Abstract
Penaeid shrimp aquaculture is impacted by various diseases. However, most published studies on physiological responses to pathogens have focused on the changes in one or two tissues of shrimp infected by a single pathogen, or the effects of two pathogens infecting the shrimp in a single tissue. There has been limited systematic examination on the similarities and differences of immune responses in multiple tissues under various pathogen infection. Here, the transcriptomic changes of three immune tissues (gill, hepatopancreas and hemocytes) under the infection of white spot syndrome virus (WSSV), Vibrio parahaemolyticus acute hepatopancreatic necrosis disease (VPAHPND), and decapod iridovirus 1 (DIV1) were examined to provide new insights regarding the immune responses of the most important cultured shrimp, Penaeus vannamei. The results showed tissue-specific differences in the immune responses of shrimp tissues. The significant differentially expressed genes (DEGs) in gill are mainly related to environmental information processing and cellular processes. The DEGs in hemocytes are mostly involved in cellular processes, while those in hepatopancreas are primarily associated with metabolism. In addition, cytoskeleton-related proteins, MAPK signaling pathway, complement and coagulation level pathway, and thermogenesis may play key roles in the shrimp–pathogen interactions across tissues. These findings shed light on the typical immune responses of Penaeus vannamei under the infection of pathogens and contribute to the sustainable development of penaeid shrimp farming.
1. Introduction
Penaeus vannamei, commonly known as the Pacific white shrimp, is a commercially important aquaculture species. It is currently the most cultured crustacean species worldwide [1], accounting for about 80% of the total production of cultured penaeid shrimp in Asia [2,3]. However, the frequent outbreaks of infectious diseases have caused large-scale mortality of cultured shrimp, seriously impacting this aquaculture industry as the main obstacle to its development [4,5,6]. The pathogens that cause large-scale mortality of shrimp are mainly viral or bacterial. White spot syndrome virus (WSSV), yellow head virus (YhV), and decapod iridescent virus 1 (DIV1) are the common viral pathogens, while Vibrio parahaemolyticus (VP) which causes acute hepatopancreatic necrosis disease (AHPND) is the common bacterial pathogen [7,8,9] (Table 1). Among these infectious diseases, white spot syndrome (WSS) and AHPND are most destructive, causing huge losses of up to 10 billion USD per year [3]. Recently, the newly emerged DIV1 has also caused substantial economic loss to P. vannamei aquaculture in China [10].

Table 1.
Pathogens involved in this study.
Like other invertebrates, crustaceans do not have acquired immunity. They rely entirely on innate immunity to fight against pathogen infections. Their innate immune system includes physical defense, cellular immunity, and humoral immunity [20,21], which can be stimulated by certain feed additives [22,23]. Gills, hemocytes, and hepatopancreas are the three immune tissues that play crucial roles in penaeids’ defense against pathogens [24,25,26]. Hemocytes play a crucial role in cellular and humoral immune responses [27,28]. Gills are vital tissues of aquatic organisms, responsible for controlling osmotic pressure, ion balance, and gas exchange [29]. Immune-related genes, such as phenol oxidase, are expressed in gill tissues during the infection of pathogenic bacteria [30]. The hepatopancreas plays a pivotal role in the immune defense of crustaceans. It is an essential organ for storage, metabolism, and detoxification, and produces immune proteins, such as hemocyanin and lectin [31].
While there have been many transcriptome studies on the immune tissues of P. vannamei infected by WSSV, VPAHPND, and DIV1 for reviews on the common immune genes and signal pathways in [5,32,33,34], these studies mainly focused on a few key genes that are highly expressed in the tissues under pathogen infection. Genes that are relatively low in expression but differentially expressed are often ignored, but they may be related to important pathways common in multiple tissues or may shed light on the different roles played by various tissues in immune responses. Therefore, it is necessary to investigate in greater detail the differentially expressed genes and pathways through transcriptome analysis of different tissues under the influence of pathogens.
In this study, transcriptome profiles from nine studies on the differential gene expression in the gills, hemocytes, or hepatopancreas of P. vannamei infected by WSSV, VPAHPND, or DIV1 were collected and re-analyzed. The differentially expressed genes in the three tissues of P. vannamei under different pathogen infections were compared. The overarching aim is to identify genes fundamental to the immune response of P. vannamei, and to illuminate the different roles of the immune tissues. The results would improve our understanding on the interactions between pathogens and shrimp, as well as the innate immune mechanisms of the shrimp, which would be conducive to disease prevention and treatment in penaeid shrimp farming.
2. Materials and Methods
2.1. Data Collection
All available RNAseq BioProject data relating to differential gene expression in the three target tissues of Penaeus vannamei in experimental studies on WSSV, VPAHPND, or DIV1 infection were downloaded from NCBI SRA. Their accession numbers are PRJNA233549, PRJNA413606, PRJNA421143, PRJNA428228, PRJNA448614, PRJNA524934, PRJNA554075, PRJNA612147, and PRJNA716175 (Table 2) [4,32,33,34,35,36,37,38]. A total of 109 transcriptome expression profiles were obtained. Profiles from similar treatments were homogenized during analyses (see Table 2 for details).

Table 2.
Transcriptome data analyzed in this study. N: Number of transcriptome expression profiles.
2.2. Transcriptome Assembly and Differential Expression Gene Analysis
Trim Galore ver. 0.6.8 was used to filter low-quality reads and to remove adaptor sequences. After filtering, FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 19 June 2022) was used to check the quality of the reads. Using Hisat2 ver. 2.2.1 [39] with default parameter, clean reads were mapped to the genome of P. vannamei (GenBank accession number ASM397254v1) to obtain the corresponding transcripts. Then, featureCounts [40] were used for read summarization.
To identify differentially expressed genes in each BioProject, we used R/Bioconductor limma [41], EdgeR [42], and DESeq2 [43] for differential expression analysis, and log2FoldChange (log2FC) values and their respective p-values were calculated. Genes with absolute log2FC greater than 2 and the p-value less than or equal to 0.05 were considered differentially expressed candidate genes (DEGC). Genes identified as DEGC in at least two of the three analyses were considered significantly differentially expressed genes (DEGs) in each BioProject (See Supplementary Materials for details).
The amino acid sequences of the DEGs were searched against the non-redundant protein public database of eggNOG [44] for GO and KEGG pathway annotation using eggnog-mapper ver. 2.1.9 [45] with default parameters and an E-value threshold of 1e-5. Matching sequences from non-eukaryotes and chordates were considered contaminants and were eliminated. Clusterprofiler ver. 4.0 [46] was used to conduct GO and KEGG pathway enrichment analyses of the DEGs. GO terms and KEGG pathway descriptions with p-value ≤ 0.05 and ≤ 0.1, respectively, were considered significantly enriched.
The R package GOSemSim [47] was used to measure the functional similarity between GO terms or gene products, and R package ggplot2 was used to visualize the topological structure diagram of functional clustering. As a large number of processes were annotated in gills and hepatopancreas, to reveal key processes involved in the shrimp’s immunity, we removed branches that shared low functional similarity with the others.
3. Results
3.1. Significant Differentially Expressed Genes (DEGs)
In gills, 2195 and 809 DEGC were detected by EdgeR and DESeq2, respectively, and 400 unigenes were DEGs (Table 3). In hemocytes, 388 and 377 DEGC were detected by EdgeR and limma, respectively, and 84 unigenes were DEGs (Table 3). In hepatopancreas, 767 and 1854 DEGC were detected by EdgeR and limma respectively, and 434 unigenes were DEGs (Table 3). We mapped DEGs to the KEGG level1 category (Figure 1). When compared with the other two tissues, DEGs in gills were enriched in “Environmental Information Processing” and “Cellular Processes”, DEGs in hemocytes were mainly enriched in “Cellular Processes”, while those in hepatopancreas were mainly enriched in “Metabolism”.

Table 3.
The number of differentially expressed genes (DEGs) found in gills, hemocytes, and hepatopancreas of P. vannamei under WSSV, VPAHPND, or DIV1 infection compared to control group. Hyphen: the specific method could not identify any DEGC or was not suitable for the dataset.
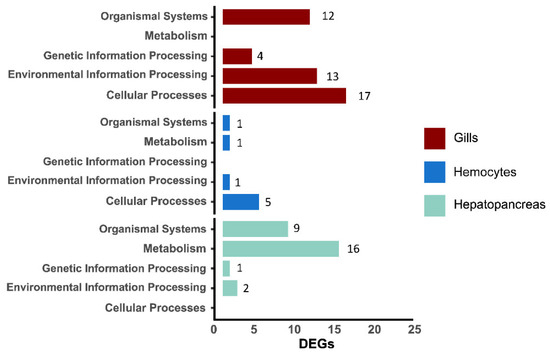
Figure 1.
Histogram of the number of DEGs in the gills, hemolymph, and hepatopancreas of P. vannamei under three pathogens infection assigned to five KEGG level 1 categories, i.e., organismal systems, metabolism, genetic information processing, environmental information processing, and cellular processes.
3.2. GO Analysis of DEGs
GO analysis was conducted to obtain an overview of the molecular responses invoked during pathogen infection in P. vannamei (Figure 2A). While many of the enriched GO terms were tissue-specific and pathogen-specific, some were shared across tissues and pathogens. The GO term “Extracellular Region”, containing 23 DEGs enriched in gills, hemocytes, and hepatopancreas under both WSSV and DIV1 infection, appeared to be the most affected molecular response (Figure 2B). Under the infection of WSSV and VPAHPND, “Gated Channel Activity” (eight DEGs) and “Response to Growth Factor” (five DEGs) were affected in gills and hepatopancreas. “Epithelial Cell Differentiation” (total of seven DEGs) appeared to be markedly affected by all pathogens in hepatopancreas, while 10 DEGs of gills under WSSV and VPAHPND infection were annotated to “R7 Cell Differentiation”.
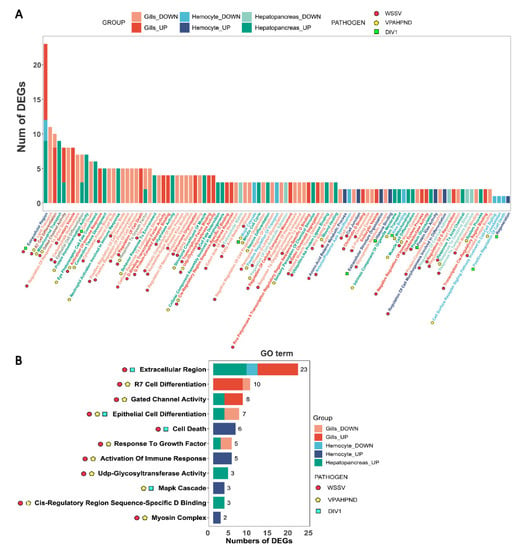
Figure 2.
(A) Distribution of GO terms in all DEGs annotated in the gills, hemocytes, and hepatopancreas of P. vannamei under WSSV, VPAHPND, or DIV1 infection. (B) GO terms of DEGs in the gills, hemolymph, and hepatopancreas that are shared between at least two of the three pathogen infections. The colors of the bars represent the tissue types and expressional changes (down- or up-regulated). The icons next to the KEGG level 3 categories indicate the types of infection.
To reveal the broad categories of responses under different pathogen infection and to compare the response across tissues, we calculated the functional similarity between GO terms or gene products for constructing the functional clustering diagram of DEGs using the GOSemSim algorithm (Figure 3). With respect to WSSV’s influence on the three tissues, the main GO terms of DEGs in the gills could be divided into seven broad categories, namely cell division, differentiation, regeneration, signal transduction, cell connection, and response to external stimuli (Figure 3A). In contrast, the GO terms of DEGs in the hepatopancreas could be divided into only three categories, namely extracellular tissue, cell connection, and cell differentiation (Figure 3B). In the hemocytes, GO terms were mainly related to cell connection and adhesion (Figure 3C). Overall, the commonly enriched DEGs of the three tissues were related to cell differentiation and cell connection. DEGs in the gills were enriched in functions related to external stimulation (Figure 3A).
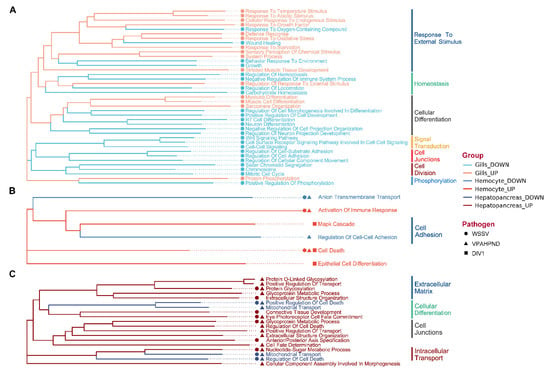
Figure 3.
GO clustering diagram based on DEGs functional similarity in (A) gills, (B) hemocytes, and (C) hepatopancreas, constructed using the R package GOSemSim (branches that shared low functional similarity with the others were removed to reveal critical processes involved in the shrimp’s immunity). DEGs functional categories are grouped according to functional themes. The colors of the branches indicate tissue types and expressional changes (down- or up-regulated), while icons at the tip indicate the types of infection.
3.3. Pathway Enrichment Analysis of DEGs
We analyzed the KEGG pathway of DEGs in the three tissues under different pathogen infections. Regardless of the pathogen infected, the MAPK pathway was affected in all three tissues, though it was up-regulated in hemocytes infected by WSSV and DIV1 but down-regulated in gills and hepatopancreas under WSSV infection (Figure 4). Under WSSV infection, several immune pathways, including phagosome and actin, which are related to the cytoskeleton or cell connection, were enriched in both gills and hemocytes, while the thermogenic pathway was enriched in gills and hepatopancreas (Figure 4A,C). Regarding the tissue-specific response under WSSV infection, it is noteworthy that up-regulated DEGs in gills were annotated to pathways related to immunity (e.g., Rap1 signaling pathway) and cell junction, those in hemocytes were annotated to pathways related to tight junctions, phagosome, and platelet activation, while those in hepatopancreas were mostly associated with glycosaminoglycan biosynthesis.

Figure 4.
Results of KEGG enrichment analysis of DEGs in (A) gill, (B) hemocytes, and (C) hepatopancreas in WSSV, VPAHPND, or DIV1 infected P. vannamei, showing the pathway identifiers, the numbers of up-regulated DEGs (GEH, in red) and down-regulated DEGs (DEL, in green), and the corresponding KEGG level 3, 2 and 1 functional categories. The icons next to the KEGG level 3 categories indicate the types of infection, with grey diamond indicating the KEGG level 3 categories enriched in two of the tissues (gills and hemocytes or hepatopancreas), while the black diamond indicates categories enriched in all three tissues. DEGs were first assigned to a specific KEGG pathway level 2, then assigned to either one of the five KEGG level 1 category (i.e., organismal systems, metabolism, genetic information processing, environmental information processing, and cellular processes).
The KEGG pathways of DEGs from hemocytes infected by WSSV and VPAHPND were almost identical. Lipid metabolism, tight junction, cytoskeleton, phagosome, and platelet activation pathways were enriched in both cases, while MAPK signaling pathway and lysosome pathway were enriched in hemocytes in DIV1-infected shrimp (Figure 4B). In contrast, VPAHPND and WSSV elicited different responses in hepatopancreas, and only the glycosaminoglycan biosynthesis related pathway was invoked in both cases.
4. Discussion
With the global demand for seafood, the aquaculture production of penaeid shrimp has been increasing exponentially. However, shrimp farming is plagued by diseases, which seriously restricts the further development of this industry [48]. There is still a lack of multi-tissue studies on penaeid shrimp infected by various pathogens. In the present study, we investigated the gene expression profiles in multiple tissues of P. vannamei infected with three pathogens of WSSV, VPAHPND, and DIV1 to explore the shrimp’s immune responses to pathogen attack at the molecular level.
4.1. Tissue-Specific Responses
Penaeid shrimp can only protect themselves from pathogens through the innate immune system [49], in which the hepatopancreas, hemocytes, and gills play crucial roles [50,51,52]. In this study, we found tissue-specific immune responses. DEGs in the gills are mainly related to environmental information processing and cellular processes, while DEGs in hemocytes are mainly associated with cellular processes. In contrast, DEGs in the hepatopancreas are primarily related to metabolism. Apparently, the structural and functional differences between the three tissues lead to the tissue-specific responses under pathogen’s attack. The gills are in direct contact with the external environment and are mainly responsible for gas exchange, osmoregulation, and ion balance [25,29]. Thus, the gills may play a role in protection, blocking, and signal transmission against pathogen infection. They receive external stimuli and transmit signals to various tissues of the body. Hemocytes play a role in maintaining homeostasis, immune regulation, and wound healing, while the hepatopancreas is an essential tissue for storage, metabolism, and detoxification [24,26]. Hence, upon pathogen infection, hemocytes and hepatopancreas may play important roles in killing and removing the pathogens to achieve recovery.
Hemolymph contains many immune cells, such as hemocytes which are involved in the phagocytosis of apoptotic cells, as well as immune active substances, such as phenol oxidase, complement factor, lectin, and antimicrobial peptides. They represent one of the critical barriers to pathogen invasion in invertebrates [53]. Some studies have shown that VPAHPND and WSSV stimulate different immune responses in penaeid shrimp lymphoid tissues: VPAHPND causes more extensive immune responses in lymphoid tissues, while WSSV affects metabolic processes [14,54]. However, the DEGs upon infection of both pathogens have been annotated to similar pathways, such as the pro activation system, lysosomes, extracellular matrix, cytoskeleton proteins, and chitin-binding proteins [14]. In our analyses, the pathways annotated by DEGs of hemocytes under WSSV and VPAHPND infection were similar, mainly related to fatty acid metabolism, tight junction, actin cytoskeleton, and phagosome. Moreover, compared with WSSV or DIV1, more upregulated pathways were caused by VPAHPND stimulation, which may indicate that the hemolymph plays a more important role in the anti-bacterial immune responses.
In addition, we found that the complement and coagulation cascade pathways in gill and hepatopancreas tissues infected with WSSV were down-regulated. Previous study indicated that WSSV-infected P. vannamei lost blood coagulation ability after 48 h of infection [55], and the coagulation system seems to be a target of viral and bacterial attacks in crustaceans [56]. A prolonged exposure viral attack may reduce hemocytes abundance, as indicated by a study on P. indicus [57], further causing the loss of blood coagulation ability [55].
Hence, the present study reveals markedly difference responses of gills, hepatopancreas, and hemocytes under pathogen infection. This may be attributable to their different structural attributes, roles in immunity, and their unique interaction with the pathogens.
4.2. Immune Responses Shared among Tissues
MAPK signaling pathway plays a vital role in the responses of cells to extracellular stimuli. In vertebrates, the MAPK signaling pathway plays a crucial role in the innate immune system, especially in the fight against pathogenic infections [53,58]. The differential expression of proteins related to MAPK signaling pathway, Ras signaling pathway, phagosome, and Hippo signaling pathway was also found in mud crabs infected with WSSV and VPAHPND [59]. However, some studies have also reported that virus may activate hosts’ MAPK pathway in cytoskeleton, which might help viral replication in cells, and thus promote pathogen infection [60,61]. Hence, a lowered expression of genes involved in MAPK signaling pathways could indicate that the virus is inefficient in activating the said pathway, or that the host actively down-regulate the pathway to hamper viral replication. Our study shows that the MAPK signaling pathway was down-regulated in the gills and hepatopancreas of WSSV challenged, but up-regulated in the hemocytes of DIV1 and VPAHPND challenged shrimps, which may reflect complex interactions between the host and pathogens, warranting further investigation.
The present study shows that WSSV elicited thermogenic responses in the hepatopancreas and gills. Chen et al. found that in the early stage of WSSV infection, slightly warming the penaeid shrimp pond could effectively increase shrimp’s resistance to WSSV and reduced the cumulative mortality [21]. Lin et al. found that the fever response can positively regulate the immune response through heat shock protein [62]. Yuan et al. found that the activation of heat shock protein can reduce the cumulative mortality of penaeid shrimp infected with WSSV [21,62,63]. Thus, under the attack of WSSV, thermogenic changes may be part of the shrimp’s immune responses which may entail the activation of heat shock protein. The exact mechanism in this respect needs to be further studied.
Genes related to cytoskeleton or cell junction, including actin and focal adhesion, are generally upregulated upon pathogen infection, and it is believed that the cytoskeleton plays an essential role in the innate immunity of invertebrates through cell adhesion and phagocytosis [64,65,66]. However, pathogens might have evolved to regulate the host’s cytoskeleton structure to gain easy passage to tissues, as evidenced in a study of gill, gut, and cuticular epithelium of P. vannamei under WSSV infection [4]. This study showed that cytoskeleton-related proteins such as actin and focal adhesion were significantly upregulated in gills, hemocytes, and hepatopancreas of shrimp infected by the three pathogens, but the underlying cause and the consequence are yet to be determined in future studies.
5. Conclusions
To improve the understanding of the immune responses of P. vannamei upon pathogen infection, we compared the transcriptome data from 109 transcriptome expression profiles of P. vannamei infected by three common pathogens. Though shrimps exhibit various responses under the attack of different pathogens, we also identify common molecular responses potentially related to immunity, including the upregulation of cytoskeletal transcripts related to viral trafficking or cell phagocytosis during infection, MAPK signaling pathway, complement and coagulation level communication, and thermogenesis. The differences in the functions of the three tissues may lead to their tissue-specific differences under pathogen infection. Compared with WSSV and DIV1, VPAHPND caused more immune responses in the hemolymph, which may indicate that the hemolymph plays an essential role in the anti-bacterial immune responses. In addition, there may be a relationship between thermogenic response and the activation of heat shock proteins. The MAPK signaling pathway may be a double-edged sword in host–pathogen interactions. In conclusion, our findings provide new insights into the interactions between P. vannamei and pathogens. The results improve the understanding of the immune response mechanisms of penaeid shrimp, provide a theoretical basis for the prevention and control of diseases in cultured penaeid shrimp, and are important for promoting their health for the sustainable development of shrimp farming.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jmse11020389/s1, Figure S1: Transcriptomic changes of gills infected by WSSV based on two studies (BioProject accession number: (A) PRJNA524934; (B) PRJNA716175), showing results from principal component analysis (PCA), gene abundance heat map, Wien diagrams showing number of differentially expressed candidate genes; Figure S2: Transcriptomic changes of haemolymph infected by WSSV, VPAHPND and DIV1 based on two studies (BioProject accession number: (A) PRJNA233549; (B) PRJNA448614; (C) PRJNA612147), showing results from principal component analysis (PCA), gene abundance heat map, Wien diagrams showing number of differentially expressed candidate genes; Figure S3: Transcriptomic changes of hepatopancreas infected by WSSV and VPAHPND based on two studies (BioProject accession number: (A) PRJNA413606; (B) PRJNA421143; (C) PRJNA428228; (D) PRJNA554075), showing results from principal component analysis (PCA), gene abundance heat map, Wien diagrams showing number of differentially expressed candidate genes.
Author Contributions
Conceptualization, Z.W. and K.Y.M.; data curation, Z.W.; formal analysis, Z.W.; investigation, Z.W.; methodology, Z.W.; project administration, K.Y.M.; resources, Z.W. and K.Y.M.; supervision, K.Y.M.; validation, Z.W. and K.Y.M.; visualization, Z.W.; writing—original draft, Z.W. and K.Y.M.; writing—review & editing, Z.W., K.H.C., and K.Y.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data in this study are openly available in Dryad at doi:10.5061/dryad.sxksn0376.
Acknowledgments
Computation in this work was performed on the Secevo HPC cluster of the School of Ecology, Sun Yat-sen University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cai, J.; Galli, G. Top 10 Species Groups in Global Aquaculture 2019; Food and Agriculture Organization of the United Nations: Rome, Italy, 2021. [Google Scholar]
- Zhang, S.; Fang, S.; Song, S.; Zheng, Y.; Tan, B.; Shi, L. Effect of changes in the activity of Wnt/β-catenin signalling pathway on the growth performance, immunity and transcriptome response in Litopenaeus vannamei. Aquac. Rep. 2021, 20, 100774. [Google Scholar] [CrossRef]
- Flegel, T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012, 110, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Millard, R.; Bickley, L.; Bateman, K.; Farbos, A.; Minardi, D.; Moore, K.; Ross, S.; Stentiford, G.; Tyler, C.; van Aerle, R.; et al. Global mRNA and miRNA Analysis reveal key processes in the initial response to infection with WSSV in the Pacific whiteleg shrimp. Viruses 2021, 13, 1140. [Google Scholar] [CrossRef]
- Yan, X.; Liao, X.; He, J.; Yin, B.; Li, C. Effects of WSSV and DIV1 infection on hemolymph coagulation in Litopenaeus vannamei. Shengwuxuezazhi 2022, 39, 18. [Google Scholar]
- Caro, L.F.A.; Mai, H.N.; Noble, B.; Dhar, A.K. Acute hepatopancreatic necrosis disease (VPAHPND), a chronic disease in shrimp (Penaeus vannamei) population raised in latin America. J. Invertebr. Pathol. 2020, 174, 107424. [Google Scholar] [CrossRef]
- Lightner, D.V. The penaeid shrimp viruses TSV, IHHNV, WSSV, and YHV: Current status in the Americas, available diagnostic methods, and management strategies. J. Appl. Aquac. 1999, 9, 27–52. [Google Scholar] [CrossRef]
- Chen, X.; Qiu, L.; Wang, H.; Zou, P.; Dong, X.; Li, F.; Huang, J. Susceptibility of Exopalaemon carinicauda to the infection with shrimp hemocyte iridescent virus (SHIV 20141215), a strain of Decapod Iridescent Virus 1 (DIV1). Viruses 2019, 11, 387. [Google Scholar] [CrossRef] [PubMed]
- Sangpo, P.; Thitamadee, S.; Dong, H.T.; Senapin, S. Aeromonas schubertii, a novel bacterium recovered from AHPND affected farm is lethal to whiteleg shrimp, Penaeus vannamei. Microb. Pathog. 2020, 149, 104501. [Google Scholar] [CrossRef]
- Qiu, L.; Chen, M.-M.; Wan, X.-Y.; Li, C.; Zhang, Q.-L.; Wang, R.-Y.; Cheng, D.-Y.; Dong, X.; Yang, B.; Wang, X.-H.; et al. Characterization of a new member of Iridoviridae, Shrimp hemocyte iridescent virus (SHIV), found in white leg shrimp (Litopenaeus vannamei). Sci. Rep. 2017, 7, 11834. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, B.; Rai, P.; Mohan, S.A.; Shekhar, M.S.; Karunasagar, I. Biology, host range, pathogenesis and diagnosis of white spot syndrome virus. Indian J. Virol. 2012, 23, 161–174. [Google Scholar] [CrossRef]
- Yan, D.C.; Huang, J.; Yang, B.; Sun, H.S.; Wang, Y.Y.; Liu, X. Competition of infectious hypodermal and haematopoietic necrosis virus (IHHNV) with white spot syndrome virus (WSSV) for binding to shrimp cellular membrane. J. Fish Dis. 2016, 39, 1225–1229. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Söderhäll, K.; Söderhäll, I. Characterization of white spot syndrome virus replication in in vitro-cultured haematopoietic stem cells of freshwater crayfish, Pacifastacus leniusculus. J. Gen. Virol. 2006, 87, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, S.; Li, F. Different immune responses of the lymphoid organ in shrimp at early challenge stage of Vibrio parahaemolyticus and WSSV. Animals 2021, 11, 2160. [Google Scholar] [CrossRef]
- Escobedo-Bonilla, C.M.; Alday-Sanz, V.; Wille, M.; Sorgeloos, P.; Pensaert, M.B.; Nauwynck, H.J. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 2007, 31, 1–18. [Google Scholar] [CrossRef]
- Gong, H.-Y.; Li, Q.-Y.; Zhang, H.; Ye, L.; Shi, L.; Feng, Y.-H. Development and comparison of qPCR and qLAMP for rapid detection of the decapod iridescent virus 1 (DIV1). J. Invertebr. Pathol. 2021, 182, 107567. [Google Scholar] [CrossRef]
- Qiu, L.; Chen, X.; Gao, W.; Li, C.; Guo, X.-M.; Zhang, Q.-L.; Yang, B.; Huang, J. Molecular epidemiology and histopathological study of a natural infection with decapod iridescent virus 1 in farmed white leg shrimp, Penaeus vannamei. Aquaculture 2020, 533, 736105. [Google Scholar] [CrossRef]
- Srisala, J.; Sanguanrut, P.; Thaiue, D.; Laiphrom, S.; Siriwattano, J.; Khudet, J.; Powtongsook, S.; Flegel, T.W.; Sritunyalucksana, K. Infectious myonecrosis virus (IMNV) and Decapod iridescent virus 1 (DIV1) detected in captured, wild Penaeus monodon. Aquaculture 2021, 545, 737262. [Google Scholar] [CrossRef]
- Thitamadee, S.; Prachumwat, A.; Srisala, J.; Jaroenlak, P.; Salachan, P.V.; Sritunyalucksana, K.; Flegel, T.W.; Itsathitphaisarn, O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture 2016, 452, 69–87. [Google Scholar] [CrossRef]
- Tassanakajon, A.; Somboonwiwat, K.; Supungul, P.; Tang, S. Discovery of immune molecules and their crucial functions in shrimp immunity. Fish Shellfish Immunol. 2013, 34, 954–967. [Google Scholar] [CrossRef]
- Chen, Y.-H.; He, J.-G. Effects of environmental stress on shrimp innate immunity and white spot syndrome virus infection. Fish Shellfish Immunol. 2018, 84, 744–755. [Google Scholar] [CrossRef]
- Abdel-Latif, H.M.; Dawood, M.A.; Alagawany, M.; Faggio, C.; Nowosad, J.; Kucharczyk, D. Health benefits and potential applications of fucoidan (FCD) extracted from brown seaweeds in aquaculture: An updated review. Fish Shellfish Immunol. 2022, 122, 115–130. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Latif, H.M.; Abdel-Daim, M.M.; Shukry, M.; Nowosad, J.; Kucharczyk, D. Benefits and applications of Moringa oleifera as a plant protein source in Aquafeed: A review. Aquaculture 2021, 547, 737369. [Google Scholar] [CrossRef]
- Liu, X.-L.; Xi, Q.-Y.; Yang, L.; Li, H.-Y.; Jiang, Q.-Y.; Shu, G.; Wang, S.-B.; Gao, P.; Zhu, X.-T.; Zhang, Y.-L. The effect of dietary Panax ginseng polysaccharide extract on the immune responses in white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2011, 30, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-L.; Hsieh, Y.-T. Immunostimulation of tiger shrimp (Penaeus monodon) hemocytes for generation of microbicidal substances: Analysis of reactive oxygen species. Dev. Comp. Immunol. 1994, 18, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.-F.; Yao, C.-L.; Wang, Z.-Y. Immune response and gene expression in shrimp (Litopenaeus vannamei) hemocytes and hepatopancreas against some pathogen-associated molecular patterns. Fish Shellfish Immunol. 2009, 27, 563–570. [Google Scholar] [CrossRef]
- Söderhäll, I. Crustacean hematopoiesis. Dev. Comp. Immunol. 2016, 58, 129–141. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, S.-C.; Li, Y.-L.; Li, J.; Liu, H.-P. Hemocyte-Mediated Phagocytosis in Crustaceans. Front. Immunol. 2020, 11, 268. [Google Scholar] [CrossRef]
- Clavero-Salas, A.; Sotelo-Mundo, R.R.; Gollas-Galván, T.; Hernández-López, J.; Peregrino-Uriarte, A.B.; Muhlia-Almazán, A.; Yepiz-Plascencia, G. Transcriptome analysis of gills from the white shrimp Litopenaeus vannamei infected with White Spot Syndrome Virus. Fish Shellfish Immunol. 2007, 23, 459–472. [Google Scholar] [CrossRef]
- Frischer, M.E.; Landers, S.C.; Walker, A.N.; Powell, S.A.; Lee, R.F. Black gill in marine decapod crustaceans: A review. Rev. Fish. Sci. Aquac. 2022, 30, 498–519. [Google Scholar] [CrossRef]
- Rao, R.; Zhu, Y.B.; Alinejad, T.; Tiruvayipati, S.; Thong, K.L.; Wang, J.; Bhassu, S. RNA-seq analysis of Macrobrachium rosenbergii hepatopancreas in response to Vibrio parahaemolyticus infection. Gut Pathog. 2015, 7, 6. [Google Scholar] [CrossRef]
- Maralit, B.A.; Jaree, P.; Boonchuen, P.; Tassanakajon, A.; Somboonwiwat, K. Differentially expressed genes in hemocytes of Litopenaeus vannamei challenged with Vibrio parahaemolyticus AHPND (VPAHPND) and VPAHPND toxin. Fish Shellfish Immunol. 2018, 81, 284–296. [Google Scholar] [CrossRef]
- Peruzza, L.; Shekhar, M.S.; Kumar, K.V.; Swathi, A.; Karthic, K.; Hauton, C.; Vijayan, K.K. Temporal changes in transcriptome profile provide insights of White Spot Syndrome Virus infection in Litopenaeus vannamei. Sci. Rep. 2019, 9, 13509. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, X.; Zhou, Z.; Zhang, T.; Yi, Q.; Liu, R.; Wang, M.; Song, L. The promotion of cytoskeleton integration and redox in the haemocyte of shrimp Litopenaeus vannamei after the successive stimulation of recombinant VP28. Dev. Comp. Immunol. 2014, 45, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Peruzza, L.; Thamizhvanan, S.; Vimal, S.; Kumar, K.V.; Shekhar, M.; Smith, V.; Hauton, C.; Vijayan, K.; Hameed, A.S. A comparative synthesis of transcriptomic analyses reveals major differences between WSSV-susceptible Litopenaeus vannamei and WSSV-refractory Macrobrachium rosenbergii. Dev. Comp. Immunol. 2019, 104, 103564. [Google Scholar] [CrossRef] [PubMed]
- Tinwongger, S.; Thawonsuwan, J.; Kondo, H.; Hirono, I. Identification of an anti-lipopolysaccharide factor AV-R isoform (LvALF AV-R) related to Vp_PirAB-like toxin resistance in Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 84, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Andrade, S.; Fernandes, J.; Freitas, P. Shedding the light on Litopenaeus vannamei differential muscle and hepatopancreas immune responses in White Spot Syndrome Virus (WSSV) Exposure. Genes 2020, 11, 805. [Google Scholar] [CrossRef]
- Liao, X.; Wang, C.; Wang, B.; Qin, H.; Hu, S.; Wang, P.; Sun, C.; Zhang, S. Comparative transcriptome analysis of Litopenaeus vannamei reveals that triosephosphate isomerase-like genes play an important role during Decapod Iridescent Virus 1 Infection. Front. Immunol. 2020, 11, 1904. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.V.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Yu, G. Gene ontology semantic similarity analysis using GOSemSim. In Stem Cell Transcriptional Networks: Methods and Protocols; Kidder, B.L., Ed.; Springer US: New York, NY, USA, 2020; pp. 207–215. [Google Scholar]
- Kidane, D.G.; Brækkan, E.H. Global Seafood demand growth differences across regions, income levels, and time. Mar. Resour. Econ. 2021, 36, 289–305. [Google Scholar] [CrossRef]
- Sarathi, M.; Ahmed, V.I.; Venkatesan, C.; Balasubramanian, G.; Prabavathy, J.; Hameed, A.S. Comparative study on immune response of Fenneropenaeus indicus to Vibrio alginolyticus and white spot syndrome virus. Aquaculture 2007, 271, 8–20. [Google Scholar] [CrossRef]
- Habib, Y.J.; Wan, H.; Sun, Y.; Shi, J.; Yao, C.; Lin, J.; Ge, H.; Wang, Y.; Zhang, Z. Genome-wide identification of toll-like receptors in Pacific white shrimp (Litopenaeus vannamei) and expression analysis in response to Vibrio parahaemolyticus invasion. Aquaculture 2021, 532, 735996. [Google Scholar] [CrossRef]
- Burnett, K.G.; Burnett, L.E. Respiratory and metabolic impacts of crustacean immunity: Are there implications for the insects? Integr. Comp. Biol. 2015, 55, 856–868. [Google Scholar] [CrossRef]
- Habib, Y.J.; Zhang, Z. The involvement of crustaceans toll-like receptors in pathogen recognition. Fish Shellfish Immunol. 2020, 102, 169–176. [Google Scholar] [CrossRef]
- Iwanaga, S.; Lee, B.-L. Recent advances in the innate immunity of invertebrate animals. BMB Rep. 2005, 38, 128–150. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Yao, C.; Wang, Z. Reactive oxygen system plays an important role in shrimp Litopenaeus vannamei defense against Vibrio parahaemolyticus and WSSV infection. Dis. Aquat. Org. 2011, 96, 9–20. [Google Scholar] [CrossRef]
- Karthick, S.; Sunil Kumar, K.; Meiyalagan, V.; Arumugam, M. Comparative study on immunological responses in Scylla serrata and Litopenaeus vannamei upon WSSV infection. Aquac. Res. 2022, 53, 2249–2262. [Google Scholar] [CrossRef]
- Liu, C.-H.; Chang, C.-C.; Chiu, Y.-C.; Cheng, W.; Yeh, M.-S. Identification and cloning of a transglutaminase from giant freshwater prawn, Macrobrachium rosenbergii, and its transcription during pathogen infection and moulting. Fish Shellfish Immunol. 2011, 31, 871–880. [Google Scholar] [CrossRef]
- Yoganandhan, K.; Thirupathi, S.; Hameed, A.S. Biochemical, physiological and hematological changes in white spot syndrome virus-infected shrimp, Penaeus indicus. Aquaculture 2003, 221, 1–11. [Google Scholar] [CrossRef]
- Cheesman, H.K.; Feinbaum, R.L.; Thekkiniath, J.; Dowen, R.H.; Conery, A.L.; Pukkila-Worley, R. Aberrant activation of p38 MAP kinase-dependent innate immune responses is toxic to Caenorhabditis elegans. G3 Genes Genomes Genet. 2016, 6, 541–549. [Google Scholar] [CrossRef]
- Sun, B.; Wang, Z.; Wang, Z.; Ma, X.; Zhu, F. A Proteomic Study of hemocyte proteins from mud crab (Scylla paramamosain) infected with white spot syndrome virus or Vibrio alginolyticus. Front. Immunol. 2017, 8, 468. [Google Scholar] [CrossRef]
- Kumar, R.; Khandelwal, N.; Thachamvally, R.; Tripathi, B.N.; Barua, S.; Kashyap, S.K.; Maherchandani, S.; Kumar, N. Role of MAPK/MNK1 signaling in virus replication. Virus Res. 2018, 253, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Zhang, Y.; Liu, P.; Li, J. Comparative proteomic investigation of Marsupenaeus japonicus hepatopancreas challenged with Vibrio parahaemolyticus and white spot syndrome virus. Fish Shellfish Immunol. 2019, 93, 851–862. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, Y.; Zhang, K.; Zheng, Y.; Lu, L.; Chang, H.; Yang, H.; Yang, Y.; Wan, Y.; Wang, S.; et al. Fever promotes t lymphocyte trafficking via a thermal sensory pathway involving heat shock protein 90 and α4 integrins. Immunity 2019, 50, 137–151.e6. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Yuan, F.-H.; He, H.-H.; Bi, H.-T.; Weng, S.-P.; He, J.-G.; Chen, Y.-H. Heat shock 70 kDa protein cognate 5 involved in WSSV toleration of Litopenaeus vannamei. Dev. Comp. Immunol. 2017, 72, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, F.; Sun, Z.; Zhang, X.; Xiang, Z. Differentially proteomic analysis of the Chinese shrimp at WSSV latent and acute infection stages by iTRAQ ap-proach. Fish Shellfish Immunol. 2016, 54, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Wang, Z.; Wang, X. Characterization of myosin light chain in shrimp hemocytic phagocytosis. Fish Shellfish Immunol. 2010, 29, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, F. Different roles of a novel shrimp microRNA in white spot syndrome virus (WSSV) and Vibrio alginolyticus infection. Dev. Comp. Immunol. 2018, 79, 21–30. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).