presence of organic matter and it is well-known that fine sediment contains a higher proportion of organic matter compared to coarse sediments [27]. Furthermore, even slight alterations in the availability of organic substances can lead to a change in the composition of the microbial community [33,34]. The RDA biplot (Figure 4) further demonstrated that microbial community structure was spatially segregated according to organic matter variables (TOC, LOI), supporting the hypothesis that sediment composition and nutrient enrichment shape microbial assemblages across intertidal zones.
The sediment analysis revealed that Thermodesulfobacteriota, which are mostly thermophilic sulfate-reducing bacteria (SRB), was the second most abundant microbial group (Table 3). These bacteria perform anaerobic respiration, utilizing sulfate (SO42−) as the terminal electron acceptor and reducing it to hydrogen sulfide (H2S) [35,36,37,38]. In this study, it was observed a decrease in the abundance of these microbes at the Phylum level in the sediment of the high tidal zone (IH, OH; Table 3). This may be due to the reduced concentration of AVS (Figure 3d) and the exposure of the sediment to oxygen at high tide. The outer zone sediments show a higher percentage of microbes at the species level compared to the inner zone sediments (Figure 6). This difference may be due to the warmer temperatures and lower oxygen availability in the outer zone sediments.
The phylum Bacteroidetes is very prevalent among bacterial populations in marine environments, ranking third in abundance behind Proteobacteria and Cyanobacteria [39]. As shown in Figure 6, Portibacter lacus was found predominant in the inner zone sediment. This is because Bacteroidetes are heterotrophic and associated with high organic substances [33,40]. In contrast, it was observed that Cyanobacterial oxygenic photoautotrophic species, including Cyanothece aeruginosa and Pleurocapsa fuliginosa, were most prevalent in the intertidal zone. Also, Dulcicalothrix necridiiformans were found across Gomso Bay, indicating a relatively greater abundance compared to other species within the Cyanobacteriota group. Cyanobacteria are widely accepted as oxygenic photoautotrophs that contribute to the oxidation of organic substances and play a significant part in the carbon cycle within marine sediment [41].
The inner and middle zone sediments, particularly the high tidal sediments (IH, MH; Figure 6), showed a dominance of aerobic or obligatory aerobic microbial species belonging to the phylum Actinomycetota, specifically Actinomarinicola tropica [42], as well as aerobic species of the Phylum Verrucomicrobiota, including Haloferula helveola [43] and Luteolibacter algae [44]. Similarly, the outer zone sediment was shown to be dominated by species of Phylum Chloroflexota, including Ornatilinea apprima [45] and Thermomarinilinea lacunifontana [46], both are anaerobic and obligatory anaerobic organotrophic bacteria. These findings clearly show that tidal flat sediments exposed to atmospheric oxygen are more likely to support the growth of aerobic bacteria, whereas sediment in the outer zone is immersed in seawater, which has lower oxygen concentrations or may be anoxic. These distributional patterns were consistent with the RDA ordination, which revealed the separation of aerobic and anaerobic taxa along organic matter and redox-related gradients, confirming that tidal zonation and sediment properties jointly drive microbial community differentiation in Gomso Bay.
5. Conclusions
The findings of this research indicate that the surface sediment texture, TOC, AVS, and bacterial community composition in the intertidal zone are influenced by the tidal flat area. The sediment in the inner zone mostly consists of silt and sand, whereas the sediment within the outer zone is predominantly formed of sand. The middle zone has a mixed composition consisting of a combination of sand, silt, and clay. Water content, LOI, TOC, and AVS concentrations show a decreasing trend from the inner zone to the middle and outer zones, suggesting a linear association with sediment texture. A gradient in the overall concentration of heavy metals was also observed from the inner to the outer and middle zones.
Intertidal zonation was a key factor influencing microbial diversity. The inner zone, particularly the high tidal area exposed to the atmosphere, was dominated by aerobic microbes belonging to the phyla Pseudomonadota and Cyanobacteriota. In contrast, the outer zone, especially the low tidal sediments, was dominated by anaerobic microbes of the phylum Chloroflexota, likely due to low-oxygen conditions in seawater-submerged sediments.
These findings highlight the ecological significance of tidal zonation in shaping distinct microbial niches that support biodiversity and biogeochemical processes in coastal environments. Aerobic communities in high tidal zones may facilitate organic matter decomposition and nutrient cycling, while anaerobic communities in low tidal zones contribute to carbon sequestration.
The baseline data provided by this study on the UNESCO World Heritage site of Gomso Bay will be valuable for monitoring anthropogenic impacts and environmental changes in this ecologically important area. Future research should focus on seasonal variations and the functional roles of key bacterial taxa to better understand sediment–microbe interactions and ecosystem responses to environmental changes.
Author Contributions
Conceptualization, M.P.P., J.-O.K. and K.K.; Methodology, M.P.P., S.O.R., H.-E.W., J.-O.K. and K.K.; Formal analysis, M.P.P., H.-E.W., C.-G.L., H.N.O. and S.Y.J.; Software, J.-O.K.; Validation, K.K.; Investigation, S.O.R. and J.-O.K.; Resources, S.O.R. and K.K.; Data curation, S.O.R., H.-E.W., C.-G.L., H.N.O., S.Y.J. and J.-O.K.; Visualization, M.P.P. and H.-E.W.; Writing—original draft preparation, M.P.P.; Writing—review and editing, S.O.R., H.-E.W., C.-G.L., H.N.O., S.Y.J., J.-O.K. and K.K.; Supervision, K.K.; Project administration, J.-O.K. and K.K.; Funding acquisition, K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the World Heritage Promotion Team of Korean Tidal Flats, Tidal Flat Biodiversity Survey as Part of the Integrated Monitoring of Korea Tidal Flats World Heritage (2024-11).
Data Availability Statement
The 16S rRNA gene amplicon sequence that supports the findings of this study is available in NCBI (http://www.ncbi.nlm.nih.gov/) under the BioProject and Sequence Read Archive (SRA) numbers PRJNA1039572 and SRP471432, respectively.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ortega-Morales, B.O.; Chan-Bacab, M.J.; De la Rosa, S.D.C.; Camacho-Chab, J.C. Valuable processes and products from marine intertidal microbial communities. Curr. Opin. Biotechnol. 2010, 21, 346–352. [Google Scholar] [CrossRef]
- Rios-Yunes, D.; Tiano, J.C.; van Rijswijk, P.; De Borger, E.; van Oevelen, D.; Soetaert, K. Long-term changes in ecosystem functioning of a coastal bay expected from a shifting balance between intertidal and subtidal habitats. Cont. Shelf Res. 2023, 254, 104904. [Google Scholar] [CrossRef]
- Rios-Yunes, D.; Grandjean, T.; di Primio, A.; Tiano, J.; Bouma, T.J.; van Oevelen, D.; Soetaert, K. Sediment resuspension enhances nutrient exchange in intertidal mudflats. Front. Mar. Sci. 2023, 10, 1155386. [Google Scholar] [CrossRef]
- Hwang, D.W.; Kim, G.; Yang, H.S. Active exchange of water and nutrients between seawater and shallow pore water in intertidal sandflats. Ocean Sci. J. 2008, 43, 223–232. [Google Scholar] [CrossRef]
- Jiang, H.; Huang, J.; Li, L.; Huang, L.; Manzoor, M.; Yang, J.; Wu, G.; Sun, X.; Wang, B.; Egamberdieva, D.; et al. Onshore soil microbes and endophytes respond differently to geochemical and mineralogical changes in the Aral Sea. Sci. Total Environ. 2021, 765, 142675. [Google Scholar] [CrossRef]
- Yue, Y.; Rong, H.; Yang, Z.; Pan, X.; Chen, Y.; Yang, M. Microbial diversity and functional profiling in coastal tidal flat sediment with pollution of nutrients and potentially toxic elements. J. Soils Sediment. 2023, 23, 2935–2950. [Google Scholar] [CrossRef]
- Zhang, X.; Lan, T.; Jiang, H.; Ye, K.; Dai, Z. Bacterial community driven nitrogen cycling in coastal sediments of intertidal transition zone. Sci. Total Environ. 2024, 908, 168299. [Google Scholar] [CrossRef]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef]
- Köpke, B.; Wilms, R.; Engelen, B.; Cypionka, H.; Sass, H. Microbial diversity in coastal subsurface sediments: A cultivation approach using various electron acceptors and substrate gradients. Appl. Environ. Microbiol. 2005, 71, 7819–7830. [Google Scholar] [CrossRef]
- Acosta-González, A.; Marqués, S. Bcterial diversity in oil-polluted marine coastal sediments. Curr. Opin. Biotechnol. 2016, 38, 24–32. [Google Scholar] [CrossRef]
- Webb, S.J.; Rabsatt, T.; Erazo, N.; Bowman, J.S. Impacts of Zostera eelgrasses on microbial community structure in San Diego coastal waters. Elem. Sci. Anth. 2019, 7, 11. [Google Scholar] [CrossRef]
- Degenhardt, J.; Merder, J.; Heyerhoff, B.; Simon, H.; Engelen, B.; Waska, H. Cross-shore and depth zonations in bacterial diversity are linked to age and source of dissolved organic matter across the intertidal area of a sandy beach. Microorganisms 2021, 9, 1720. [Google Scholar] [CrossRef]
- Lee, H.J. Preliminary results on suspended sediment transport by tidal currents in Gomso Bay, Korea. Ocean Sci. J. 2010, 45, 187–195. [Google Scholar] [CrossRef]
- Yang, B.; Dalrymple, R.W.; Gingras, M.K.; Chun, S.; Lee, H. Up-estuary variation of sedimentary facies and ichnocoenoses in an open-mouthed, macrotidal, mixed-energy estuary, Gomso Bay, Korea. J. Sediment. Res. 2007, 77, 757–771. [Google Scholar] [CrossRef]
- Chang, J.H.; Ryu, S.O.; Jo, Y.J. Long-term variation of tidal-flat sediments in Gomso bay, west coast of Korea. J. Korean Earth Sci. Soc. 2007, 28, 357–366. [Google Scholar] [CrossRef]
- Ingram, R.L. Sieve analysis. In Procedures in Sedimentary Petrology; Carver, R.E., Ed.; Wiley-Interscience: New York, NY, USA, 1971; pp. 49–67. [Google Scholar]
- Patil, M.P.; Jeong, I.; Woo, H.E.; Kim, J.O.; Lee, D.I.; Kim, K. Natural variations in the benthic environment and bacterial communities of coastal sediments around aquaculture farms in South Korea. Indian J. Microbiol. 2023, 63, 100–105. [Google Scholar] [CrossRef]
- Song, Y.H.; Choi, M.S. REE Geochemistry of fine-grained sediments from major rivers around the Yellow Sea. Chem. Geol. 2009, 266, 328–342. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- Wilson, A.M.; Huettel, M.; Klein, S. Grain size and depositional environment as predictors of permeability in coastal marine sands. Estuar. Coast. Shelf Sci. 2008, 80, 193–199. [Google Scholar] [CrossRef]
- Bryant, W.R. Permeability of clays, silty-clays and clayey-silts. Gulf Coast Assoc. Geol. Soc. Trans. 2002, 52, 1069–1077. [Google Scholar]
- Cinco-Castro, S.; Herrera-Silveira, J.; Comín, F. Sedimentation as a support ecosystem service in different ecological types of mangroves. Front. For. Glob. Change 2022, 5, 733820. [Google Scholar] [CrossRef]
- Kim, C.S.; Kim, S.H.; Lee, W.C.; Lee, D.H. Spatial variability of water quality and sedimentary organic matter during winter season in coastal aquaculture zone of Korea. Mar. Pollut. Bull. 2022, 182, 113991. [Google Scholar] [CrossRef]
- Wu, S.; Tao, S.; Ye, X.; Wang, A.; Liu, Z.; Ran, C.; Liang, H.; Li, H.; Yang, Y.; Zhang, W.; et al. Characteristics of sedimentary organic matter in tidal estuaries: A case study from the Minjiang river estuary. Water 2023, 15, 1682. [Google Scholar] [CrossRef]
- LaRowe, D.E.; Arndt, S.; Bradley, J.A.; Estes, E.R.; Hoarfrost, A.; Lang, S.Q.; Lloyd, K.G.; Mahmoudi, N.; Orsi, W.D.; Walter, S.S.; et al. The fate of organic carbon in marine sediments—New insights from recent data and analysis. Earth-Sci. Rev. 2020, 204, 103146. [Google Scholar] [CrossRef]
- Hwang, D.W.; Kim, P.J.; Kim, S.G.; Sun, C.I.; Koh, B.S.; Ryu, S.O.; Kim, T.H. Spatial distribution and pollution assessment of metals in intertidal sediments, Korea. Environ. Sci. Pollut. Res. 2019, 26, 19379–19388. [Google Scholar] [CrossRef] [PubMed]
- National Fisheries Research and Development Institute (NFRDI). Technical Report of National Fisheries Research and Development Institute in 2014; Report No. TR-2015-PM-002; NFRDI: Busan, Republic of Korea, 2015; p. 1366. (In Korean)
- Patil, M.P.; Woo, H.E.; Kim, J.O.; Kim, K. Field study on short-term changes in benthic environment and benthic microbial communities using pyrolyzed oyster shells. Sci. Total Environ. 2022, 824, 153891. [Google Scholar] [CrossRef]
- Lee, J.H.; Patil, M.P.; Kim, J.O.; Woo, H.E.; Kim, K. Diversity of microbial communities in sediment from Yeosu bay, republic of Korea, as determined by 16s rRNA gene amplicon sequencing. Microbiol. Resour. Announc. 2022, 11, e00363-22. [Google Scholar] [CrossRef]
- Cottrell, M.T.; Kirchman, D.L. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 2000, 66, 1692–1697. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, N.; Beaupré, S.R.; Steen, A.D.; Pearson, A. Sequential bioavailability of sedimentary organic matter to heterotrophic bacteria. Environ. Microbiol. 2017, 19, 2629–2644. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Van Kuijk, B.L.; Plugge, C.M.; Akkermans, A.D.; De Vos, W.M.; Stams, A.J. Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 1998, 48, 1383–1387. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Tourova, T.P.; Panteleeva, A.N.; Muyzer, G. Desulfonatronobacter acidivorans gen. nov., sp. nov. and Desulfobulbus alkaliphilus sp. nov., haloalkaliphilic heterotrophic sulfate-reducing bacteria from Soda Lakes. Int. J. Syst. Evol. Microbiol. 2012, 62, 2107–2113. [Google Scholar] [CrossRef]
- Pérez-Rodríguez, I.; Choi, J.K.; Abuyen, K.; Tyler, M.; Ronkowski, C.; Romero, E.; Trujillo, A.; Tremblay, J.; Viney, I.; Savalia, P.; et al. Geothermobacter hydrogeniphilus sp. nov., a mesophilic, Iron (III)-reducing bacterium from seafloor/subseafloor environments in the Pacific Ocean, and emended description of the genus Geothermobacter. Int. J. Syst. Evol. Microbiol. 2021, 71, 004739. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Shimamura, S.; Tame, A.; Sawayama, S.; Miyazaki, J.; Takai, K.; Nakagawa, S. Physiological and comparative proteomic characterization of Desulfolithobacter dissulfuricans gen. nov., sp. nov., a novel mesophilic, sulfur-disproportionating chemolithoautotroph from a deep-sea hydrothermal vent. Front. Microbiol. 2022, 13, 1042116. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Gómez, B.; Richter, M.; Schüler, M.; Pinhassi, J.; Acinas, S.G.; González, J.M.; Pedrós-Alió, C. Ecology of marine Bacteroidetes: A comparative genomics approach. ISME J. 2013, 7, 1026–1037. [Google Scholar] [CrossRef]
- Yoon, J.; Matsuo, Y.; Kasai, H.; Yokota, A. Portibacter lacus gen. nov., sp. nov., a new member of the family Saprospiraceae isolated from a Saline Lake. J. Gen. Appl. Microbiol. 2012, 58, 191–197. [Google Scholar] [CrossRef]
- Golubic, S.; Seong-Joo, L.; Browne, K.M. Cyanobacteria: Architects of sedimentary structures. In Microbial Sediments; Springer: Berlin/Heidelberg, Germany, 2000; pp. 57–67. [Google Scholar] [CrossRef]
- He, Y.Q.; Chen, R.W.; Li, C.; Shi, S.B.; Cui, L.Q.; Long, L.J.; Tian, X.P. Actinomarinicola tropica gen. nov. sp. nov., a new marine actinobacterium of the family Iamiaceae, isolated from South China Sea sediment environments. Int. J. Syst. Evol. Microbiol. 2020, 70, 3852–3858. [Google Scholar] [CrossRef]
- Yoon, J.; Matsuo, Y.; Katsuta, A.; Jang, J.H.; Matsuda, S.; Adachi, K.; Kasai, H.; Yokota, A. Haloferula rosea gen. nov., sp. nov., Haloferula harenae sp. nov., Haloferula phyci sp. nov., Haloferula helveola sp. nov. and Haloferula sargassicola sp. nov., five marine representatives of the family Verrucomicrobiaceae within the phylum ‘Verrucomicrobia’. Int. J. Syst. Evol. Microbiol. 2008, 58, 2491–2500. [Google Scholar] [CrossRef]
- Yoon, J.; Matsuo, Y.; Adachi, K.; Nozawa, M.; Matsuda, S.; Kasai, H.; Yokota, A. Description of Persicirhabdus sediminis gen. nov., sp. nov., Roseibacillus ishigakijimensis gen. nov., sp. nov., Roseibacillus ponti sp. nov., Roseibacillus persicicus sp. nov., Luteolibacter pohnpeiensis gen. nov., sp. nov. and Luteolibacter algae sp. nov., six marine members of the phylum ‘Verrucomicrobia’, and emended descriptions of the class Verrucomicrobiae, the order Verrucomicrobiales and the family Verrucomicrobiaceae. Int. J. Syst. Evol. Microbiol. 2008, 58, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Podosokorskaya, O.A.; Bonch-Osmolovskaya, E.A.; Novikov, A.A.; Kolganova, T.V.; Kublanov, I.V. Ornatilinea apprima gen. nov., sp. nov., a cellulolytic representative of the Class Anaerolineae. Int. J. Syst. Evol. Microbiol. 2013, 63, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Nunoura, T.; Hirai, M.; Miyazaki, M.; Kazama, H.; Makita, H.; Hirayama, H.; Furushima, Y.; Yamamoto, H.; Imachi, H.; Takai, K. Isolation and characterization of a thermophilic, obligate anaerobic and heterotrophic marine Chloroflexi bacterium from a Chloroflexi-dominated microbial community associated with a Japanese shallow hydrothermal system, and proposal for Thermomarinilinea lacunofontalis gen. nov., sp. nov. Microbes Environ. 2013, 28, 228–235. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).