Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Catching and Maintaining the Animals
2.2. Bleeding Procedure and Preparation of the CFE
2.3. Proteomic Analysis
2.4. Cell Cultures
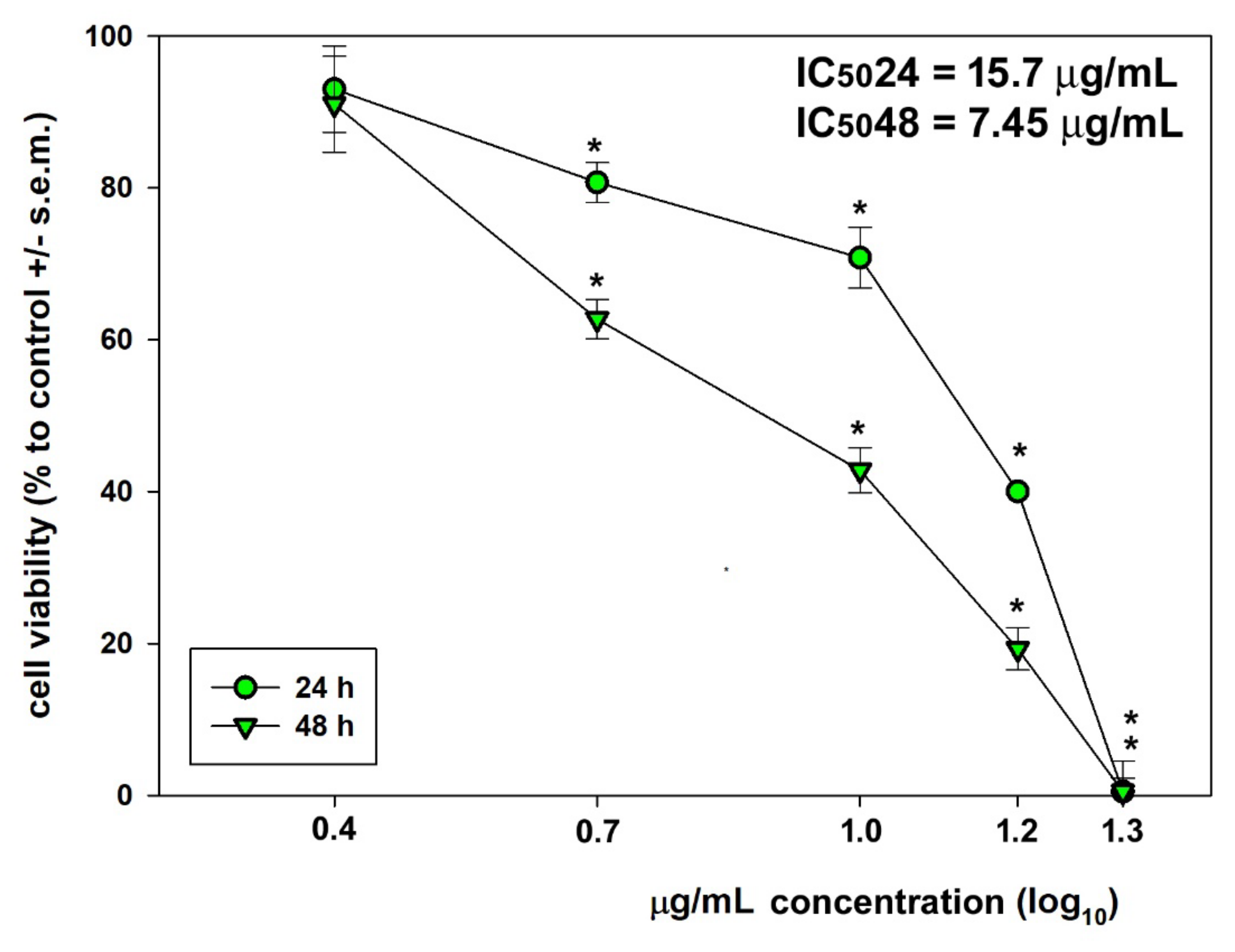
2.5. Viability Assays
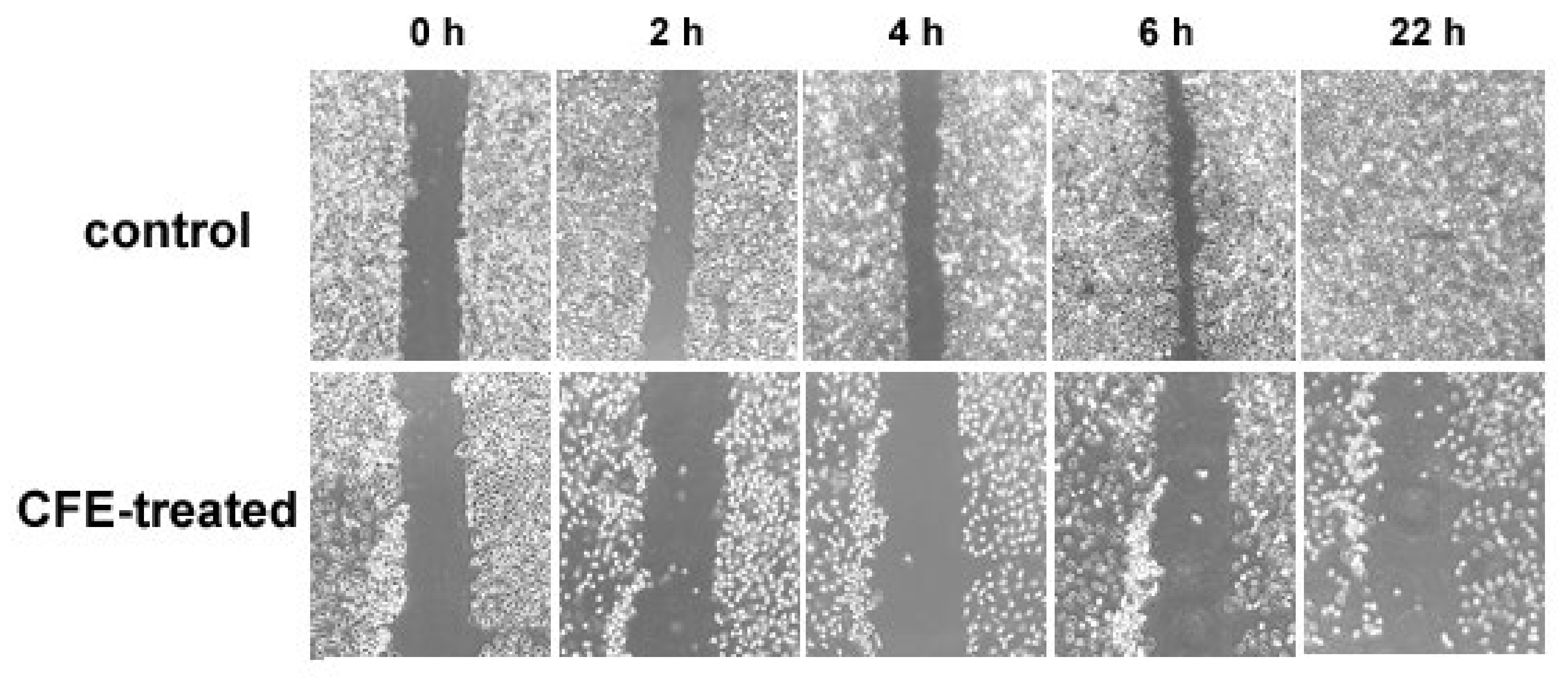
2.6. Wound Healing Assay
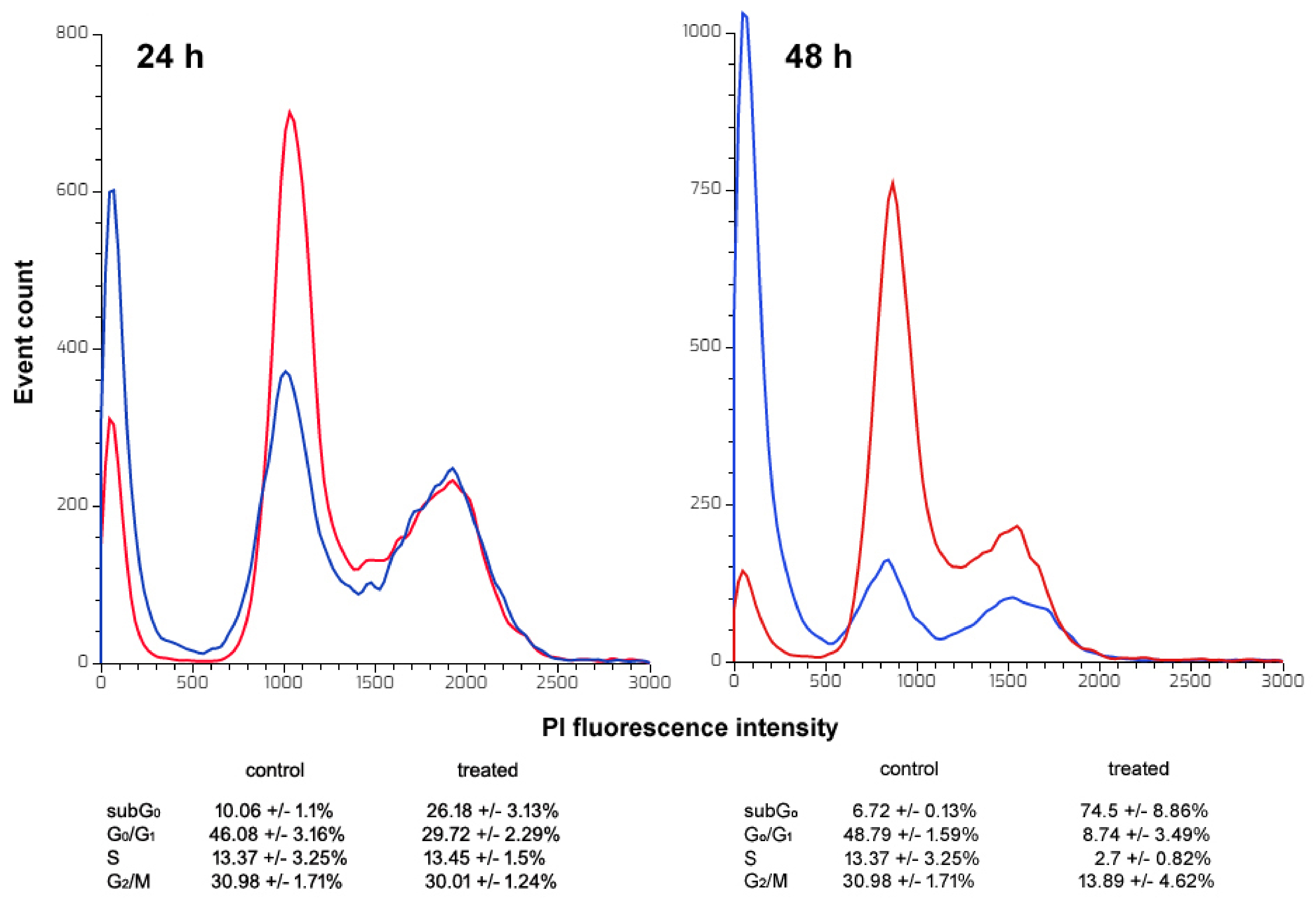
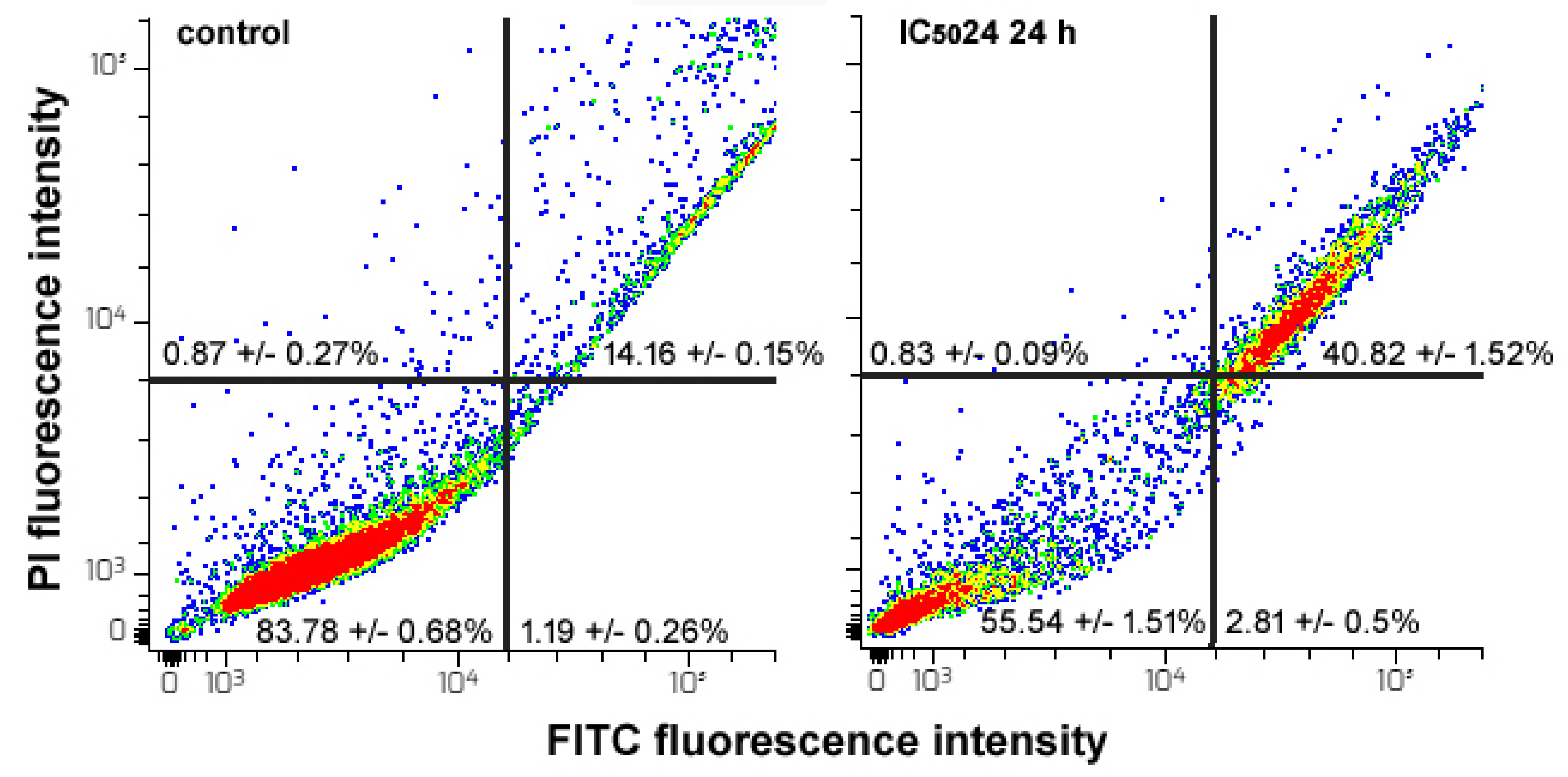
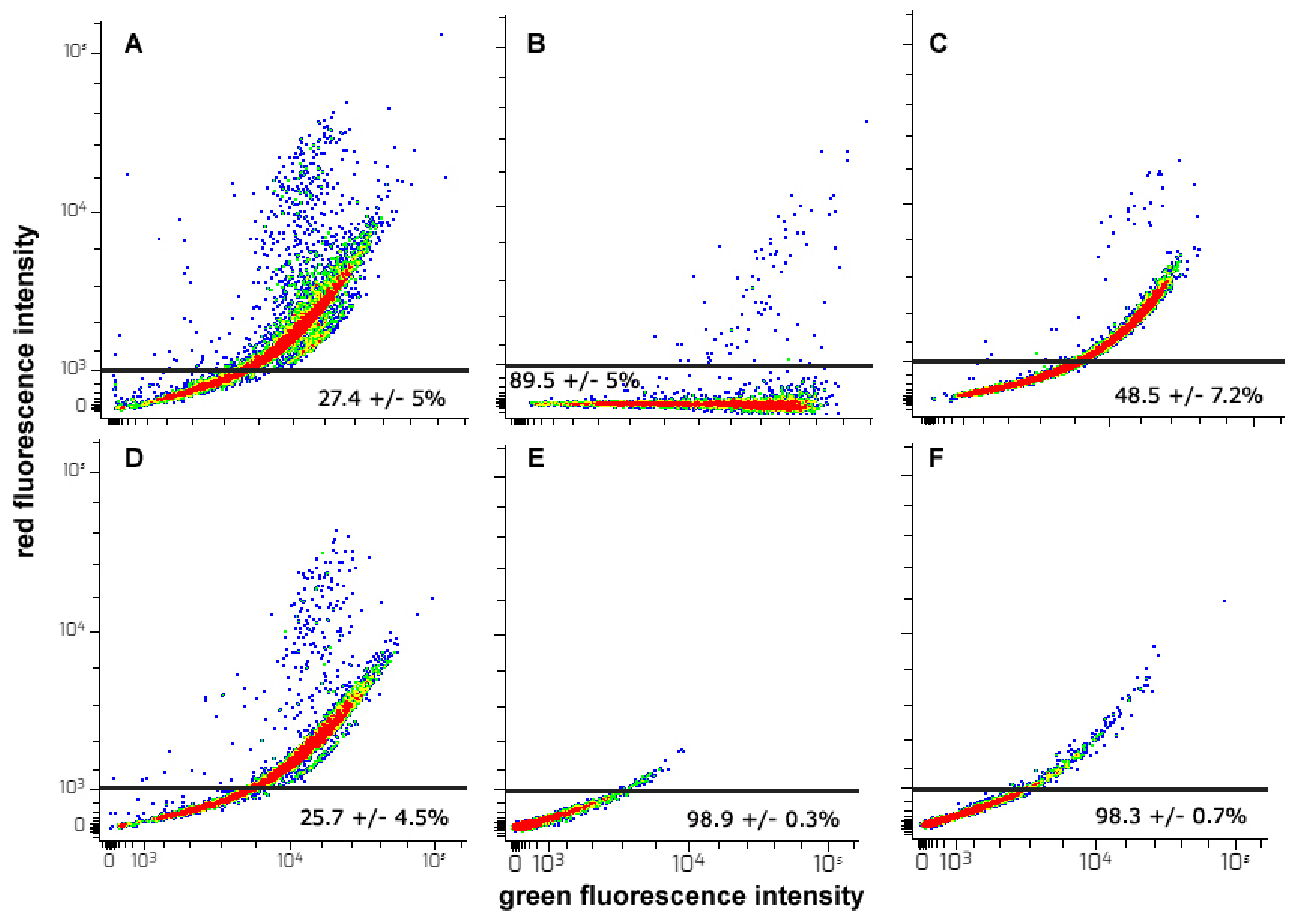
2.7. Flow Cytometry Assays
2.7.1. Cell Cycle Distribution Assay
2.7.2. Apoptosis Assay
2.7.3. Analysis of Mitochondrial Transmembrane Potential (MMP)
2.7.4. ROS Assay
2.7.5. AVO Accumulation Assay
2.8. Statistics
3. Results
4. Discussion
- (1)
- For HepG2 cells, anti-cancer agent-triggered growth arrest at the G2/M phase is an event commonly leading up to apoptosis. This impairment was ascribed to different causes, such as the inhibition of cyclin-dependent kinase-2A, the activation of p38 MAPK signalization or the inhibition of tubulin polymerization, determining the failure of the assembly and dynamics of mitotic spindle structure [80,81,82]. Therefore, the molecular basis of CFE-stimulated cell cycle block is an interesting aspect that remains to be clarified and can bring to light new intracellular targets for anti-HCC treatment options;
- (2)
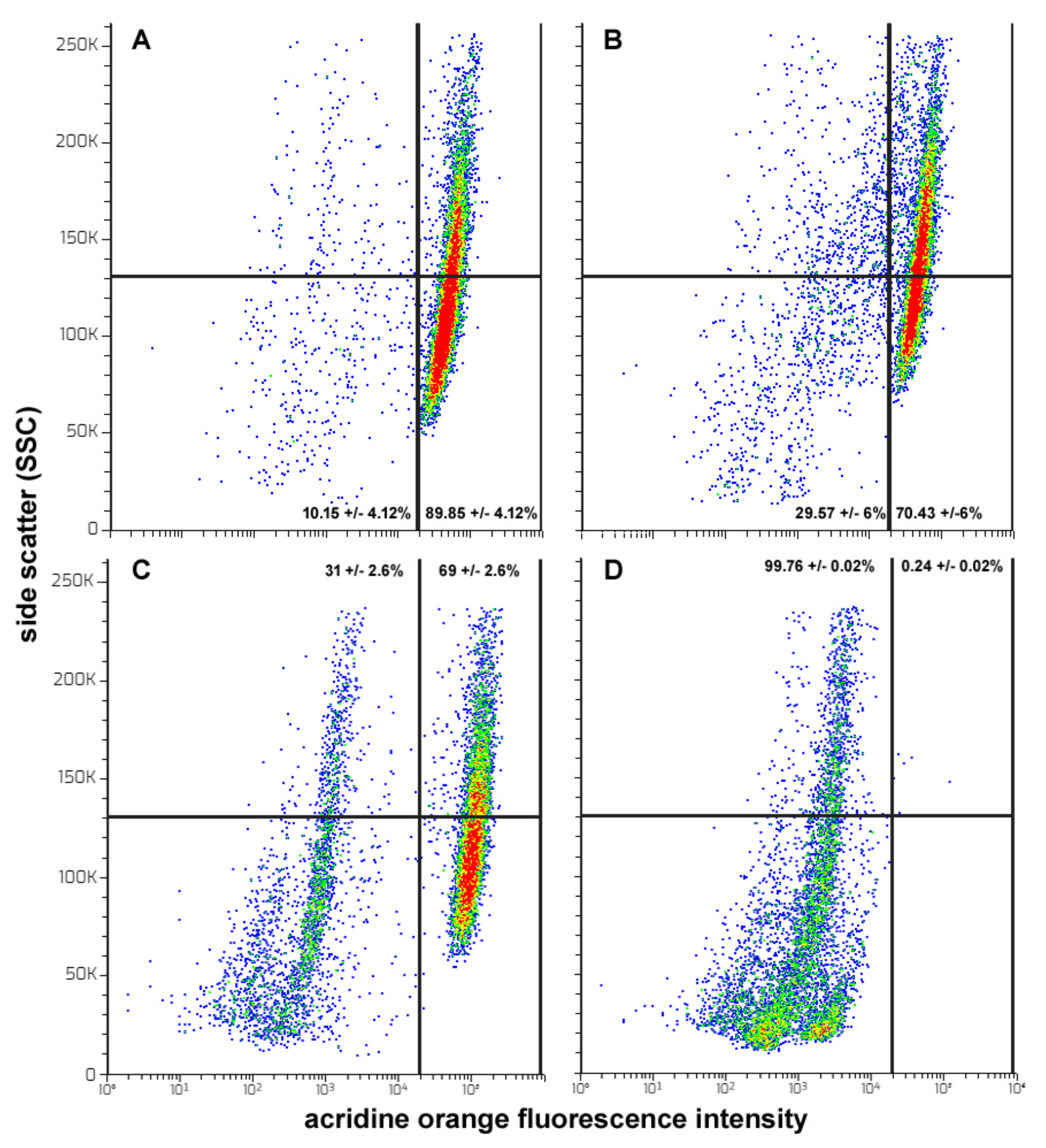
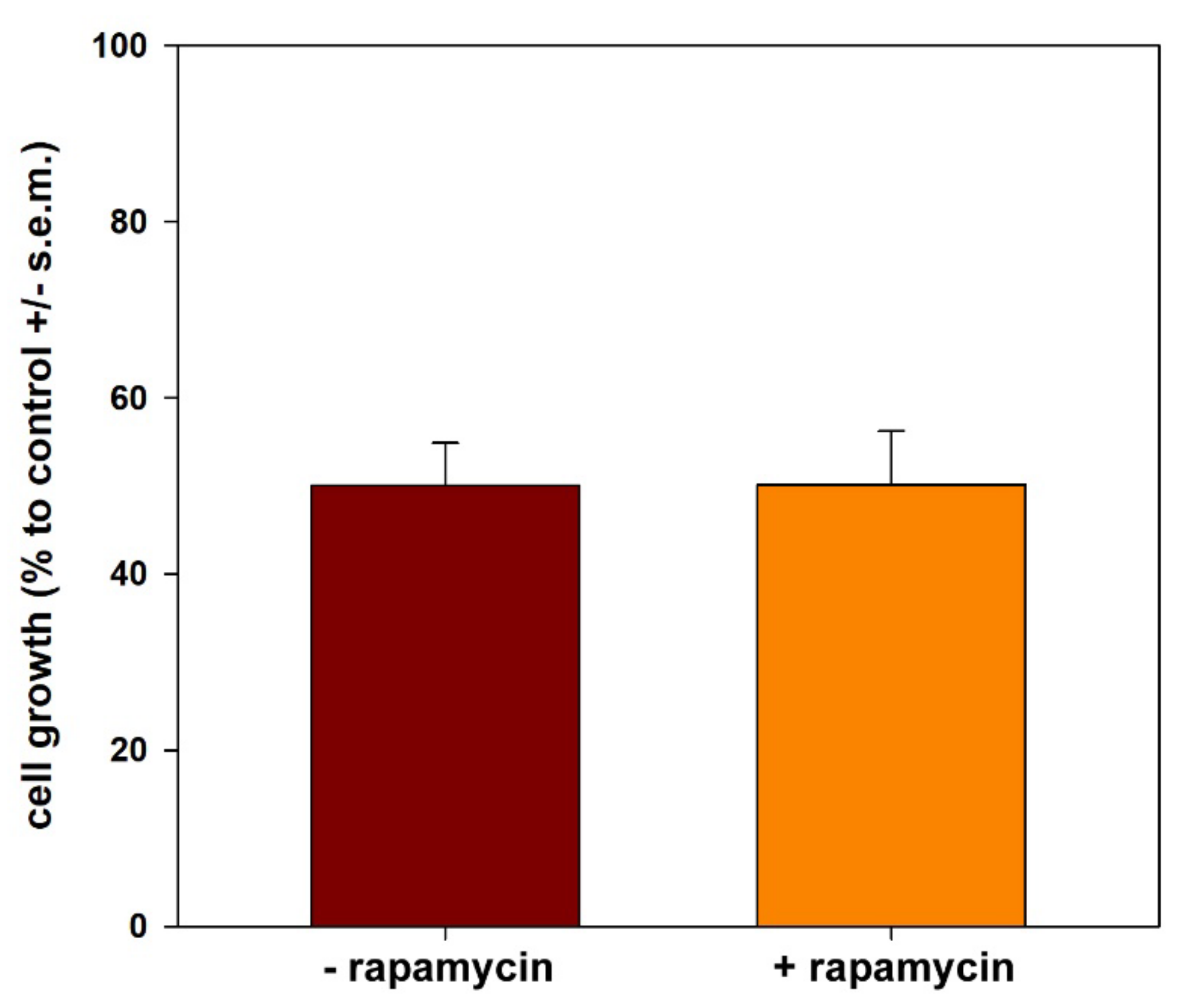
- It is known that HepG2 cells are endowed with elevated levels of basal autophagy [23,56]. This allows their survival, active growth and locomotion by fulfilling the related high metabolic and energetic demands. Thus, it is conceivable that the suppression of “protective” autophagy may contribute to CFE’s cytotoxic activity. Interestingly, in line with [83], autophagy down-regulation may lead to the inhibition of cell proliferation and the onset of apoptosis via the intrinsic pathway, as suggested in our model system by the derangement of mitochondrial function. On the other hand, the failure of rapamycin co-treatment to restore cell viability suggests that autophagy inhibition is secondary to the other CFE exposure-linked aspects of cell damage, i.e., mitochondrial dysfunction, ROS depletion and apoptosis promotion, the latter determining a massive impairment of cell functions that cannot be rescued by the sole autophagy reactivation;
- (3)
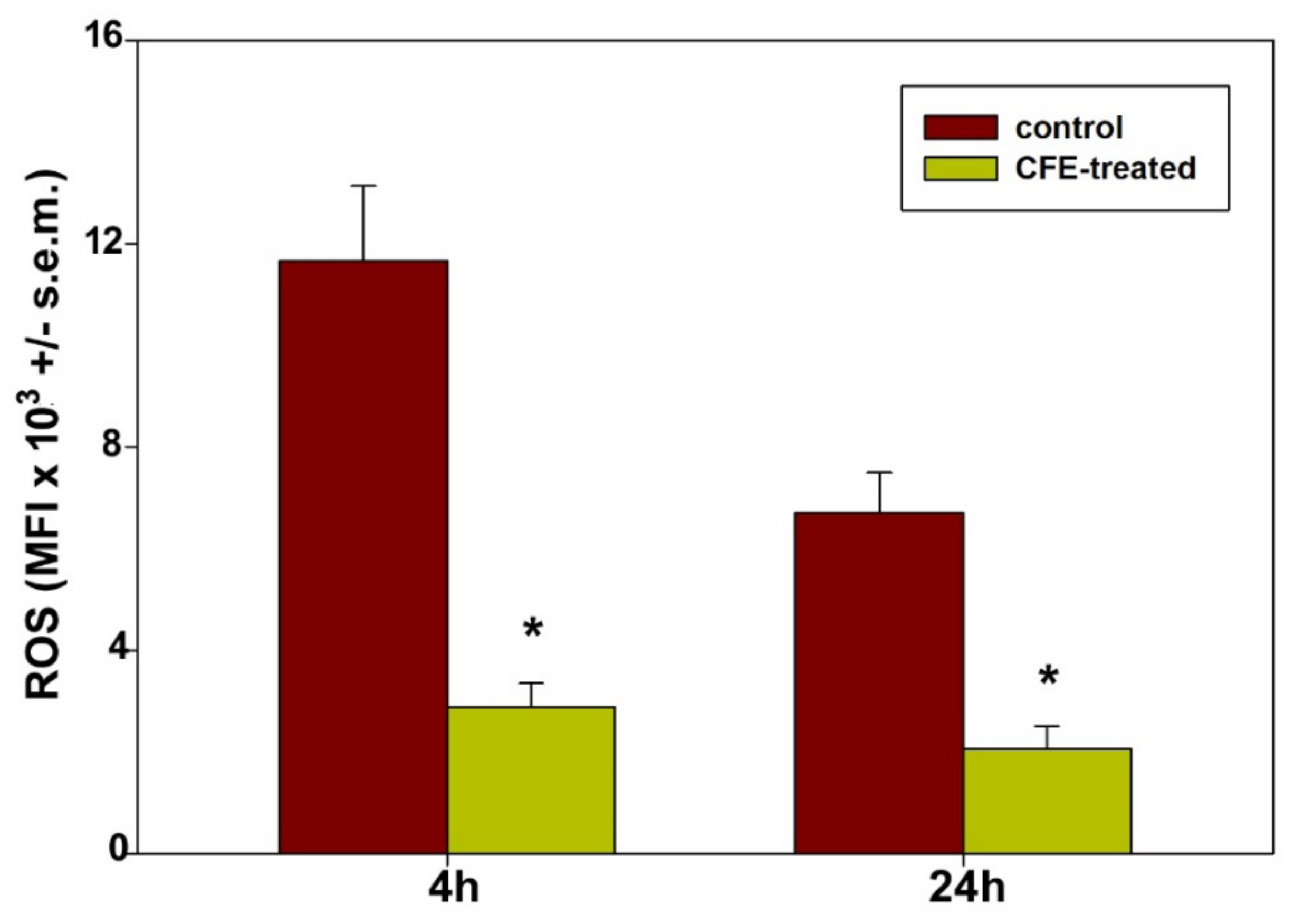
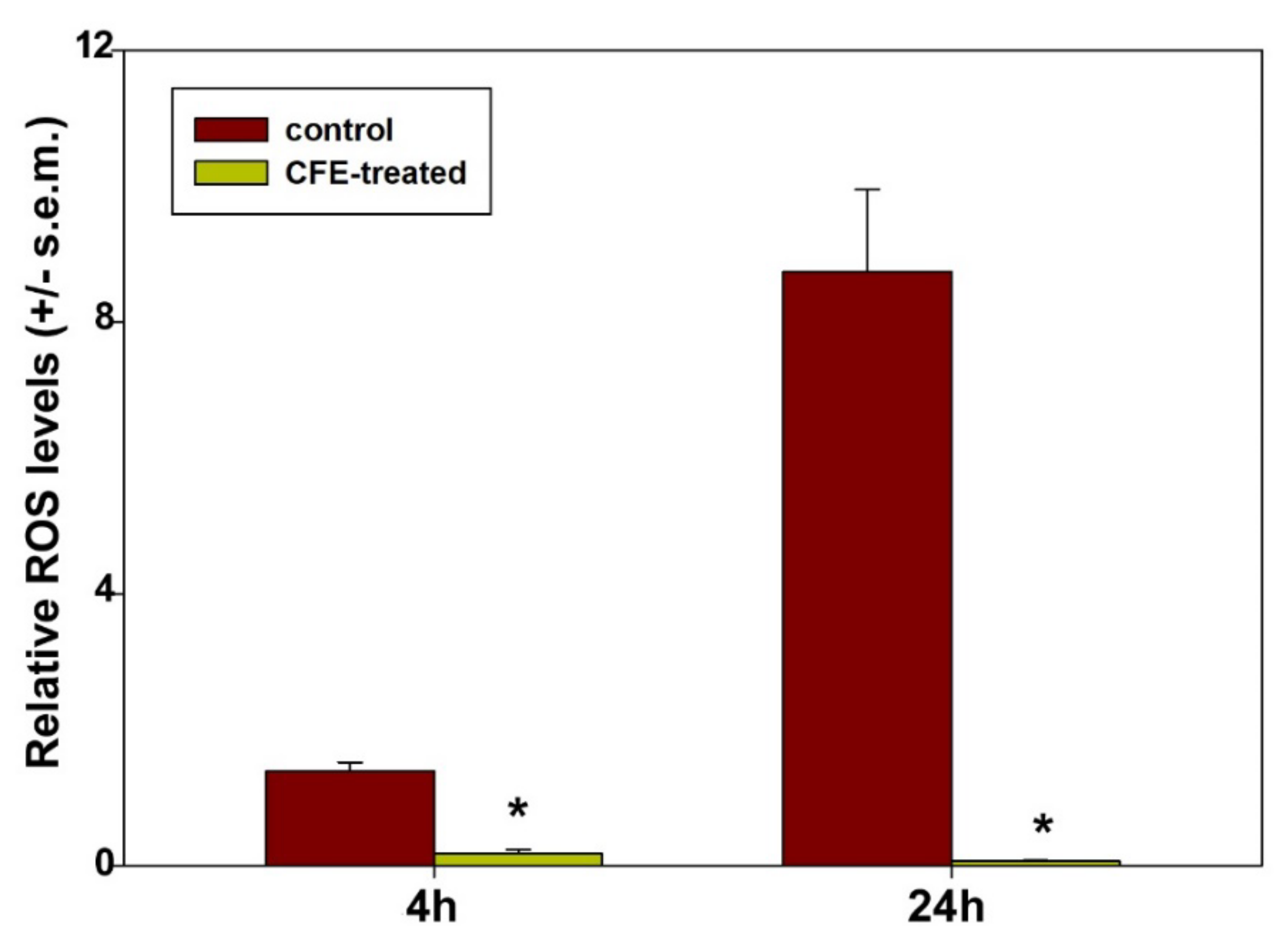
- It is worth mentioning that the HepG2 cell line is characterized by low levels of expression of the cytochrome P450 family 2 subfamily E member 1 (CYP2E1) protein, a ROS-generating enzyme of the endoplasmic reticulum, and therefore is considered a helpful model to test the formation of ROS mainly from mitochondrial sources [84]. Our results show a drastic reduction in the generation of mitochondrial ROS in exposed cells. It is known that low levels of ROS act as redox-active signaling messengers necessary for cell proliferation and functions and that cancer cells require a greater supply of these molecules; otherwise, they become unable to grow normally [85]. Thus, by analogy with the literature data, it is conceivable that the redox imbalance occurring in CFE-treated HepG2 cells may also contribute to the cytotoxicity-leading vicious cycle by promoting mitochondrial dysfunction and triggering mitochondria-mediated apoptosis.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, C.Q.; Ma, Q.Y.; Gao, X.Z.; Wang, X.; Zhang, B.L. Research Progress in Anti-Inflammatory Bioactive Substances Derived from Marine Microorganisms, Sponges, Algae, and Corals. Mar. Drugs 2021, 19, 572. [Google Scholar] [CrossRef] [PubMed]
- Lindequist, U. Marine-Derived Pharmaceuticals? Challenges and Opportunities. Biomol. Therap. 2016, 24, 561–571. [Google Scholar] [CrossRef]
- Liu, L.; Zheng, Y.Y.; Shao, C.L.; Wang, C.Y. Metabolites from marine invertebrates and their symbiotic microorganisms: Molecular diversity discovery, mining, and application. Mar. Life Sci. Technol. 2019, 1, 60–94. [Google Scholar] [CrossRef]
- Lazzara, V.; Arizza, V.; Luparello, C.; Mauro, M.; Vazzana, M. Bright Spots in The Darkness of Cancer: A Review of Starfishes-Derived Compounds and Their Anti-Tumor Action. Mar. Drugs 2019, 17, 617. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Mauro, M.; Lazzara, V.; Vazzana, M. Collective Locomotion of Human Cells, Wound Healing and Their Control by Extracts and Isolated Compounds from Marine Invertebrates. Molecules 2020, 25, 2471. [Google Scholar] [CrossRef]
- Luparello, C.; Mauro, M.; Arizza, V.; Vazzana, M. Histone Deacetylase Inhibitors from Marine Invertebrates. Biology 2020, 9, 429. [Google Scholar] [CrossRef]
- Mauro, M.; Lazzara, V.; Punginelli, D.; Arizza, V.; Vazzana, M. Antitumoral compounds from vertebrate sister group: A review of Mediterranean ascidians. Dev. Comp. Immunol. 2020, 108, 103669. [Google Scholar] [CrossRef]
- Luparello, C. Marine Animal-Derived Compounds and Autophagy Modulation in Breast Cancer Cells. Foundations 2021, 1, 3–20. [Google Scholar] [CrossRef]
- Šimat, V.; Elabed, N.; Kulawik, P.; Ceylan, Z.; Jamroz, E.; Yazgan, H.; Čagalj, M.; Regenstein, J.M.; Özogul, F. Recent Advances in Marine-Based Nutraceuticals and Their Health Benefits. Mar. Drugs 2020, 18, 627. [Google Scholar] [CrossRef]
- Debeaufort, F. Active biopackaging produced from by-products and waste from food and marine industries. FEBS Open Bio 2021, 11, 984–998. [Google Scholar] [CrossRef]
- Lobine, D.; Rengasamy, K.R.R.; Mahomoodally, M.F. Functional foods and bioactive ingredients harnessed from the ocean: Current status and future perspectives. Crit. Rev. Food Sci. Nutr. 2021, 16, 5794–5823. [Google Scholar] [CrossRef] [PubMed]
- Wangensteen, O.S.; Turon, X.; Pérez-Portela, R.; Palacín, C. Natural or naturalized? Phylogeography suggests that the abundant sea urchin Arbacia lixula is a recent colonizer of the Mediterranean. PLoS ONE 2012, 7, e45067. [Google Scholar] [CrossRef] [PubMed]
- Cakal Arslan, O.; Parlak, H. Embryotoxic effects of nonylphenol and octylphenol in sea urchin Arbacia lixula. Ecotoxicol. 2007, 6, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Carballeira, C.; De Orte, M.R.; Viana, I.G.; Delvalls, T.A.; Carballeira, A. Assessing the toxicity of chemical compounds associated with land-based marine fish farms: The sea urchin embryo bioassay with Paracentrotus lividus and Arbacia lixula. Arch. Environ. Contam. Toxicol. 2012, 2, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Vazzana, M.; Mauro, M.; Ceraulo, M.; Dioguardi, M.; Papale, E.; Mazzola, S.; Arizza, V.; Beltrame, F.; Inguglia, L.; Buscaino, G. Underwater high frequency noise: Biological responses in sea urchin Arbacia lixula (Linnaeus, 1758). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2020, 242, 110650. [Google Scholar] [CrossRef]
- Cirino, P.; Brunet, C.; Ciaravolo, M.; Galasso, C.; Musco, L.; Vega Fernández, T.; Sansone, C.; Toscano, A. The Sea Urchin Arbacia lixula: A Novel Natural Source of Astaxanthin. Mar. Drugs 2017, 15, 187. [Google Scholar] [CrossRef]
- Galasso, C.; Orefice, I.; Toscano, A.; Vega Fernández, T.; Musco, L.; Brunet, C.; Sansone, C.; Cirino, P. Food Modulation Controls Astaxanthin Accumulation in Eggs of the Sea Urchin Arbacia lixula. Mar. Drugs 2018, 16, 186. [Google Scholar] [CrossRef]
- Stabili, L.; Acquaviva, M.I.; Cavallo, R.A.; Gerardi, C.; Narracci, M.; Pagliara, P. Screening of Three Echinoderm Species as New Opportunity for Drug Discovery: Their Bioactivities and Antimicrobial Properties. Evid. Based Complement. Alternat. Med. 2018, 2018, 7891748. [Google Scholar] [CrossRef]
- Sciani, J.M.; Emerenciano, A.K.; Cunha da Silva, J.R.; Pimenta, D.C. Initial peptidomic profiling of Brazilian sea urchins: Arbacia lixula, Lytechinus variegatus and Echinometra lucunter. J. Venom Anim. Toxins Incl. Trop. Dis. 2016, 22, 17. [Google Scholar] [CrossRef]
- Luparello, C.; Ragona, D.; Asaro, D.M.L.; Lazzara, V.; Affranchi, F.; Arizza, V.; Vazzana, M. Cell-Free Coelomic Fluid Extracts of the Sea Urchin Arbacia lixula Impair Mitochondrial Potential and Cell Cycle Distribution and Stimulate Reactive Oxygen Species Production and Autophagic Activity in Triple-Negative MDA-MB231 Breast Cancer Cells. J. Mar. Sci. Eng. 2020, 8, 261. [Google Scholar] [CrossRef]
- Donato, M.T.; Tolosa, L.; Gómez-Lechón, M.J. Culture and Functional Characterization of Human Hepatoma HepG2 Cells. Methods Mol. Biol. 2015, 1250, 77–93. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Comparability of Biotechnological/Biological Products Subject to Changes in Their Manufacturing Process. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/ich-q-5-e-comparability-biotechnological/biological-products-step-5_en.pdf (accessed on 20 November 2021).
- Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Drahos, L.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Cytotoxic capability and the associated proteomic profile of cell-free coelomic fluid extracts from the edible sea cucumber Holothuria tubulosa on HepG2 liver cancer cells. EXCLI J. 2022, 21, 722–743. [Google Scholar] [CrossRef] [PubMed]
- Longo, A.; Librizzi, M.; Chuckowree, I.S.; Baltus, C.B.; Spencer, J.; Luparello, C. Cytotoxicity of the urokinase-plasminogen activator inhibitor carbamimidothioic acid (4-boronophenyl) methyl ester hydrobromide (BC-11) on triple-negative MDA-MB231 breast cancer cells. Molecules 2015, 20, 9879–9889. [Google Scholar] [CrossRef]
- Librizzi, M.; Longo, A.; Chiarelli, R.; Amin, J.; Spencer, J.; Luparello, C. Cytotoxic effects of Jay Amin hydroxamic acid (JAHA), a ferrocene-based class I histone deacetylase inhibitor, on triple-negative MDA-MB231 breast cancer cells. Chem. Res. Toxicol. 2012, 25, 2608–2616. [Google Scholar] [CrossRef] [PubMed]
- Luparello, C.; Ragona, D.; Asaro, D.M.L.; Lazzara, V.; Affranchi, F.; Celi, M.; Arizza, V.; Vazzana, M. Cytotoxic Potential of the Coelomic Fluid Extracted from the Sea Cucumber Holothuria tubulosa against Triple-Negative MDA-MB231 Breast Cancer Cells. Biology 2019, 8, 76. [Google Scholar] [CrossRef]
- Luparello, C.; Asaro, D.M.L.; Cruciata, I.; Hassell-Hart, S.; Sansook, S.; Spencer, J.; Caradonna, F. Cytotoxic activity of the histone deacetylase 3-selective inhibitor Pojamide on MDA-MB-231 triple-negative breast cancer cells. Int. J. Mol. Sci. 2019, 20, 804. [Google Scholar] [CrossRef]
- Belyaeva, E.A.; Dymkowska, D.; Wieckowski, M.R.; Wojtczak, L. Reactive oxygen species produced by the mitochondrial respiratory chain are involved in Cd2+-induced injury of rat ascites hepatoma AS-30D cells. Biochim. Biophys. Acta 2006, 1757, 1568–1574. [Google Scholar] [CrossRef]
- Raja, S.M.; Clubb, R.J.; Ortega-Cava, C.; Williams, S.H.; Bailey, T.A.; Duan, L.; Zhao, X.; Reddi, A.L.; Nyong, A.M.; Natarajan, A.; et al. Anticancer activity of Celastrol in combination with ErbB2-targeted therapeutics for treatment of ErbB2-overexpressing breast cancers. Cancer Biol. Ther. 2011, 11, 263–276. [Google Scholar] [CrossRef]
- Chang, C.W.; Chen, Y.S.; Chou, S.H.; Han, C.L.; Chen, Y.J.; Yang, C.C.; Huang, C.Y.; Lo, J.F. Distinct subpopulations of head and neck cancer cells with different levels of intracellular reactive oxygen species exhibit diverse stemness, proliferation, and chemosensitivity. Cancer Res. 2014, 74, 6291–6305. [Google Scholar] [CrossRef]
- Gibson, S.B. Chapter Thirteen - Investigating the Role of Reactive Oxygen Species in Regulating Autophagy. Meth. Enzymol. 2013, 528, 217–235. [Google Scholar] [CrossRef]
- Sun, Y.; Zou, H.; Yang, L.; Zhou, M.; Shi, X.; Yang, Y.; Chen, W.; Zhao, Y.; Mo, J.; Lu, Y. Effect on the liver cancer cell invasion ability by studying the associations between autophagy and TRAP1 expression. Oncol. Lett. 2018, 16, 991–997. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhu, H.; Zhang, J.; Wang, M.; Zhu, L.; Guo, Z.; Shen, W.; Wang, D. Solamargine inhibits the migration and invasion of HepG2 cells by blocking epithelial-to-mesenchymal transition. Oncol. Lett. 2017, 14, 447–452. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zheng, J.; Shao, Y.; Jiang, Y.; Chen, F.; Liu, S.; Yu, N.; Zhang, D.; Liu, X.; Zou, L. Tangeretin inhibits hepatocellular carcinoma proliferation and migration by promoting autophagy-related BECLIN1. Cancer Manag. Res. 2019, 11, 5231–5242. [Google Scholar] [CrossRef] [PubMed]
- Furth, E.E.; Sprecher, H.; Fisher, E.A.; Fleishman, H.D.; Laposata, M. An in vitro model for essential fatty acid deficiency: HepG2 cells permanently maintained in lipid-free medium. J. Lipid Res. 1992, 33, 1719–1726. [Google Scholar] [CrossRef]
- Desquiret, V.; Loiseau, D.; Jacques, C.; Douay, O.; Malthièry, Y.; Ritz, P.; Roussel, D. Dinitrophenol-induced mitochondrial uncoupling in vivo triggers respiratory adaptation in HepG2 cells. Biochim. Biophys. Acta 2006, 1757, 21–30. [Google Scholar] [CrossRef]
- Salas-Rojas, M.; Galvez-Romero, G.; Anton-Palma, B.; Acevedo, R.; Blanco-Favela, F.; Aguilar-Setién, A. The coelomic fluid of the sea urchin Tripneustes depressus shows antiviral activity against Suid herpesvirus type 1 (SHV-1) and rabies virus (RV). Fish Shellfish Immunol. 2014, 36, 158–163. [Google Scholar] [CrossRef]
- Soleimani, S.; Mashjoor, S.; Mitra, S.; Yousefzadi, M.; Rezadoost, H. Coelomic fluid of Echinometra mathaei: The new prospects for medicinal antioxidants. Fish Shellfish Immunol. 2021, 117, 311–319. [Google Scholar] [CrossRef]
- Widyananda, M.H.; Pratama, S.K.; Samoedra, R.S.; Sari, F.N.; Kharisma, V.D.; Ansori, A.N.M.; Antonius, Y. Molecular docking study of sea urchin (Arbacia lixula) peptides as multi-target inhibitor for non-small cell lung cancer (NSCLC) associated proteins. J. Pharm. Pharmacogn. Res. 2021, 9, 484–496. [Google Scholar]
- D’Alessio, S.; Buckley, K.M.; Kraev, I.; Hayes, P.; Lange, S. Extracellular Vesicle Signatures and Post-Translational Protein Deimination in Purple Sea Urchin (Strongylocentrotus purpuratus) Coelomic Fluid-Novel Insights into Echinodermata Biology. Biology 2021, 10, 866. [Google Scholar] [CrossRef]
- Ko, K.L.; Mak, L.Y.; Cheung, K.S.; Yuen, M.F. Hepatocellular carcinoma: Recent advances and emerging medical therapies. F1000Research 2020, 9, 620. [Google Scholar] [CrossRef]
- Zimmermann, A. Etiology and Pathogenesis of Hepatocellular Carcinoma: Inflammatory and Toxic Causes. In Tumors and Tumor-Like Lesions of the Hepatobiliary Tract: General and Surgical Pathology; Zimmermann, A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 2931–2959. [Google Scholar]
- Wallace, M.C.; Friedman, S.L. Hepatic fibrosis and the microenvironment: Fertile soil for hepatocellular carcinoma development. Gene Expr. 2014, 16, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Alnajjar, A.M.; Elsiesy, H.A. Natural products and hepatocellular carcinoma: A review. Hepatoma Res. 2015, 1, 119–124. [Google Scholar] [CrossRef][Green Version]
- Jain, D.; Murti, Y.; Khan, W.U.; Hossain, R.; Hossain, M.N.; Agrawal, K.K.; Ashraf, R.A.; Islam, M.T.; Janmeda, P.; Taheri, Y.; et al. Roles of Therapeutic Bioactive Compounds in Hepatocellular Carcinoma. Oxid. Med. Cell Longev. 2021, 2021, 9068850. [Google Scholar] [CrossRef]
- Singh, A.; Verma, S.; Modak, S.B.; Chaturvedi, M.M.; Purohit, J.S. Extra-nuclear histones: Origin, significance and perspectives. Mol. Cell. Biochem. 2022, 477, 507–524. [Google Scholar] [CrossRef]
- Tadayoni Nia, A.; Bazi, Z.; Khosravi, A.; Oladnabi, M. WDR81 Gene Silencing Can Reduce Exosome Levels in Human U87-MG Glioblastoma Cells. J. Mol. Neurosci. 2021, 71, 1696–1702. [Google Scholar] [CrossRef]
- Morita, S.Y.; Deharu, Y.; Takata, E.; Nakano, M.; Handa, T. Cytotoxicity of lipid-free apolipoprotein B. Biochim. Biophys. Acta 2008, 1778, 2594–2603. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hall, A.K. Thymosin beta-10 accelerates apoptosis. Cell. Mol. Biol. Res. 1995, 41, 167–180. [Google Scholar]
- Rho, S.B.; Lee, K.W.; Chun, T.; Lee, S.H.; Park, K.; Lee, J.H. The identification of apoptosis-related residues in human thymosin beta-10 by mutational analysis and computational modeling. J. Biol. Chem. 2005, 280, 34003–34007. [Google Scholar] [CrossRef]
- Lee, S.H.; Son, M.J.; Oh, S.H.; Rho, S.B.; Park, K.; Kim, Y.J.; Park, M.S.; Lee, J.H. Thymosin {beta}(10) inhibits angiogenesis and tumor growth by interfering with Ras function. Cancer Res. 2005, 65, 137–148. [Google Scholar] [CrossRef]
- Rodrigo, A.P.; Mendes, V.M.; Manadas, B.; Grosso, A.R.; Alves de Matos, A.P.; Baptista, P.V.; Costa, P.M.; Fernandes, A.R. Specific Antiproliferative Properties of Proteinaceous Toxin Secretions from the Marine Annelid Eulalia sp. onto Ovarian Cancer Cells. Mar. Drugs 2021, 19, 31. [Google Scholar] [CrossRef]
- Lai, J.P.; Yu, C.; Moser, C.D.; Aderca, I.; Han, T.; Garvey, T.D.; Murphy, L.M.; Garrity-Park, M.M.; Shridhar, V.; Adjei, A.A.; et al. SULF1 inhibits tumor growth and potentiates the effects of histone deacetylase inhibitors in hepatocellular carcinoma. Gastroenterology 2006, 130, 2130–2144. [Google Scholar] [CrossRef] [PubMed]
- Frau, M.; Simile, M.M.; Tomasi, M.L.; Demartis, M.I.; Daino, L.; Seddaiu, M.A.; Brozzetti, S.; Feo, C.F.; Massarelli, G.; Solinas, G.; et al. An expression signature of phenotypic resistance to hepatocellular carcinoma identified by cross-species gene expression analysis. Cell. Oncol. 2012, 35, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Jiwen, W.; Dexiang, Z.; Bohao, Z.; Yueqi, W.; Han, L.; Xiaoling, N.; Tao, S.; Liu, H. DMBT1 suppresses progression of gallbladder carcinoma through PI3K/AKT signaling pathway by targeting PTEN. Biosci. Biotechnol. Biochem. 2019, 83, 2257–2264. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Peng, B.; Xie, Q.; Ruan, J.; Liang, X. Upregulation of ZBTB7A exhibits a tumor suppressive role in gastric cancer cells. Mol. Med. Rep. 2018, 17, 2635–2641. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.; Tang, J.; Li, L.; Peng, W.; Du, Z.; Wang, X.; Li, Q.; Xu, H.; Xiong, L.; Xu, C.; et al. Tumor suppressive BTB/POZ zinc-finger protein ZBTB28 inhibits oncogenic BCL6/ZBTB27 signaling to maintain p53 transcription in multiple carcinogenesis. Theranostics 2019, 9, 8182–8195. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wu, M.; Xiong, L.; Gong, Y.; Yu, R.; Peng, W.; Li, L.; Li, L.; Tian, S.; Wang, Y.; et al. BTB/POZ zinc finger protein ZBTB16 inhibits breast cancer proliferation and metastasis through upregulating ZBTB28 and antagonizing BCL6/ZBTB27. Clin. Epigenetics 2020, 12, 82. [Google Scholar] [CrossRef]
- Hadari, Y.R.; Arbel-Goren, R.; Levy, Y.; Amsterdam, A.; Alon, R.; Zakut, R.; Zick, Y. Galectin-8 binding to integrins inhibits cell adhesion and induces apoptosis. J. Cell Sci. 2000, 113, 2385–2397. [Google Scholar] [CrossRef]
- Autelli, R.; Ullio, C.; Prigione, E.; Crepaldi, S.; Schiavone, N.; Brunk, U.T.; Capaccioli, S.; Baccino, F.M.; Bonelli, G. Divergent pathways for TNF and C(2)-ceramide toxicity in HTC hepatoma cells. Biochim. Biophys. Acta 2009, 1793, 1182–1190. [Google Scholar] [CrossRef]
- Peng, X.; Wen, Y.; Zha, L.; Zhuang, J.; Lin, L.; Li, X.; Chen, Y.; Liu, Z.; Zhu, S.; Liang, J.; et al. TRIM45 Suppresses the Development of Non-small Cell Lung Cancer. Curr. Mol. Med. 2020, 20, 299–306. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, C.; Cui, J.; Ou, J.; Han, J.; Qin, Y.; Zhi, F.; Wang, R.F. TRIM45 functions as a tumor suppressor in the brain via its E3 ligase activity by stabilizing p53 through K63-linked ubiquitination. Cell Death Dis. 2017, 8, e2831. [Google Scholar] [CrossRef]
- Wallner, C.; Drysch, M.; Becerikli, M.; Jaurich, H.; Wagner, J.M.; Dittfeld, S.; Nagler, J.; Harati, K.; Dadras, M.; Philippou, S.; et al. Interaction with the GDF8/11 pathway reveals treatment options for adenocarcinoma of the breast. Breast 2018, 37, 134–141. [Google Scholar] [CrossRef]
- Drysch, M.; Schmidt, S.V.; Becerikli, M.; Reinkemeier, F.; Dittfeld, S.; Wagner, J.M.; Dadras, M.; Sogorski, A.; von Glinski, M.; Lehnhardt, M.; et al. Myostatin Deficiency Protects C2C12 Cells from Oxidative Stress by Inhibiting Intrinsic Activation of Apoptosis. Cells 2021, 10, 1680. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Shimbo, T.; Kikuchi, Y.; Matsuda, M.; Kaneda, Y. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int. J. Cancer 2010, 126, 1982–1991. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, Y.; Aishima, S.; Taketomi, A.; Nishihara, Y.; Iguchi, T.; Taguchi, K.; Maehara, Y.; Tsuneyoshi, M. 14-3-3sigma negatively regulates the cell cycle, and its down-regulation is associated with poor outcome in intrahepatic cholangiocarcinoma. Hum. Pathol. 2007, 38, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tian, F.; Lu, L.; Wang, Y.; Xiao, Z.; Yu, C.; Yu, X. Characterization of Cep85 - a new antagonist of Nek2A that is involved in the regulation of centrosome disjunction. J. Cell Sci. 2015, 128, 3290–3303. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, Y.; Li, Y.; Zhou, L.; Yang, G.; Wang, M.; Hong, Y. Cryptochrome 2 (CRY2) Suppresses Proliferation and Migration and Regulates Clock Gene Network in Osteosarcoma Cells. Med. Sci. Monit. 2018, 24, 3856–3862. [Google Scholar] [CrossRef]
- Sawada, G.; Ueo, H.; Matsumura, T.; Uchi, R.; Ishibashi, M.; Mima, K.; Kurashige, J.; Takahashi, Y.; Akiyoshi, S.; Sudo, T.; et al. CHD8 is an independent prognostic indicator that regulates Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol. Rep. 2013, 30, 1137–1142. [Google Scholar] [CrossRef]
- Su, J.; You, P.; Zhao, J.P.; Zhang, S.L.; Song, S.H.; Fu, Z.R.; Ye, L.W.; Zi, X.Y.; Xie, D.F.; Zhu, M.H.; et al. A potential role for the homeoprotein Hhex in hepatocellular carcinoma progression. Med. Oncol. 2012, 29, 1059–1067. [Google Scholar] [CrossRef]
- Marfil, V.; Blazquez, M.; Serrano, F.; Castell, J.V.; Bort, R. Growth-promoting and tumourigenic activity of c-Myc is suppressed by Hhex. Oncogene 2015, 34, 3011–3022. [Google Scholar] [CrossRef]
- Ni, S.; Hu, J.; Duan, Y.; Shi, S.; Li, R.; Wu, H.; Qu, Y.; Li, Y. Down expression of LRP1B promotes cell migration via RhoA/Cdc42 pathway and actin cytoskeleton remodeling in renal cell cancer. Cancer Sci. 2013, 104, 817–825. [Google Scholar] [CrossRef]
- Beer, A.G.; Zenzmaier, C.; Schreinlechner, M.; Haas, J.; Dietrich, M.F.; Herz, J.; Marschang, P. Expression of a recombinant full-length LRP1B receptor in human non-small cell lung cancer cells confirms the postulated growth-suppressing function of this large LDL receptor family member. Oncotarget 2016, 7, 68721–68733. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Liu, D.; Hao, Y.; Li, C.; Hao, J.; Lin, C.; Shi, S.; Wang, D. The expression of TET3 regulated cell proliferation in HepG2 cells. Gene 2019, 698, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Jordan, J.J.; Samson, L.D. Human ALKBH7 is required for alkylation and oxidation-induced programmed necrosis. Genes Dev. 2013, 27, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Na, J.; Jung, J.; Bang, J.; Lu, Q.; Carlson, B.A.; Guo, X.; Gladyshev, V.N.; Kim, J.; Hatfield, D.L.; Lee, B.J. Selenophosphate synthetase 1 and its role in redox homeostasis, defense and proliferation. Free Radic. Biol. Med. 2018, 127, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rong, W.; Ma, J.; Li, H.; Tang, X.; Xu, S.; Wang, L.; Wan, L.; Zhu, Q.; Jiang, B.; et al. Comprehensive Analysis of DNA 5-Methylcytosine and N6-Adenine Methylation by Nanopore Sequencing in Hepatocellular Carcinoma. Front. Cell. Dev. Biol. 2022, 10, 827391. [Google Scholar] [CrossRef]
- Zhang, Y.; Tong, G.H.; Wei, X.X.; Chen, H.Y.; Liang, T.; Tang, H.P.; Wu, C.A.; Wen, G.M.; Yang, W.K.; Liang, L.; et al. Identification of Five Cytotoxicity-Related Genes Involved in the Progression of Triple-Negative Breast Cancer. Front. Genet. 2022, 12, 723477. [Google Scholar] [CrossRef]
- Manshouri, R.; Coyaud, E.; Kundu, S.T.; Peng, D.H.; Stratton, S.A.; Alton, K.; Bajaj, R.; Fradette, J.J.; Minelli, R.; Peoples, M.D.; et al. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat. Commun. 2019, 10, 5125. [Google Scholar] [CrossRef]
- Fatahala, S.S.; Sayed, A.I.; Mahgoub, S.; Taha, H.; El-Sayed, M.-I.k.; El-Shehry, M.F.; Awad, S.M.; Abd El-Hameed, R.H. Synthesis of Novel 2-Thiouracil-5-Sulfonamide Derivatives as Potent Inducers of Cell Cycle Arrest and CDK2A Inhibition Supported by Molecular Docking. Int. J. Mol. Sci. 2021, 22, 11957. [Google Scholar] [CrossRef]
- Han, Z.; Zhao, X.; Zhang, E.; Ma, J.; Zhang, H.; Li, J.; Xie, W.; Li, X. Resistomycin Induced Apoptosis and Cycle Arrest in Human Hepatocellular Carcinoma Cells by Activating p38 MAPK Pathway In Vitro and In Vivo. Pharmaceuticals 2021, 14, 958. [Google Scholar] [CrossRef]
- Ren, Y.; Ruan, Y.; Cheng, B.; Li, L.; Liu, J.; Fang, Y.; Chen, J. Design, synthesis and biological evaluation of novel acridine and quinoline derivatives as tubulin polymerization inhibitors with anticancer activities. Bioorg. Med. Chem. 2021, 46, 116376. [Google Scholar] [CrossRef]
- Zhu, C.; Zhao, M.; Fan, L.; Cao, X.; Xia, Q.; Zhou, J.; Yin, H.; Zhao, L. Chitopentaose inhibits hepatocellular carcinoma by inducing mitochondrial mediated apoptosis and suppressing protective autophagy. Bioresour. Bioprocess. 2021, 8, 4. [Google Scholar] [CrossRef]
- Jiang, J.; Briedé, J.J.; Jennen, D.G.; Van Summeren, A.; Saritas-Brauers, K.; Schaart, G.; Kleinjans, J.C.; de Kok, T.M. Increased mitochondrial ROS formation by acetaminophen in human hepatic cells is associated with gene expression changes suggesting disruption of the mitochondrial electron transport chain. Toxicol. Lett. 2015, 234, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Tan, X.; Liang, J.; Wu, S.; Liu, J.; Zhang, Q.; Zhu, R. A reduction in reactive oxygen species contributes to dihydromyricetin-induced apoptosis in human hepatocellular carcinoma cells. Sci. Rep. 2014, 4, 7041. [Google Scholar] [CrossRef] [PubMed]

| Peptide Sequence(s) | Sequence ID (nr. of Matches/Range) | Expected | Identities (%) | Positives (%) | Protein Description | Organism (Sea Urchin) | Selected GO Annotations |
|---|---|---|---|---|---|---|---|
| AIGNIGEPVER AAIQSMR LN [+0.984] QLHYLVGMEPSATDATVFLR AYTIDAQYK LQSPMIK QGITFNVFSSYPTLNK LALIALK | BAL43173.1 (1/501–511) (1/543–549) (1/807–828) (1/1069–1077) (1/1815–1821) (1/5231–5246) BAL43173.1 (1/494–500) | 2 × 10−7 0.003 5 × 10−19 1 × 10−5 0.12 9 × 10−13 0.021 | 100 100 100 100 88 100 100 | 100 100 100 100 88 100 100 | Apolipoprotein B-like protein, partial Apolipoprotein B-like protein, partial | Mesocentrotus nudus Strongylocentrotus intermedius | -lipid transport |
| NTLPTKDTIEQEK | AAC26833.1 (1/26–39) | 1 × 10−7 | 93 | 100 | Thymosin beta | Strongylocentrotus purpuratus | -actin monomer binding -negative regulation of cell cycle G1/S phase transition -regulation of cell migration |
| RGTIEVSKYDGSYR GEQVYGITTYADTR | XP_041457866.1 (1/572–584) (1/725–738) | 2 × 10−9 9 × 10−10 | 100 94 | 100 93 | Low-density lipoprotein receptor-related protein 1B-like | Lytechinus variegatus | -integral component of membrane -receptor-mediated endocytosis |
| GNPTTVFPEIFK TSPNSLEPSTTVLSPYLK VYSPIVFGK | XP_030853362.1 (1/206–217) XP_030853363.1 (1/79–90) XP_030853362.1 (1/367–386) XP_030853363.1 (1/79–90) XP_030853362.1 (1/540–548) XP_030853363.1 (1/413–421) | 1 × 10−8 1 × 10−8 1 × 10−14 1 × 10−14 2 × 10−4 2 × 10−4 | 100 100 100 100 91 91 | 100 100 100 100 100 100 | Cryptochrome-2 isoform X1 Cryptochrome-2 isoform X2 Cryptochrome-2 isoform X1 Cryptochrome-2 isoform X2 Cryptochrome-2 isoform X1 Cryptochrome-2 isoform X2 | Strongylocentrotus purpuratus | -circadian regulation of gene expression -DNA binding |
| LAAEGLR IGTYGETR VFLPWTTTGLPK SLSWEGGHR | XP_041456451.1 (1/62–68) XP_041464311.1 (1/109–118) XP_041460692.1 (1/146–157) (1/352–360) | 0.007 3 × 10−4 1 × 10−8 1 × 10−4 | 100 100 100 91 | 100 100 100 100 | Arylsulfatase-like | Lytechinus variegatus | -sulfuric ester hydrolase activity -lysosome |
| LVDGSSSNEGR MLGYNGALSAPR LVGGSGPHEGR LVGGSNDADGR | XP_030831017.1, (20/25–35, 132–142, 239–249, 346–356, 453–463, 560–570, 667–677, 774–784, 881–891, 988–998, 1098–1108, 1353–1363, 1460–1470, 1567–1577, 1674–1684, 1781–1791, 1995–2005, 2102–2112, 2319–2329, 2426–2436) XP_030831023.1 (20/same as isoform X1 except for 1350–1360, 1457–1467, 1564–1574, 1671–1681, 1778–1788, 1992–2002, 2099–2109, 2316–2326, 2423–2433) XP_030831029.1 (20/same as isoform X2) XP_030831072.1 (19/same as isoform X1 without 2426–2436) XP_030831034.1 (19/same as isoform X9 except for 1888–1898, 2212–2222, 2319–2329) XP_030831054.1 (19/same as isoform X9) XP_030831048.1 (19/same as isoform X4 except for 1995–2005) XP_030831064.1 (19/same as isoform X6 except for 991–1001, 1246–1256) XP_030831041.1 (19/same as isoform X9 except for 2212–2222, 2319–2329) XP_030831078.1 (19/same as isoform X1 except for 1202–1212, 1309–1319, 1416–1426, 1523–1533, 1630–1640, 1844–1854, 1951–1961, 2166–2176, 2275–2285) XP_030831079.1 (19/same as isoform X9 except for 2209–2219) XP_030831086.1 (17/25–35, 132–142, 239–249, 346–356, 453–463, 560–570, 667–677, 774–784, 988–998, 1095–1105, 1202–1212, 1309–1319, 1416–1426, 1630–1640, 1737–1747, 1954–1964, 2061–2071) XP_030834718.1 (1/278–289) XP_030855705.1 (1/677–687) (1/1259–1269) (1/1917–1927) (1/2449–2459) (1/2129–2139) (1/1590–1600) (1/2553–2563) XP_030851619.1 (1/839–849) (1/1443–1453) | 2 × 10−6 2 × 10−6 2 × 10−6 2 × 10−6 2 × 10−6 2 × 10−6 2 × 10−6 2e−6 2 × 10−6 2 × 10−6 2 × 10−6 2 × 10−6 5 × 10−6 0.27 1 × 10−6 0.27 0.017 0.096 0.19 0.78 0.49 0.061 | 100 100 100 100 100 100 100 100 100 100 100 100 100 69 100 77 77 77 77 69 85 92 | 100 100 100 100 100 100 100 100 100 100 100 100 100 76 100 76 76 84 76 69 100 100 | Deleted in malignant brain tumors (DMBT) 1 protein isoform X1 DMBT1 protein isoform X2 DMBT1 protein isoform X3 DMBT1 protein isoform X9 DMBT1 protein isoform X4 DMBT1 protein isoform X7 DMBT1 protein isoform X6 DMBT1 protein isoform X8 DMBT1 protein isoform X5 DMBT1 protein isoform X10 DMBT1 protein isoform X11 DMBT1 protein isoform X12 DMBT1 protein-like DMBT1 protein DMBT1 protein-like | Strongylocentrotus purpuratus | -membrane -scavenger receptor activity -endocytosis |
| VLEQSDLVVDNEYALYTLIQPK | XP_030847432.1 (1/183–192) | 0.092 | 75 | 83 | BTB/POZ domain-containing protein At2g30600 | Strongylocentrotus purpuratus | -protein ubiquitination |
| MIFIQGFVPAGADR | CBX45521.1 (1/37–50) | 3 × 10−11 | 100 | 100 | Galectin-8 protein | Paracentrotus lividus | -membrane -integrin binding |
| NLLSVAYK | XP_780530.1 (1/20–26) | 0.35 | 100 | 100 | 14-3-3-like protein 2 | Strongylocentrotus purpuratus | -signal transduction |
| RLVELINQ | XP_030850878.1 (1/715–722) | 1 × 10−4 | 100 | 100 | Centrosomal protein of 85 kDa (CEP85) isoform X1 | Strongylocentrotus purpuratus | -spindle pole -regulation of mitotic centrosome separation |
| VLAYNVFELR | XP_041469541.1 (1/103–112) | 4 × 10−7 | 100 | 100 | Sphingomyelinase C 2-like | Lytechinus variegatus | - acid sphingomyelin phosphodiesterase activity |
| GDSTLQFKPK | XP_041482277.1 (1/359–368) | 5 × 10−6 | 100 | 100 | Tripartite motif-containing protein 45-like | Lytechinus variegatus | -ubiquitin-protein transferase activity -zinc ion binding |
| RLITRNSYE KEEELES RAAQFAALM KHQSYNVAMEWANIV | XP_030831990.1 (1/1592–1600) (1/2501–2507) (1/2775–2783) (1/3026–3040) | 4 × 10−6 0.003 2 × 10−5 2 × 10−13 | 100 100 | 100 100 | Chromodomain-helicase-DNA-binding protein 8 | Strongylocentrotus purpuratus | binding -negative regulation of canonical Wnt signaling pathway |
| DN [+0.984] IVIGGQAGVYDPNR | XP_001198520.2 (1/164–179) | 1 × 10−12 | 100 | 100 | Hemicentin-2 | Strongylocentrotus purpuratus | -calcium ion binding -response to stimulus |
| RVASGPLGLI | XP_030854745.1 (1/1903–1912) | 1 × 10−5 | 100 | 1000 | WD repeat-containing protein 81 | Strongylocentrotus purpuratus | -cytoplasmic vesicle |
| QINLDLLR | XP_041482078.1 (1/665–674) | 2 × 10−4 | 100 | 100 | TBC1 domain family member 2B-like isoform X1 | Lytechinus variegatus | -activation of GTPase activity |
| YPHTQLISQMDR | XP_030846390.1 (1/372–383) | 1 × 10−9 | 100 | 100 | Growth/differentiation factor 8-like | Strongylocentrotus purpuratus | -signaling receptor binding -negative regulation of skeletal muscle satellite cell proliferation |
| MQVQ [+0.984] SGVPK | XP_003726849.3 (1/1–9) | 1 × 10−4 | 100 | 100 | Selenide, water dikinase | Strongylocentrotus purpuratus | -selenocysteine biosynthetic process |
| KYSEPTVEDISPVEHVE | XP_030841466.1 (1/132–148) | 3 × 10−10 | 100 | 100 | Kv channel-interacting protein 4-like | Strongylocentrotus purpuratus | -potassium channel regulator activity -calcium ion binding |
| DFIVIQNFITEEEEDSLLK | XP_011661956.1 (1/144–162) | 2 × 10−16 | 100 | 100 | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 7, mitochondrial | Strongylocentrotus purpuratus | -mitochondrial matrix -regulation of mitochondrial membrane permeability involved in programmed necrotic cell death |
| REVPVASL | XP_011668608.2 (1/721–728) | 0.001 | 100 | 100 | Sterile alpha motif domain-containing protein 9-like | Strongylocentrotus purpuratus | -intracellular membrane-bounded organelle |
| LAKALQLSE | XP_030841717.1 (1/221–229) | 0.003 | 91 | 90 | Hematopoietically expressed homeobox protein HHEX homolog | Strongylocentrotus purpuratus | -Wnt signaling pathway -negative regulation of cyclin-dependent protein serine/threonine kinase activity |
| SQIPTK | XP_041485363.1 (1/1915–1920) XP_041485364.1 (1/1424–1431) | 0.024 0.024 | 100 100 | 100 100 | Methylcytosine dioxygenase TET3-like isoform X1 Methylcytosine dioxygenase TET3-like isoform X2 | Lytechinus variegatus | -methylcytosine dioxygenase activity -chromatin organization |
| QVTGTGATGR GGSSAQAIR | XP_796458.1 (1/107–116) NP_999723.1 (1/36–44) | 2 × 10−5 4 × 10−4 | 100 100 | 100 100 | Histone H1, gonadal-like Histone H1-beta, late embryonic | Strongylocentrotus purpuratus | |
| HLQLAIR NDEELNKLLGGVTIAQGGVLPNIQAVLLPK KLLSGVTIAQGGVLPNIQAVLLPK GDEELDSLIK | XP_030843320.1 (1/83–89) XP_001175793.2 (1/89–118) NP_999718.1 (1/97–119) NP_001116980.1 (1/93–102) | 0.002 6 × 10−26 3 × 10−18 7 × 10−6 | 100 100 100 100 | 100 100 100 100 | Histone H2A-like, partial Histone H2A, embryonic-like Late histone H2A.L3 Histone H2A.V | Strongylocentrotus purpuratus | |
| RKESYGIYIYKV VLK QVHPDTGISSR AMSIMNSFVNDVFER STITSR IAAEASR RLLLPGELAKH HAVSEGTK | P022892 (1/33–42) (1/43–45) (1/46–56) | 0.002 | 100 | 100 | Histone H2B, embryonic | Strongylocentrotus purpuratus | |
| NP_001229615.1 (1/56–70) XP_030836142.1 (1/84–89) AAB48832.1 (1/75–81) (102–110) (110–118) | 6 × 10−12 0.10 0.016 6 × 10−5 0.002 | 100 100 100 100 100 | 100 100 100 100 100 | Histone H2B-like Histone H2B Cleavage stage histone H2B | Psammechinus miliaris | ||
| YRPGTVALR STELLIR EIAQDFKTELR FQSSAVMALQEASEAYLVGLFEDTN [+0.984] LC [+57.021] AIH-AK | XP_030830328.1 (1/42–50) (1/58–64) (1/74–84) (1/85–112) | 2 × 10−5 0.008 7 × 10−8 3 × 10−22 | 100 100 100 94 | 100 100 100 93 | Histone H3, embryonic-like | Strongylocentrotus purpuratus | |
| DNIQGITKPAIR RISGLIYEETR VFLENVIR TLYGFGG | CAA75404.1 (1/1–12) (1/22–32) (1/37–44) XP_011664585.2 (1/97–102) | 2 × 10−7 3 × 10−6 2 × 10−4 0.051 | 100 100 | 100 100 | Histone H4, partial Histone H4-like | Arbacia lixula Strongylocentrotus purpuratus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luparello, C.; Branni, R.; Abruscato, G.; Lazzara, V.; Sugár, S.; Arizza, V.; Mauro, M.; Di Stefano, V.; Vazzana, M. Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma. J. Mar. Sci. Eng. 2022, 10, 1292. https://doi.org/10.3390/jmse10091292
Luparello C, Branni R, Abruscato G, Lazzara V, Sugár S, Arizza V, Mauro M, Di Stefano V, Vazzana M. Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma. Journal of Marine Science and Engineering. 2022; 10(9):1292. https://doi.org/10.3390/jmse10091292
Chicago/Turabian StyleLuparello, Claudio, Rossella Branni, Giulia Abruscato, Valentina Lazzara, Simon Sugár, Vincenzo Arizza, Manuela Mauro, Vita Di Stefano, and Mirella Vazzana. 2022. "Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma" Journal of Marine Science and Engineering 10, no. 9: 1292. https://doi.org/10.3390/jmse10091292
APA StyleLuparello, C., Branni, R., Abruscato, G., Lazzara, V., Sugár, S., Arizza, V., Mauro, M., Di Stefano, V., & Vazzana, M. (2022). Biological and Proteomic Characterization of the Anti-Cancer Potency of Aqueous Extracts from Cell-Free Coelomic Fluid of Arbacia lixula Sea Urchin in an In Vitro Model of Human Hepatocellular Carcinoma. Journal of Marine Science and Engineering, 10(9), 1292. https://doi.org/10.3390/jmse10091292








