New Record of Geoduck Clam Collected from the East Coast of South Korea and Its Reproductive Characteristics
Abstract
1. Introduction
2. Materials and Methods
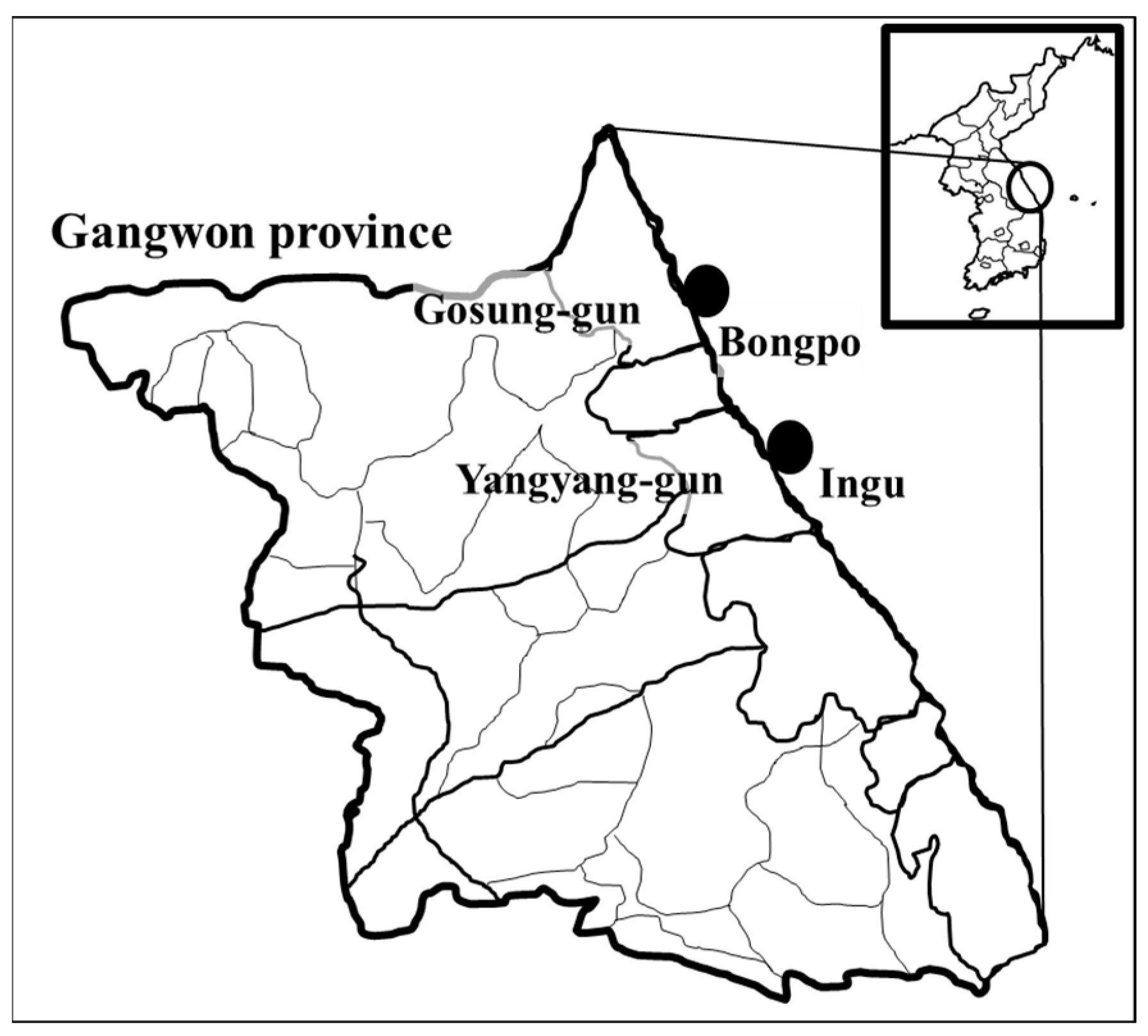
2.1. Sampling Collection
2.2. DNA Extraction and Sequencing
2.3. Sequence Alignment and Phylogenetic Analysis
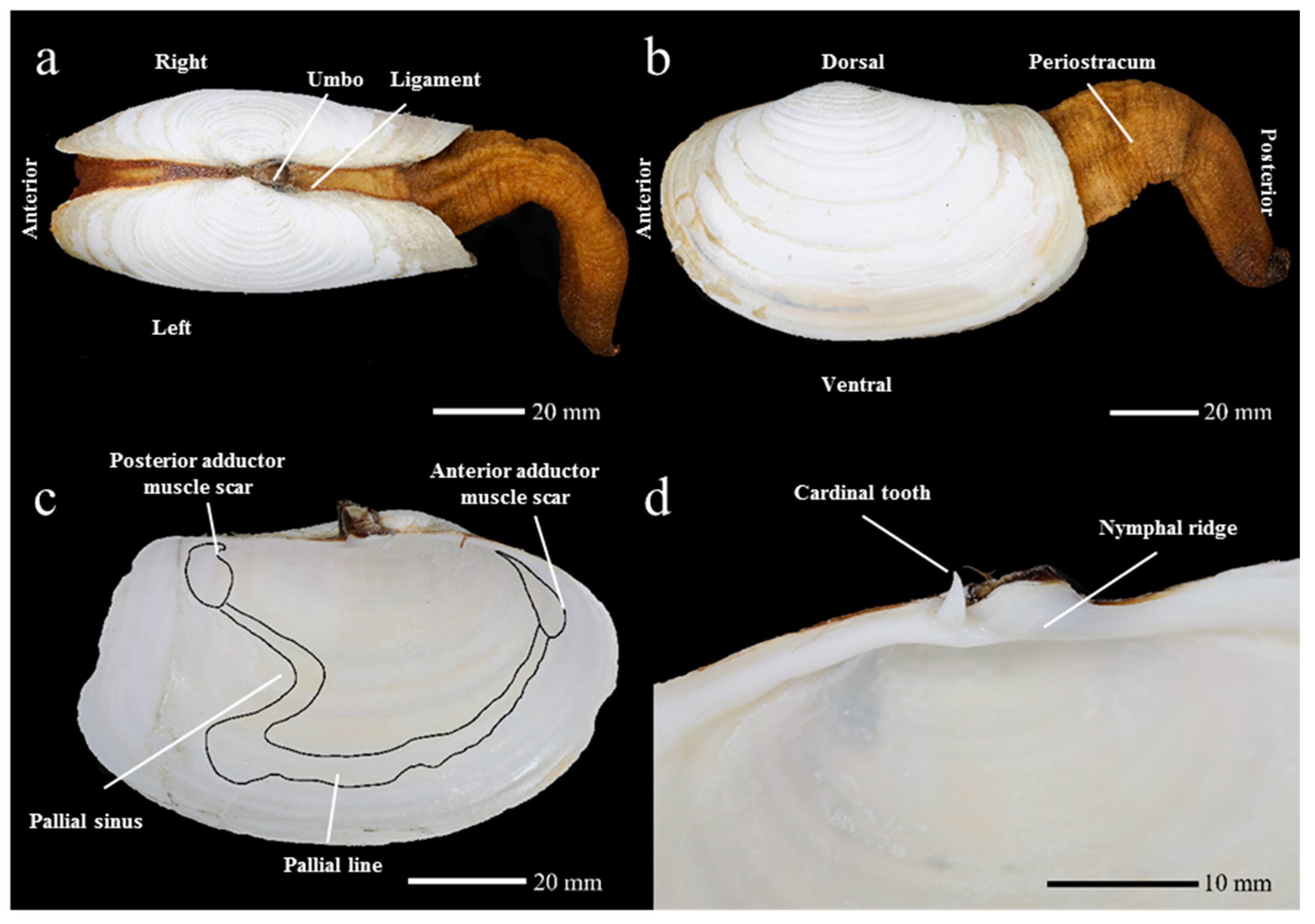
2.4. Morphological Characteristics
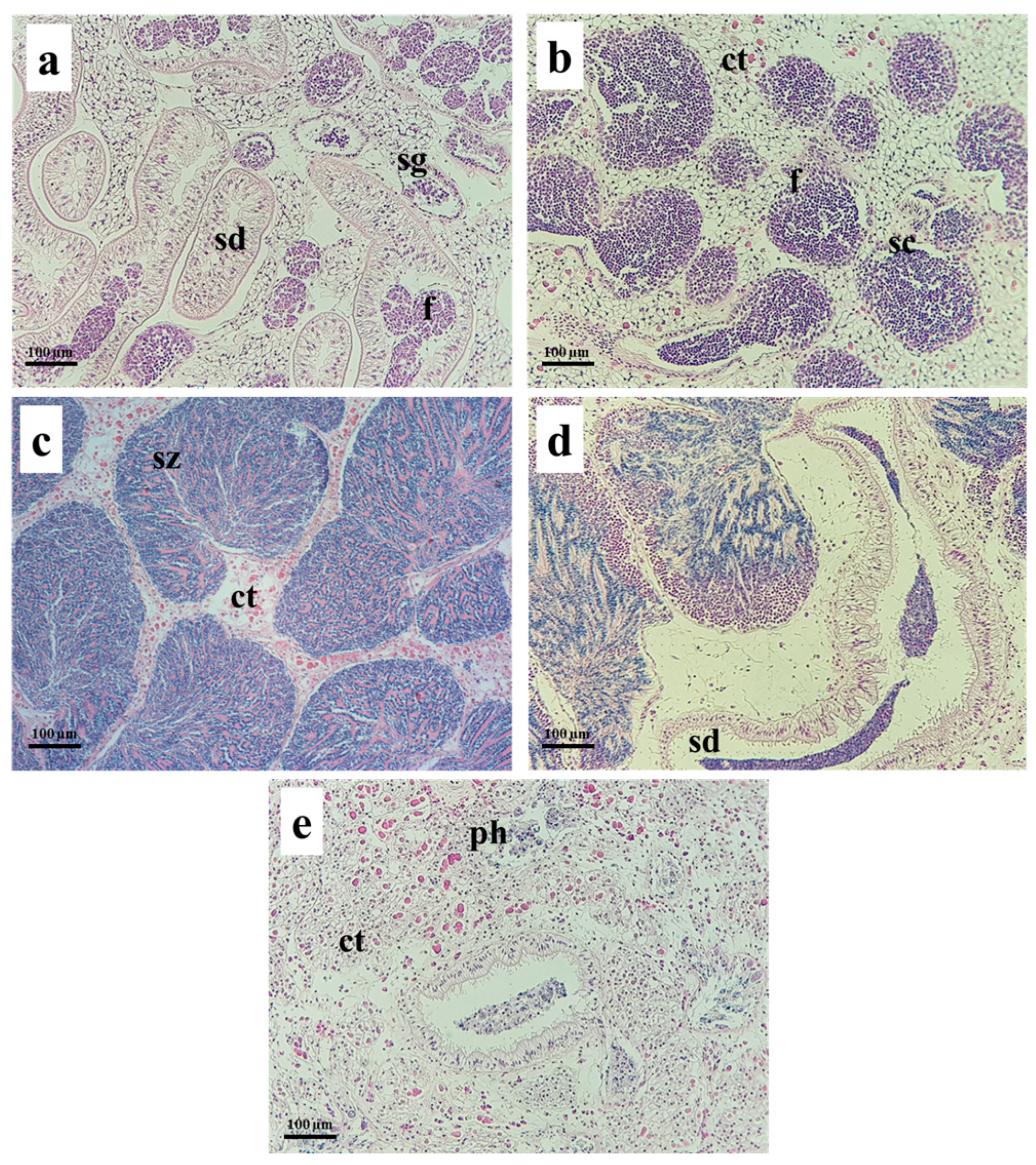
2.5. Histological Analysis
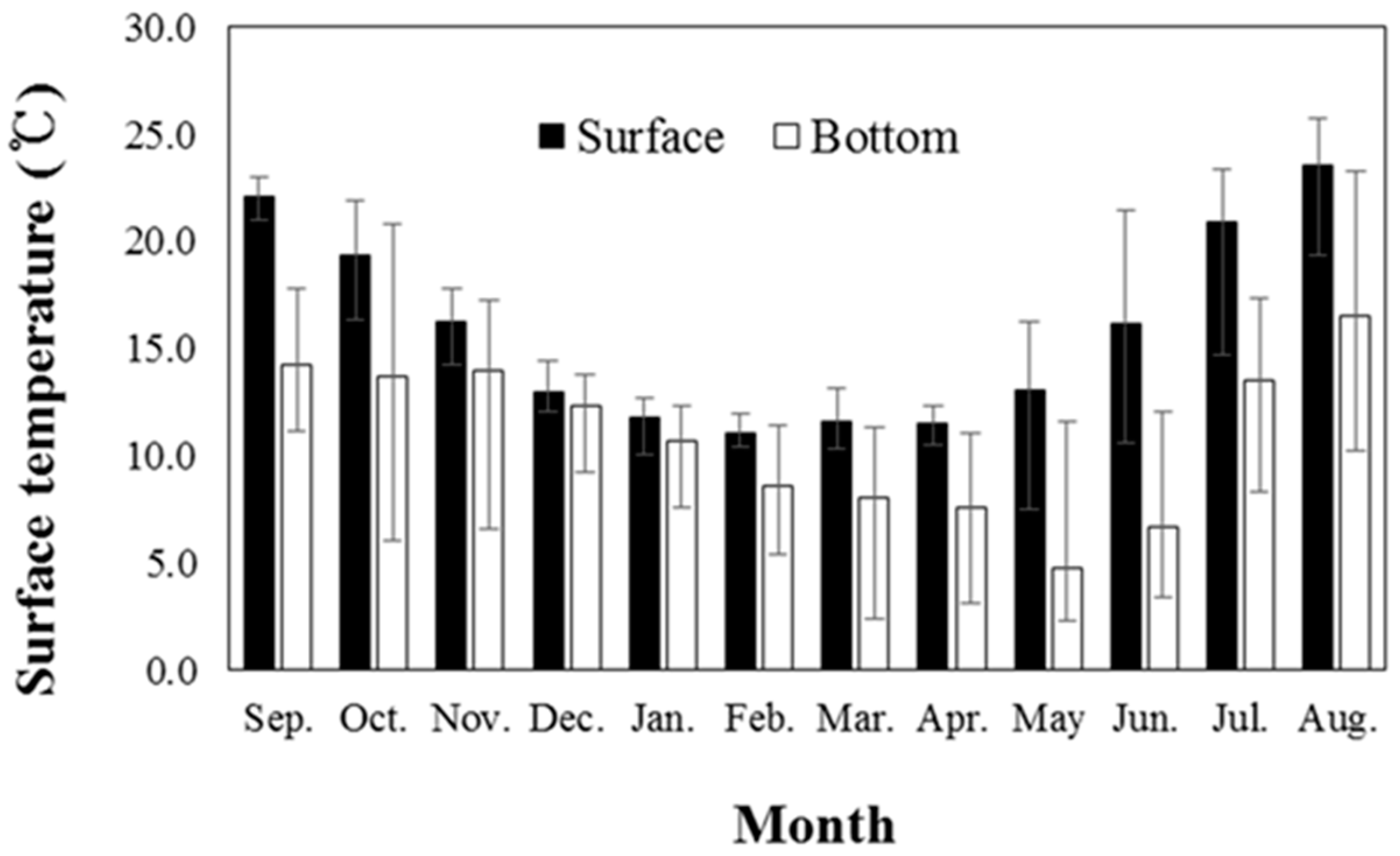
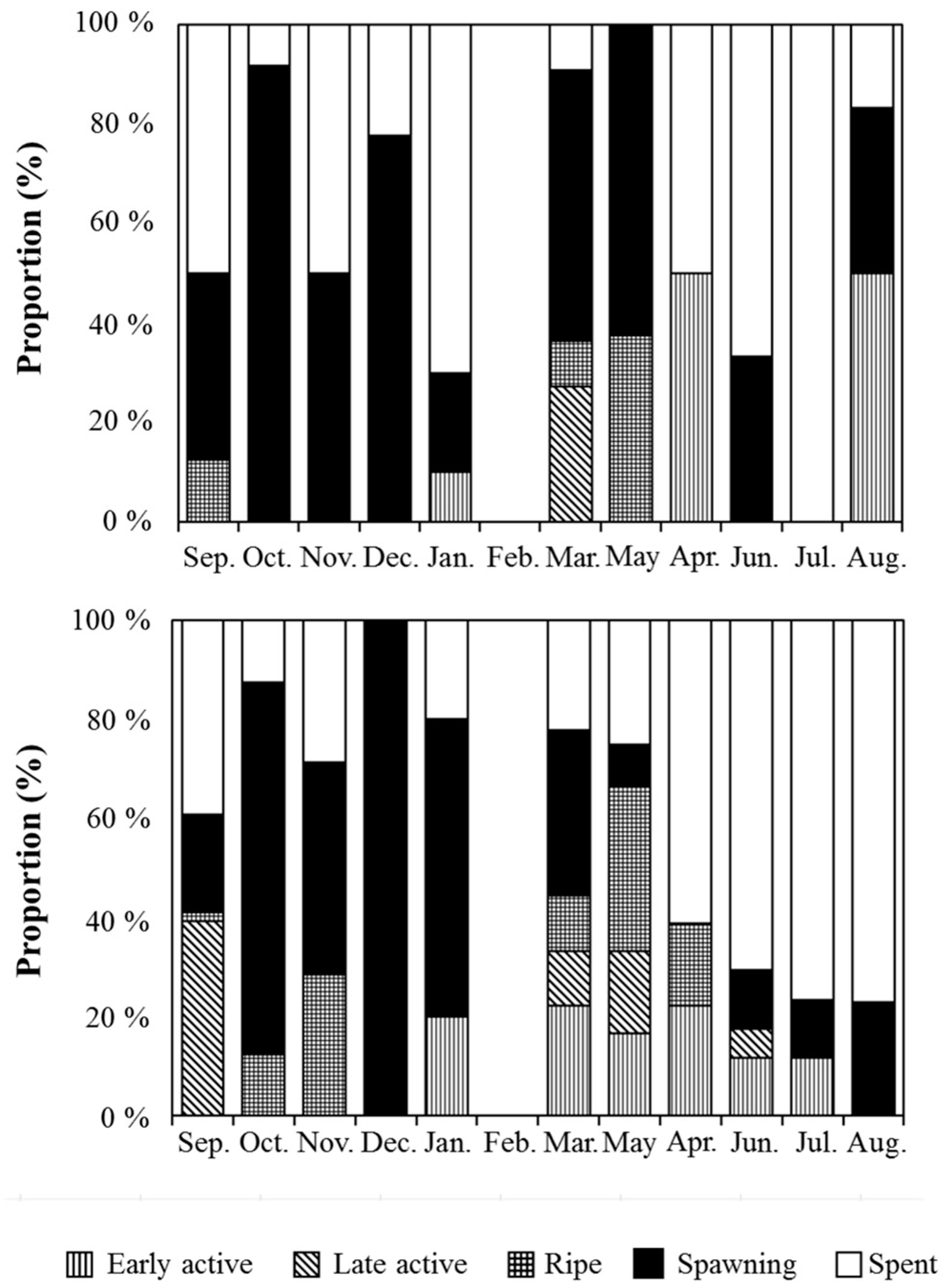
3. Results and Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bureau, D.; Hajas, W.; Surry, N.W.; Hand, C.M.; Dovey, G.; Campbell, A. Age, Size Structure, and Growth Parameters of Geoducks (Panopea abrupt Conrad, 1849) from 34 Locations in British Columbia Sampled between 1993 and 2000; Canadian Technical Report of Fisheries and Aquatic Sciences Nanaimo 2413; Fisheries and Oceans Canada: Nanaimo, BC, Canada, 2002; pp. 1–84. [Google Scholar]
- Leyva-Valencia, I.; Cruz-Hernández, P.; Álvarez-Castañeda, S.T.; Rojas-Posadas, D.I.; Correa-Ramírez, M.M.; Vadopalas, B.; Lluch-Cota, D.B. Phylogeny and Phylogeography of the Geoduck Panopea (Bivalvia: Hiatellidae). J. Shellfish Res. 2015, 34, 11–20. [Google Scholar] [CrossRef]
- Yu, M.; Zhong, S.; Yang, S.; Chen, J.; Saha, T.T. The complete mitochondrial genome of Panopea abrupta (Myoida: Hiatellidae). Mitochondrial DNA Part A 2016, 27, 711–712. [Google Scholar] [CrossRef] [PubMed]
- Habe, T. Fauna of Akkeshi Bay XXI. Pelecypoda and Scaphopoda. Public Akkeshi Mar. Biol. Stn. 1955, 4, 20–27. [Google Scholar]
- Min, D.-K.; Lee, J.-S.; Koh, D.-B.; Je, J.-G. Mollusks in Korea; Min Molluscan Research Institute: Seoul, Republic of Korea, 2004; p. 566. [Google Scholar]
- Welch, C. China’s Demand for Geoducks Send Prices, Profifits Soaring. 2012. The Bellingham Herald Online. Available online: https://www.heraldandnews.com/china-s-demand-for-geoducks-sends-prices-profits-soaring/image_612787da-9bf5-11e1-84c8-0019bb2963f4.html (accessed on 15 May 2012).
- Fan, X.B. A model for sustainable fisheries: Canadian geoduck fishery management. Ocean Fish. 2012, 12, 78–87. (In Chinese) [Google Scholar]
- Shamshak, G.L.; King, J.R. From cannery to culinary luxury: The evolution of the global geoduck market. Mar. Policy 2015, 55, 81–89. [Google Scholar] [CrossRef]
- Lee, C.S. Studies on feeding activity and environmental tolerance of geoduck clam, Panopea generosa. J. Aquac. 1997, 10, 213–218. (In Korean) [Google Scholar]
- Lee, C.S.; Rho, S.; Park, Y.J. Studies on the artificial seedling production of geoduck clam, Panope japonica. I. Spawning induction and hatching. J. Aquac. 1997, 10, 113–121. (In Korean) [Google Scholar]
- Lee, C.S.; Rho, S. Studies on the artificial seedling production of geoduck clam, Panope japonica. II. Development of egg and larvae. J. Aquac. 1997, 10, 25–32. (In Korean) [Google Scholar]
- Lee, C.S.; Baik, K.K.; Hong, K.E. Ecological studies on the habitat of geoduck clam, Panope japonica. J. Aquac. 1998, 11, 105–111. (In Korean) [Google Scholar]
- Elson, J.L.; Lightowlers, R.N. Mitochondrial DNA clonality in the dock: Can surveillance swing the case? Trends Genet. 2006, 22, 603–607. [Google Scholar] [CrossRef]
- Botero-Castro, F.; Tilak, M.; Justy, F.; Catzeflis, F.; Delsuc, F.; Douzery, E.J. Next-generation sequencing and phylogenetic signal of complete mitochondrial genomes for resolving the evolutionary history of leaf-nosed bats (Phyllostomidae). Mol. Phylogenet. Evol. 2013, 69, 728–739. [Google Scholar] [CrossRef]
- Kurbalija Novicic, Z.; Immonen, E.; Jeli, M.; AnDelkovic, M.; Stamenkovic-Radak, M.; Arnqvist, G. Within-population genetic effects of mtDNA on metabolic rate in Drosophila subobscura. J. Evol. Biol. 2015, 28, 338–346. [Google Scholar] [CrossRef]
- Qiu, F.; Kitchen, A.; Beerli, P.; Miyamoto, M.M. A possible explanation for the population size discrepancy in tuna (genus Thunnus) estimated from mitochondrial DNA and microsatellite data. Mol. Phylogenet. Evol. 2013, 66, 463–468. [Google Scholar] [CrossRef]
- Bisbal-Pardo, C.I.; Del Río-Portilla, M.A.; Rocha-Olivares, A. The complete mitochondrial DNA of the Pacific Geoduck clam (Panopea generosa). Mitochondrial DNA 2014, 27, 1955–1956. [Google Scholar] [CrossRef]
- Bisbal-Pardo, C.I.; Del Rio-Portilla, M.A.; Rocha-Olivares, A. Novel gene arrangement in the complete mitochondrial genome of the Cortes Geoduck (Panopea globosa). Mitochondrial DNA Part A 2016, 27, 1957–1958. [Google Scholar] [CrossRef]
- Parmesan, C. Ecological and Evolutionary Responses to Recent Climate Change. Annu. Rev. Ecol. Evol. Syst. 2006, 37, 637–669. [Google Scholar] [CrossRef]
- Paaijmans, K.P.; Heinig, R.L.; Seliga, R.A.; Blanford, J.I.; Blanford, S.; Murdock, C.C.; Thomas, M.B. Temperature variation makes ectotherms more sensitive to climate change. Glob. Chang. Biol. 2013, 19, 2373–2380. [Google Scholar] [CrossRef]
- Li, Q.; Liu, W.; Shirasu, K.; Chen, W.; Jiang, S. Reproductive cycle and biochemical composition of the Zhe oyster Crassostrea plicatula Gmelin in an eastern coastal bay of China. Aquaculture 2006, 261, 75–759. [Google Scholar] [CrossRef]
- Arag´on-Noriega, E.A.; Ch´avez-Villalba, J.; Gribben, P.E.; Alc´antara-Razo, E.; Maeda-Martínez, A.N.; Arambula-Pujol, E.M.; García-Juárez, A.R.; Maldonado-Amparo, R. Morphometric relationships, gametogenic development and spawning of the geoduck clam Panopea globosa (bivalvia: Hiatellidae) in the central gulf of California. J. Shellfish Res. 2007, 26, 423–431. [Google Scholar] [CrossRef]
- Calderon-Aguilera, L.E.; Aragón-Noriega, E.A.; Morales-Bojórquez, E.; Alcántara-Razo, E.; Chávez-Villalba, J. Reproductive cycle of the geoduck clam Panopea generosa at its southernmost distribution limit. Mar. Biol. Res. 2013, 10, 61–72. [Google Scholar] [CrossRef]
- Hamli, H.; Idris, M.H.; Hena, M.K.A.; Rajaee, A.H. Fisheries assessment, gametogenesis and culture practice of local bivalve: A review. Pertanika J. Trop. Agric. Sci. 2019, 42, 103–124. [Google Scholar]
- Han, J.; Kim, H.-J.; Oh, S.-Y.; Choi, Y.-U. Reproductive Characteristics of the Flat Oyster Ostrea denselamellosa (Bivalvia, Ostreidae) Found on the Southern Coast of South Korea. J. Mar. Sci. Eng. 2022, 10, 1326. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, J.H.; Alfaya, J.E.F. Panopea abbreviate (Bivalvia: Hiatellidae) in the Southwestern Atlantic Ocean, taxonomic revision and anatomy. Malacologia 2014, 57, 279–293. [Google Scholar] [CrossRef]
- Molen, S.V.D.; Kroeck, M.; Ciocco, N. Reproductive cycle of the southern geoduck clam, Panopea abbreviate (Bivalvia: Hiatellidae), in north Patagonia, Argentina. Invertebr. Reprod. Dev. 2007, 50, 75–84. [Google Scholar] [CrossRef]
- Vadopalas, B.; Pietsch, T.W.; Friedman, C.S. The proper name for the geoduck: Resurrection of Panopea generosa Gould, 1850, from the synonymy of Panopea abrupta (Conrad, 1849) (Bivalvia: Myoida: Hiatellidae). Malacologia 2010, 52, 169–173. [Google Scholar] [CrossRef]
- Andersen, A.M. Spawning, Growth, and Spatial Distribution of the Geoduck Clam, Panopea generosa Gould, in Hood Canal, Washington. Ph.D. Thesis, University of Washington, Seattle, WA, USA, 1971. [Google Scholar]
- Campbell, A.; Ming, M.D. Maturity and growth of the Pacific geoduck clam, Panopea abrupta, in southern British Columbia, Canada. J. Shellfish Res. 2003, 22, 85–90. [Google Scholar]
- Solan, N.A.; Robinson, S.M.C. Age and gonad development in the geoduck clam Panope abrupta (Conrad) from southern British Columbia. J. Shellfish Res. 1984, 4, 131–137. [Google Scholar]
- Goodwin, L. Observations on spawning and growth of subtidal geoducks (Panope generosa, Gould). Proc. Natl. Shellfish. Assoc. 1976, 65, 49–58. [Google Scholar]
- Borozone, C.C. El ciclo gonadal de Venus antiqua King and Brodrip, 1935 (Veneridae, Bivalvia) en el golfo San José. Physis 1989, 47, 61–72. [Google Scholar]
- Barón, P.J.; Real, L.E.; Ciocci, N.; Ré, M.E. Mophometry, growth and reproduction of an Atlantic popuation of the razor clam Ensis macha (Molina, 1782). Sci. Mar. 2004, 68, 211–217. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Li, F.X.; Ke, C.H. On the sex gonad development and reproductive cycle of clam Paphia undulata. J. Fish. China Shuichan Xuebao 1991, 15, 1–8. [Google Scholar] [CrossRef]


| Family | Species | Size (bp) | Accession No. |
|---|---|---|---|
| Arcticidae | Arctica islandica | 18,289 | NC_022709 |
| Cardiidae | Acanthocardia tuberculata | 16,104 | NC_008452 |
| Cardiidae | Fulvia mutica | 19,110 | NC_022194 |
| Cardiidae | Tridacna squamosa | 20,930 | NC_026558 |
| Hiatellidae | Panopea abrupta | 15,381 | KX494111 |
| Hiatellidae | Panopea generosa | 15,585 | NC_025635 |
| Hiatellidae | Panopea globosa | 15,469 | NC_025636 |
| Lucinidae | Loripes lacteus | 17,321 | NC_013271 |
| Lucinidae | Lucinella divaricata | 18,940 | NC_013275 |
| Myidae | Mya arenaria | 17,947 | NC_024738 |
| Full Gene Name | Location | Size (bp) | Start Codon | Stop Codon | Intergenic Region * |
|---|---|---|---|---|---|
| Cytochrome c oxidase subunit I (cox1) | 1–1563 | 1563 | GTG | TAG | 0 |
| D-lopp (DR) | 1564–2401 | 838 | - | - | 0 |
| Cytochrome c oxidase subunit II (cox2) | 2402–3139 | 738 | ATG | TAA | 10 |
| tRNA-Val (trnV) | 3150–3214 | 65 | - | - | 2 |
| tRNA-Thr (trnT) | 3217–3282 | 66 | - | - | 69 |
| tRNA-Tyr (trnY) | 3352–3413 | 62 | - | - | 10 |
| NADH dehydrogenase subunit 4L (nad4l) | 3424–3714 | 291 | ATG | TAG | 52 |
| ATP synthase F0 subunit 8 (atp8) | 3767–3880 | 114 | ATG | TAA | 163 |
| NADH dehydrogenase subunit 4 (nad4) | 4044–5231 | 1188 | ATA | TAG | 0 |
| tRNA-His (trnH) | 5232–5295 | 64 | - | - | 0 |
| tRNA-Glu (trnE) | 5296–5359 | 64 | - | - | −5 |
| tRNA-Ser2 (trnS2) | 5355–5417 | 63 | - | - | 3 |
| NADH dehydrogenase subunit 3 (nad3) | 5421–5786 | 366 | ATG | TAG | −1 |
| tRNA-Ile (trnI) | 5786–5852 | 67 | - | - | 6 |
| tRNA-Asp (trnD) | 5859–5922 | 64 | - | - | 4 |
| tRNA-Lys (trnK) | 5927–5989 | 63 | - | - | 2 |
| tRNA-Leu2 (TrnL2) | 5992–6056 | 65 | - | - | 0 |
| NADH dehydrogenase subunit 1 (nad1) | 6057–6980 | 924 | ATG | TAG | 1 |
| tRNA-Leu1 (TrnL1) | 6982–7046 | 65 | - | - | 32 |
| tRNA-Asn (TrnN) | 7079–7146 | 68 | - | - | 1 |
| NADH dehydrogenase subunit 5 (nad5) | 7148–8872 | 1725 | ATG | TAA | −1 |
| NADH dehydrogenase subunit 6 (nad6) | 8872–9399 | 528 | ATG | TAA | −1 |
| tRNA-Arg (trnR) | 9399–9462 | 64 | - | - | 2 |
| Cytochrome b (cob) | 9465–10,618 | 1154 | GTG | TAA | 13 |
| tRNA-Trp (trnW) | 10,632–10,698 | 67 | - | - | 0 |
| 16S ribosomal RNA (trnL) | 10,699–11,931 | 1233 | - | - | 0 |
| ATP synthase F0 subunit 6 (atp6) | 11,932–12,639 | 708 | ATG | TAA | 7 |
| tRNA-Met (trnM) | 12,647–12,710 | 64 | - | - | 1 |
| 12S ribosomal RNA (rrnS) | 12,712–13,574 | 863 | - | - | 0 |
| Cytochrome c oxidase subunit III (cox3) | 13,575–14,360 | 786 | ATG | TAG | 4 |
| tRNA-Ser1 (trnS1) | 14,365–14,431 | 67 | - | - | 0 |
| NADH dehydrogenase subunit 2 (nad2) | 14,432–15,478 | 1047 | ATG | TAA | 13 |
| tRNA-Gln (trnQ) | 15,492–15,557 | 66 | - | - | 8 |
| tRNA-Phe (trnF) | 15,566–15,628 | 63 | - | - | 14 |
| tRNA-Cys (trnC) | 15,643–15,707 | 65 | - | - | 22 |
| tRNA-Pro (trnP) | 15,730–15,796 | 67 | - | - | 10 |
| tRNA-Gly (trnG) | 15,807–15,872 | 66 | - | - | 15 |
| tRNA-Ala (trnA) | 15,888–15,952 | 65 | - | - | - |
| Month | Sex Categories | Sex Ratio | |
|---|---|---|---|
| Female | Male | ||
| September | 8 (85.9–111.3) | 9 (91.7–113.8) | 0.88:1 |
| October | 12 (101.5–127.0) | 8 (103.5–129.0) | 1.50:1 |
| November | 6 (82.4–120.7) | 14 (80.5–129.0) | 0.42:1 |
| January | 9 (104.0–117.2) | 10 (101.5–134.4) | 1:1 |
| February | nd | nd | nd |
| March | 11 (80.5–130.5) | 9 (80.5–128.5) | 1.22:1 |
| April | 8 (80.5–125.1) | 12 (82.4–127.0) | 0.66:1 |
| May | 2 (109.4–117.2) | 18 (82.4–122.6) | 0.11:1 * |
| June | 3 (105.5–125.1) | 17 (78.5–112.8) | 0.17:1 * |
| July | 2 (103.3–118.2) | 17 (74.6–115.5) | 0.11:1 * |
| August | 6 (81.6–135.9) | 13 (86.3–112.8) | 0.46:1 |
| Total | 77 | 137 | 0.56:1 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Kim, J.G.; Kwon, O.-N.; Choi, Y.-U. New Record of Geoduck Clam Collected from the East Coast of South Korea and Its Reproductive Characteristics. J. Mar. Sci. Eng. 2023, 11, 776. https://doi.org/10.3390/jmse11040776
Han J, Kim JG, Kwon O-N, Choi Y-U. New Record of Geoduck Clam Collected from the East Coast of South Korea and Its Reproductive Characteristics. Journal of Marine Science and Engineering. 2023; 11(4):776. https://doi.org/10.3390/jmse11040776
Chicago/Turabian StyleHan, Jeonghoon, Jong Guk Kim, O-Nam Kwon, and Young-Ung Choi. 2023. "New Record of Geoduck Clam Collected from the East Coast of South Korea and Its Reproductive Characteristics" Journal of Marine Science and Engineering 11, no. 4: 776. https://doi.org/10.3390/jmse11040776
APA StyleHan, J., Kim, J. G., Kwon, O.-N., & Choi, Y.-U. (2023). New Record of Geoduck Clam Collected from the East Coast of South Korea and Its Reproductive Characteristics. Journal of Marine Science and Engineering, 11(4), 776. https://doi.org/10.3390/jmse11040776







