Evaluating eDNA for Use within Marine Environmental Impact Assessments
Abstract
1. Introduction
“… the process of identifying the future consequences of a current or proposed action. The “impact” is the difference between what would happen with the action and what would happen without it.” [27]
2. The Main Elements of Environmental Impact Assessment
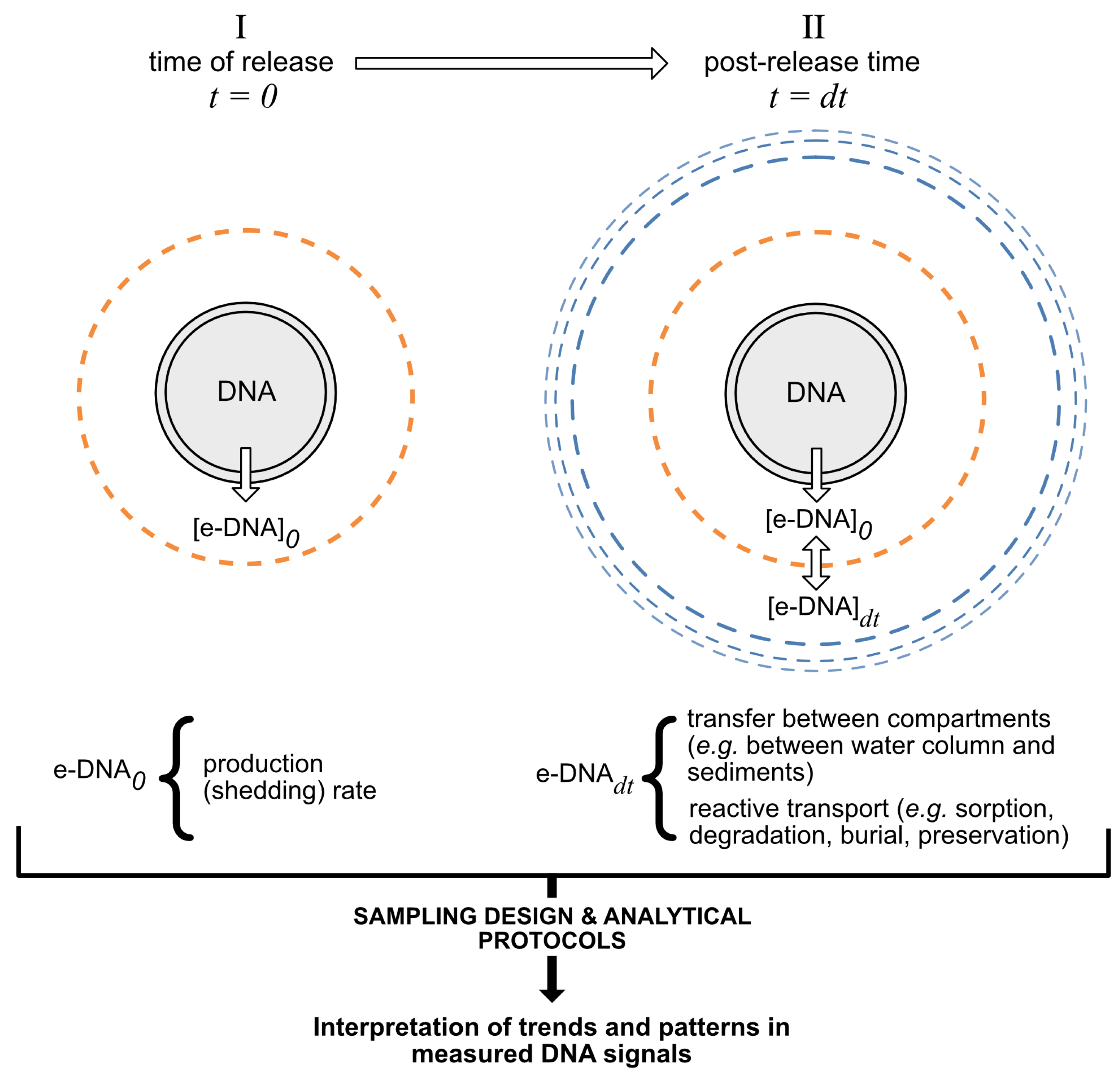
3. The Fate of DNA in Aquatic and Marine Environments
3.1. Sources of DNA: Shedding Rates
3.2. DNA Degradation
3.3. Transport and Dispersion
3.4. DNA Burial and Preservation
4. Exploring eDNA as a Proxy for an EIA Receptor
5. The EIA eDNA Sampling Conundrum
6. The Puzzling Statistics of eDNA Results
- , is the probability of the species to be detected when really present in the medium (true positive),
- the associated probability is defined as the probability to not be detected while absent in the medium (true negative),
- the third probability defines the probability of a species to be detected in the sample when not present, hence being a false positive result, and
- defines the probability of a species to not be detected in the sample while actually being present in the medium and is a false negative result.
7. Opportunities and Challenges for EIA with eDNA
7.1. New Solutions That May Facilitate eDNA Use within EIA
- the disconnection between the receptor state and the corresponding eDNA signal;
- ensuring that a sampling strategy imposed for a given environment will also sample the eDNA targeted; and
- the statistical analysis of eDNA information for which only probabilities can be estimated and not certainties.
7.2. Persistent Need for Taxonomic Expertise
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Topical Bibliography
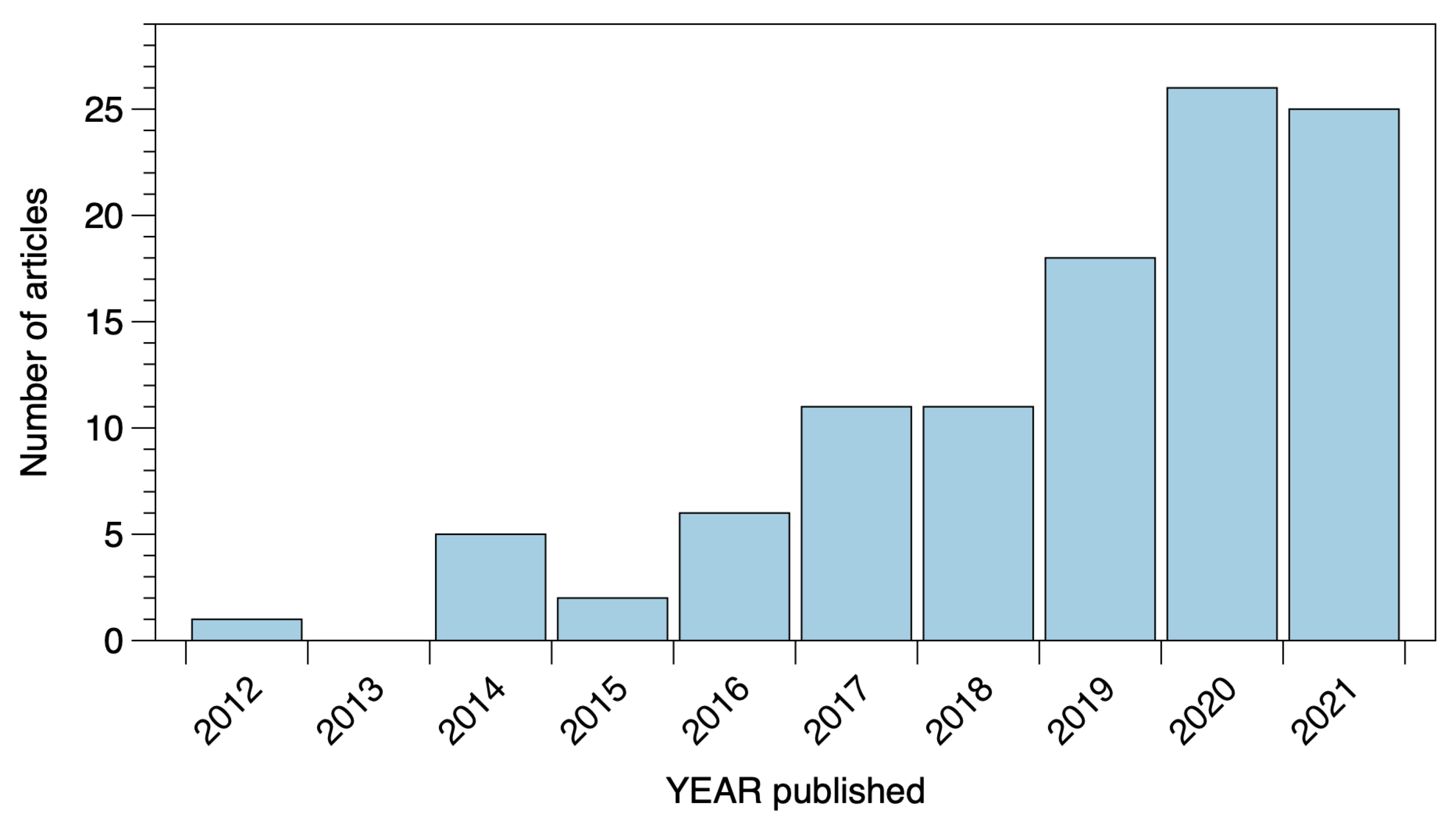
Appendix A.1. Search Method Used with Web of Science Database
Appendix A.2. Historical Overview of Important Conceptual Breakthroughs
- -
- Aardema, B.W.; Lorenz, M.G.; Krumbein, W.E. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Applied and Environmental Microbiology 1983, 46, 417–420. doi:10.1128/aem.46.2.417-420.1983.
- -
- DeFlaun, M.F.; Paul, J.H.; Jeffrey, W.H. Distribution and molecular weight of dissolved DNA in subtropical estuarine and oceanic environments. Marine Ecology Progress Series 1987, 38, 65–73.
- -
- Lorenz, M.G.; Wackernagel, W. Adsorption of DNA to sand and variable degradation rates of adsorbed DNA. Applied and Environmental Microbiology 1987, 53, 2948–2952. doi:10.1128/aem.53.12.2948-2952.1987.
- -
- Paul, J.H.; Jeffrey, W.H.; David, A.W.; DeFlaun, M.F.; Cazares, L.H. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Applied and Environmental Microbiology 1989, 55, 1823–1828.
- -
- Mullis, K.B.; Faloona, F.A.; Scharf, S.J.; Saiki, R.; Horn, G.T.; Erlich, H.A. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. doi:10.1101/sqb.1986.051.01.032.
- -
- Mullis, K.B.; Faloona, F.A. Specific Synthesis of DNA in vitro via a Polymerase-Catalyzed Chain Reaction. Methods in Enzymology 1987, 155(F), 335–350. doi: 10.1016/ 0076-6879(87)55023-6.
- -
- Saiki, R.; Gelfand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988, 239, 487–491. doi:10.1126/science.2448875.
- -
- Verlaan-de Vries, M.; Bogaard, M.E.; van den Elst, H.; van Boom, J.H.; van der Eb, A.J.; Bos, J.L. A dot- blot screening procedure for mutated ras oncogenes using synthetic oligodeoxynucleotides. Gene1986, 50, 313–320. doi:10.1016/0378-1119(86)90335-5.
- -
- Bhadury, P.; Austen, M.C.; Bilton, D.T.; Lambshead, P.J.D.; Rogers, A.D.; Smerdon, G.R. Molecular detection of marine nematodes from environmental samples: overcoming eukaryotic interference. Aquatic Microbial Ecology 2006, 44, 97–103. doi:10.3354/ ame044097.
- -
- Strickler, K.M.; Fremier, A.K.; Goldberg, C.S. Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biological Conservation 2014. doi:10.1016/j.biocon.2014.11.038.
- -
- Buxton, A.S.; Groombridge, J.J.; Griffiths, R.A. Is the detection of aquatic environmental DNA influenced by substrate type? PLoS One 2017, 12, e0183371. doi: 10.1371/journal.pone.0183371.
- -
- Guillera-Arroita, G.; Lahoz-Monfort, J.J.; van Rooyen, A.R.; Weeks, A.R.; Tingley, R. Dealing with false- positive and false-negative errors about species occurrence at multiple levels. Methods in Ecology and Evolution 2017, 8, 1081–1091. doi:10.1111/2041-210X.12743.
- -
- Corsaro, D.; Venditti, D. An apparent Acanthamoeba genotype is the product of a chimeric 18S rDNA artifact. Parasitology Research 2018, 117, 571–577.doi:10.1007/s00436-017-5690-9.
- -
- Kelly, R.P.; Shelton, A.O.; Gallego, R. Understanding PCR Processes to Draw Meaningful Conclusions from Environmental DNA Studies. Scientific Reports 2019, 9. doi:10.1038/s41598-019-48546-x.
- -
- Furlan, E.M.; Davis, J.; Duncan, R.P. Identifying error and accurately interpreting environmental DNA metabarcoding results: A case study to detect vertebrates at arid zone waterholes. Molecular Ecology Resources 2020, 20, 1259–1276. doi:10.1111/1755-0998.13170.
- -
- Sales, N.G.; McKenzie, M.B.; Drake, J.; Harper, L.R.; Browett, S.S.; Coscia, I.; Wangensteen, O.S.; Baillie, C.; Bryce, E.; Dawson, D.A.; Ochu, E.; Haenfling, B.; Handley, L.L.; Mariani, S.; Lambin, X.; Sutherland, C.; McDevitt, A.D. Fishing for mammals: Landscape-level monitoring of terrestrial and semi- aquatic communities using eDNA from riverine systems. Journal of Applied Ecology 2020, 57, 707–716. doi:10.1111/1365-2664.13592.
- -
- Pawlowski, J.; Lejzerowicz, F.; Apothéloz-Perret-Gentil, L.; Visco, J.; Esling, P. Protist metabarcoding and environmental biomonitoring: Time for change. European Journal of Protistology 2016, pp. 12–25. doi:10.1016/j.ejop.2016.02.003.
- -
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Molecular Ecology 2012, 21, 1789–1793. doi:10.1111/j.1365-294X.2012.05542.x.
- -
- Thomsen, P.F.; Kielgast, J.O.S.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; et al. Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology 2012, 21, 2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x.
Appendix A.3. Methodological Issues Relevant for EIA
Evaluating Sampling Strategies
- -
- de Souza, L.S.; Godwin, J.C.; Renshaw, M.A.; Larson, E. Environmental DNA (eDNA) detection probability is influenced by seasonal activity of organisms. PLoS One 2016, 11, e0165273. doi:10.1371/journal.pone.0165273.
- -
- Cornman, R.S.; McKenna, James E., J.; Fike, J.; Oyler-McCance, S.J.; Johnson, R. An experimental comparison of composite and grab sampling of stream water for metagenetic analysis of environmental DNA. PeerJ 2018, 6. doi:10.7717/peerj5871.
- -
- Abrams, J.F.; Hoerig, L.A.; Brozovic, R.; Axtner, J.; Crampton-Platt, A.; Mohamed, A.; Wong, S.T.; Sollmann, R.; Yu, D.W.; Wilting, A. Shifting up a gear with iDNA: From mammal detection events to standardised surveys. Journal of Applied Ecology 2019, 56, 1637–1648. doi:10.1111/1365-2664.13411.
- -
- Fukaya, K.; Kondo, N.I.; Matsuzaki, S.I.S.; Kadoya, T. Multispecies site occupancy modelling and study design for spatially replicated environmental DNA metabarcoding. Methods in Ecology and Evolution. 2021, doi:10.1111/2041-210X.13732.
Evaluating the Quality and Limits of Species Identifications
- -
- Galtier, N.; Nabholz, B.; Glémin, S.; Hurst, G.D. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Molecular Ecology 2009, 18, 4541–4550. doi:10.1111/ j.1365-294X.2009.04380.x.
- -
- Yao, H.; Song, J.; Liu, C.; Luo, K.; Han, J.; Li, Y.; Pang, X.; Xu, H.; Zhu, Y.; Xiao, P.; Chen, S. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One 2010, 5, e13102. doi: 10.1371/journal.pone.0013102.
- -
- Pilgrim, E.M.; Jackson, S.A.; Swenson, S.; Turcsanyi, I.; Friedman, E.; Weigt, L.; Bagley, M. Incorporation of DNA barcoding into a large-scale biomonitoring. Journal of North American Benthol. Society 2011, 30, 217–231. doi:10.1899/10-012.1.
- -
- Costa, F.; Landi, M.; Martins, R.; Costa, M.; Carneiro, M.; Alves, M.J.; Steinke, D.; Carvalho, G. A ranking system for reference libraries of DNA barcodes: Application to marine fish species from Portugal. PLoS One 2012.doi:10.1371/journal.pone.0035858.
- -
- Bourlat, S.J.; Borja, A.; Gilbert, J.; Taylor, M.I.; Davies, N.; Weisberg, S.B.; et al.. Genomics in marine monitoring: new opportunities for assessing marine health status. Marine Pollution Bulletin 2013, 74, 19– 31. doi:10.1016/j.marpolbul. 2013.05.042.
- -
- Stoeck, T.; Breiner, H.; Filker, S.; Ostermaier, V.; Kammerlander, B.; Sonntag, B. A morphogenetic survey on ciliate plankton from a mountain lake pinpoints the necessity of lineage-specific barcode markers in microbial ecology. Environmental Biology 2013, 16, 430–444. doi:10.1111/1462-2920.12194.
- -
- de Barba, M.; Miquel, C.; Boyer, F.; Mercier, C.; Rioux, D.; Coissac, E.; Taberlet, P. DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet. Molecular Ecology Resources 2014, 14, 306–323. doi:10.1111/1755-0998.12188.
- -
- Deagle, B.E.; Jarman, S.N.; Coissac, E.; Pompanon, F.; Taberlet, P. DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biology Letters 2014, 10, 20140562.
- -
- Schnell, I.B.; Bohmann, K.; Gilbert, M.T.P. Tag jumps illuminated - reducing sequence-to-sample misidentifications in metabarcoding studies. Molecular Ecology Resources2015, 15, 1289–1303. doi:10.1111/1755- 0998.12402.
- -
- Bhattacharya, M.; Sharma, A.; Patra, B.; Sharma, G.; Seo, E.; Nam, J.; Chakraborty, C.; Lee, S.S. DNA barcoding to fishes: current status and future directions. Mitochondrial DNA Part A 2016, 27, 2744–2752. doi: 10.3109/19401736.2015.1046175.
- -
- Weltz, K.; Lyle, J.M.; Ovenden, J.; Morgan, J.A.T.; Moreno, D.A.; Semmens, J.M. Application of environmental DNA to detect an endangered marine skate species in the wild. PLoS One2017, 12. doi:10.1371/journal.pone.0178124.
- -
- Hajibabaei, M.; Porter, T.M.; Robinson, C.V.; Baird, D.J.; Shokralla, S.; Wright, M.T.G. Watered-down biodiversity? A comparison of metabarcoding results from DNA extracted from matched water and bulk tissue biomonitoring samples. PLoS One 2019, 14. doi:10.1371/journal.pone.0225409.
- -
- Specchia, V.; Tzafesta, E.; Marini, G.; Scarcella, S.; D’Attis, S.; Pinna, M. Gap Analysis for DNA Barcode Reference Libraries for Aquatic Macroinvertebrate Species in the Apulia Region (Southeast of Italy). Journal of Marine Science and Engineering 2020, 8. doi:10.3390/jmse8070538.
Environmental Forensics
- -
- Allwood, J.S.; Fierer, N.; Dunn, R.R. The Future of Environmental DNA in Forensic Science. Applied and Environmental Microbiology 2020, 86. doi:10.1128/AEM.01504-19.
- -
- Bourret, V.; Albert, V.; April, J.; Cote, G.; Morissette, O. Past, present and future contributions of evolutionary biology to wildlife forensics, management and conservation. Evolutionary Applications2020, 13, 1420–1434. doi:10.1111/eva.12977.
Appendix A.4. Future Developments
New Sampling Approaches and Uses
- -
- Siegenthaler, A.; Wangensteen, O.S.; Soto, A.Z.; Benvenuto, C.; Corrigan, L.; Mariani, S. Metabarcoding of shrimp stomach content: Harnessing a natural sampler for fish biodiversity monitoring. Molecular Ecology Resources2019, 19, 206–220. doi:10.1111/1755-0998.12956.
- -
- Turon, M.; Angulo-Preckler, C.; Antich, A.; Praebel, K.; Wangensteen, O.S. More Than Expected From Old Sponge Samples: A Natural Sampler DNA Metabarcoding Assessment of Marine Fish Diversity in Nha Trang Bay (Vietnam). Frontiers in Marine Science2020, 7. doi:10.3389/fmars.2020.605148.
New Technologies
- -
- Priestley, V.; Allen, R.; Binstead, M.; Arnold, R.; Savolainen, V. Quick detection of a rare species: Forensic swabs of survey tubes for hazel dormouse Muscardinus avellanarius urine. Methods in Ecology and Evolution 2021, 12, 818–827. doi:10.1111/2041-210X.13573.
- -
- Williams, M.A.; O’Grady, J.; Ball, B.; Carlsson, J.; de Eyto, E.; McGinnity, P.; Jennings, E.; Regan, F.; Parle-McDermott, A. The application of CRISPR-Cas for single species identification from environmental DNA. Molecular Ecology Resources 2019, 19, 1106–1114. doi:10.1111/1755-0998.13045.
- -
- Wong, M.K.S.; Nakao, M.; Hyodo, S. Field application of an improved protocol for environmental DNA extraction, purification, and measurement using Sterivex filter. Scientific Reports 2020, 10. doi:10.1038/s41598-020-77304-7.
- -
- Doi, H.; Watanabe, T.; Nishizawa, N.; Saito, T.; Nagata, H.; Kameda, Y.; Maki, N.; Ikeda, K.; Fukuzawa, T. On-site environmental DNA detection of species using ultrarapid mobile PCR. Molecular Ecology Resources2021, 21, 2364–2368. doi:10.1111/1755-0998.13448.
References
- Wilcox, T.; McKelvey, K.; Young, M.; Jane, S.; Lowe, W.; Whiteley, A.; Schwartz, M. Robust Detection of Rare Species Using Environmental DNA: The Importance of Primer Specificity. PLoS ONE 2013, 8, e59520. [Google Scholar] [CrossRef] [PubMed]
- Holm-Hansen, O.; Sutcliffe, W.H.; Sharp, J. Measurement of deoxyribonucleic acid in the ocean and its ecological significance. Limnol. Oceanogr. 1968, 13, 507–514. [Google Scholar] [CrossRef]
- Maeda, M.; Taga, N. Deoxyribonuclease activity in seawater and sediment. Mar. Biol. 1973, 20, 58–63. [Google Scholar] [CrossRef]
- Cristescu, M.E.; Hebert, P.D.N. Uses and Misuses of Environmental DNA in Biodiversity Science and Conservation. In Annual Review of Ecology, Evolution, and Systematics; Futuyma, D., Ed.; Palo Alto: Santa Clara Co., CA, USA, 2018; Volume 49, pp. 209–230. [Google Scholar] [CrossRef]
- Torsvik, V.L. Isolation of bacterial DNA from soil. Soil Biol. Biochem. 1980, 12, 15–21. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Barkay, T. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Bailiff, M.D.; Karl, D.M. Dissolved and particulate DNA dynamics during a spring bloom in the Antarctic Peninsula region, 1986–1987. Deep. Sea Res. Part A Oceanogr. Res. Pap. 1991, 38, 1077–1095. [Google Scholar] [CrossRef]
- Henne, A.S. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 3113–3116. [Google Scholar] [CrossRef]
- Michotey, V.M. Comparison of methods for quantification of cytochrome cd 1-denitrifying bacteria in environmental marine samples. Appl. Environ. Microbiol. 2000, 66, 1564–1571. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Herbert, P.D.N. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Rey, A.; Basurko, O.C.; Rodriguez-Ezpeleta, N. Considerations for metabarcoding-based port biological baseline surveys aimed at marine nonindigenous species monitoring and risk assessments. Ecol. Evol. 2020, 10, 2452–2465. [Google Scholar] [CrossRef] [PubMed]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Jiang, L.; Yang, Y. Visualization of international environmental DNA research. Curr. Sci. 2017, 112, 1659–1664. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Machler, E.; Seymour, M.; Lacoursiere-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Fernandes, K.; van der Heyde, M.; Bunce, M.; Dixon, K.; Harris, R.J.; Wardell-Johnson, G.; Nevill, P.G. DNA metabarcoding-a new approach to fauna monitoring in mine site restoration. Restor. Ecol. 2018, 26, 1098–1107. [Google Scholar] [CrossRef]
- Bani, A.; De Brauwer, M.; Creer, S.; Dumbrell, A.J.; Limmon, G.; Jompa, J.; von der Heyden, S.; Beger, M. Informing marine spatial planning decisions with environmental DNA. In Tropical Ecosystems in the 21st Century; Dumbrell, A., Turner, E., Fayle, T., Eds.; Academic Press Ltd.—Elsevier Science Ltd.: London, UK, 2020; Volume 62, pp. 375–407. [Google Scholar] [CrossRef]
- Coble, A.A.; Flinders, C.A.; Homyack, J.A.; Penaluna, B.E.; Cronn, R.C.; Weitemier, K. eDNA as a tool for identifying freshwater species in sustainable forestry: A critical review and potential future applications. Sci. Total. Environ. 2019, 649, 1157–1170. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Birch, J.M.; Barnhart, E.P.; Merkes, C.M.; Yamahara, K.M.; Marin, R.; Kinsey, S.M.; Wright, P.R.; Schmidt, C. Robotic environmental DNA bio-surveillance of freshwater health. Sci. Rep. 2020, 10, 14389. [Google Scholar] [CrossRef]
- Pansu, J.; De Danieli, S.; Puissant, J.; Gonzalez, J.M.; Gielly, L.; Cordonnier, T.; Zinger, L.; Brun, J.J.; Choler, P.; Taberlet, P.; et al. Landscape-scale distribution patterns of earthworms inferred from soil DNA. Soil Biol. Biochem. 2015, 83, 100–105. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, J.; Yang, J.; Giesy, J.P.; Yu, H.; Zhang, X. Environmental DNA metabarcoding reveals primary chemical contaminants in freshwater sediments from different land-use types. Chemosphere 2017, 172, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Lodge, D.; Simonin, P.; Burgiel, S.; Keller, R.; Bossenbroek, J.; Jerde, C.; Kramer, A.; Rutherford, E.; Barnes, M.; Wittmann, M.; et al. Risk Analysis and Bioeconomics of Invasive Species to Inform Policy and Management. In Annual Review of Environment and Resources; Gadgil, A., Gadgil, T., Eds.; Palo Alto: Santa Clara Co., CA, USA, 2016; Volume 41, pp. 453–488. [Google Scholar] [CrossRef]
- Wang, P.; Yan, Z.; Yang, S.; Wang, S.; Zheng, X.; Fan, J.; Zhang, T. Environmental DNA: An Emerging Tool in Ecological Assessment. Bull. Environ. Contam. Toxicol. 2019, 103, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Decher, J.; Gray, C.R.; Garteh, J.C.; Kilpatrick, C.W.; Kuhn, H.J.; Phalan, B.; Monadjem, A.; Kadjo, B.; Jacquet, F.; Denys, C. New Evidence of the Semi-Aquatic Nimba Otter Shrew (Micropotamogale lamottei) at Mount Nimba and in the Putu Range of Liberia Uncertain Future for an Evolutionary Distinct and Globally Endangered (EDGE) Species in the Face of Recent Industrial Developments. J. Contemp. Water Res. Educ. 2016, 157, 46–57. [Google Scholar] [CrossRef]
- IAIA Publications. IAIA: What Is Impact Assessment? 2009. Available online: https://www.iaia.org/news-details.php?ID=30 (accessed on 7 December 2021).
- UN Environment. Assessing Environmental Impacts—A Global Review of Legislation; Technical Report; UN Environment World Conservation Monitoring Centre: Nairobi, Kenya, 2018; ISBN 978-92-807-3679-3. [Google Scholar]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Martone, R.G.; Lowell, N.; Thomsen, P.F.; Mach, M.E.; Bennett, M.; Prahler, E.; Caldwell, M.R.; et al. Harnessing DNA to improve environmental management. Science 2014, 344, 1455–1456. [Google Scholar] [CrossRef]
- Hering, D.; Borja, A.; Jones, J.I.; Pont, D.; Boets, P.; Bouchez, A.; Bruce, K.; Drakare, S.; Hanfling, B.; Kahlert, M.; et al. Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res. 2018, 138, 192–205. [Google Scholar] [CrossRef]
- Cordier, T.; Frontalini, F.; Cermakova, K.; Apotheloz-Perret-Gentil, L.; Treglia, M.; Scantamburlo, E.; Bonamin, V.; Pawlowski, J. Multi-marker eDNA metabarcoding survey to assess the environmental impact of three offshore gas platforms in the North Adriatic Sea (Italy). Mar. Environ. Res. 2019, 146, 24–34. [Google Scholar] [CrossRef]
- Eble, J.A.; Daly-Engel, T.S.; DiBattista, J.D.; Koziol, A.; Gaither, M.R. Marine environmental DNA: Approaches, applications, and opportunities. In Advances in Marine Biology; Sheppard, C., Ed.; Elsevier Academic Press: San Diego, CA, USA, 2020; Volume 86, pp. 141–169. [Google Scholar] [CrossRef]
- Dully, V.; Balliet, H.; Fruehe, L.; Daeumer, M.; Thielen, A.; Gallie, S.; Berrill, I.; Stoeck, T. Robustness, sensitivity and reproducibility of eDNA metabarcoding as an environmental biomonitoring tool in coastal salmon aquaculture—An inter-laboratory study. Ecol. Indic. 2021, 121, 107049. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Pochon, X.; Zaiko, A.; Keeley, N.; Bruce, K.; Hong, P.; Ruiz, G.M.; Stein, E.D.; Theroux, S.; et al. Translational Molecular Ecology in practice: Linking DNA-based methods to actionable marine environmental management. Sci. Total. Environ. 2020, 744, 140780. [Google Scholar] [CrossRef]
- Thalinger, B.; Deiner, K.; Harper, L.R.; Rees, H.C.; Blackman, R.C.; Sint, D.; Traugott, M.; Goldberg, C.S.; Bruce, K. A validation scale to determine the readiness of environmental DNA assays for routine species monitoring. Environ. DNA 2021, 3, 823–836. [Google Scholar] [CrossRef]
- Sepulveda, A.J.; Hutchins, P.R.; Forstchen, M.; Mckeefry, M.N.; Swigris, A.M. The Elephant in the Lab (and Field): Contamination in Aquatic Environmental DNA Studies. Front. Ecol. Evol. 2020, 8, 440. [Google Scholar] [CrossRef]
- Burian, A.; Mauvisseau, Q.; Bulling, M.; Domisch, S.; Qian, S.; Sweet, M. Improving the reliability of eDNA data interpretation. Mol. Ecol. Resour. 2021, 21, 1422–1433. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, J.; Bruce, K.; Panksep, K.; Aguirre, F.I.; Amafitano, S.; Apotheloz-Perret-Gentil, L.; Baussant, T.; Bouchez, A.; Carugati, L.; Cermakova, K.; et al. Environmental DNA metabarcoding for benthic monitoring: A review of sediment sampling and DNA extraction methods. Sci. Total. Environ. 2021, 151783. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018; p. 253. [Google Scholar] [CrossRef]
- Morris, P.; Therivel, R. (Eds.) Methods of Environmental Impact Assessment; Taylor & Francis: London, UK, 2001; Volume 2. [Google Scholar] [CrossRef]
- Wilson, J.; Hinz, S.; Coston-Guarini, J.; Mazé, C.; Guarini, J.M.; Chauvaud, L. System-Based Assessments—Improving the Confidence in the EIA Process. Environments 2017, 4, 95. [Google Scholar] [CrossRef]
- Morgan, R.K. Environmental impact assessment: The state of the art. Impact Assess. Proj. Apprais. 2012, 30, 5–14. [Google Scholar] [CrossRef]
- Johnson, M.D.; Cox, R.D.; Grisham, B.A.; Lucia, D.; Barnes, M.A. Airborne eDNA Reflects Human Activity and Seasonal Changes on a Landscape Scale. Front. Environ. Sci. 2021, 8, 563431. [Google Scholar] [CrossRef]
- Oldach, D.; Delwiches, C.; Jakobsen, K.; Tengs, T.; Brown, E.; Kempton, J.W.; Steidinger, K. Heteroduplex mobility assay-guided sequence discovery: Elucidation of the small subunit (18S) rDNA sequences of Pfisesteria piscicida and related dinoflagellates from complete algal culture and environmental sample DNA pools. Proc. Natl. Acad. Sci. USA 2000, 97, 4303–4308. [Google Scholar] [CrossRef]
- Popels, L.C.; Cary, S.; Hutchins, D.; Forbes, R.; Pustizzi, F.; Gobler, C.; Coyne, K. The use of quantitative polymerase chain reaction for the detection and enumeration of the harmful alga Aureococcus anophagefferens in environmental samples along the United States East coast. Limnol. Oceanogr. Methods 2003, 1, 92–102. [Google Scholar] [CrossRef]
- Peterson, K. Quality of environmental impact statements and variability of scrutiny by reviewers. Environ. Impact Assess. Rev. 2010, 30, 169–176. [Google Scholar] [CrossRef]
- Willsteed, E.A.; Birchenough, S.N.R.; Gill, A.B.; Jude, S. Structuring cumulative effects assessments to support regional and local marine management and planning obligations. Mar. Policy 2018, 98, 23–32. [Google Scholar] [CrossRef]
- Jonsson, P.R.; Hammar, L.; Wahlstrom, I.; Palsson, J.; Hume, D.; Almroth-Rosell, E.; Mattsson, M. Combining seascape connectivity with cumulative impact assessment in support of ecosystem-based marine spatial planning. J. Appl. Ecol. 2020, 58, 576–586. [Google Scholar] [CrossRef]
- Handford, P.; Bell, G.; Reimchen, T. A Gillnet Fishery Considered as an Experiment in Artificial Selection. J. Fish. Res. Board Can. 1977, 34, 954–961. [Google Scholar] [CrossRef]
- Hendry, A.P.; Gotanda, K.M.; Svensson, E.I. Human influences on evolution, and the ecological and societal consequences. Philos. Trans. R. Soc. B 2017, 372, 20160028. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, C.; Enberg, K.; Dunlop, E.S.; Arlinghaus, R.; Boukal, D.S.; Brander, K.; Ernande, B.; Gardmark, A.; Johnston, F.; Matsumura, S.; et al. Managing evolving fish stocks. Science 2007, 318, 1247–1248. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2015, 17, 1–17. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental Conditions Influence eDNA Persistence in Aquatic Systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef]
- Shelton, A.O.; O’Donnell, J.L.; Samhouri, J.F.; Lowell, N.; Williams, G.D.; Kelly, R.P. A framework for inferring biological communities from environmental DNA. Ecol. Appl. 2016, 26, 1645–1659. [Google Scholar] [CrossRef]
- Chambert, T.; Pilliod, D.S.; Goldberg, C.S.; Doi, H.; Takahara, T. An analytical framework for estimating aquatic species density from environmental DNA. Ecol. Evol. 2018, 8, 3468–3477. [Google Scholar] [CrossRef]
- Doi, H.; Fukaya, K.; Oka, S.I.; Sato, K.; Kondoh, M.; Miya, M. Evaluation of detection probabilities at the water-filtering and initial PCR steps in environmental DNA metabarcoding using a multispecies site occupancy model. Sci. Rep. 2019, 9, 3581. [Google Scholar] [CrossRef]
- Banerjee, P.; Dey, G.; Antognazza, C.M.; Sharma, R.K.; Maity, J.P.; Chan, M.W.; Huang, Y.H.; Lin, P.Y.; Chao, H.C.; Lu, C.M.; et al. Reinforcement of Environmental DNA Based Methods (Sensu Stricto) in Biodiversity Monitoring and Conservation: A Review. Biology 2021, 10, 1223. [Google Scholar] [CrossRef]
- Allan, E.A.; Zhang, W.G.; Lavery, A.C.; Govindarajan, A.F. Environmental DNA shedding and decay rates from diverse animal forms and thermal regimes. Environ. DNA 2020, 3, 492–514. [Google Scholar] [CrossRef]
- Jo, T.; Arimoto, M.; Murakami, H.; Masuda, R.; Minamoto, T. Particle Size Distribution of Environmental DNA from the Nuclei of Marine Fish. Environ. Sci. Technol. 2019, 53, 9947–9956. [Google Scholar] [CrossRef] [PubMed]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2015, 183, 77–84. [Google Scholar] [CrossRef]
- Lacoursiere-Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, A.; Nakamura, K.; Yamanaka, H.; Kondoh, M.; Minamoto, T. The release rate of environmental DNA from juvenile and adult fish. PLoS ONE 2014, 9, e114639. [Google Scholar] [CrossRef] [PubMed]
- Minamoto, T.; Fukuda, M.; Katsuhara, K.R.; Fujiwara, A.; Hidaka, S.; Yamamoto, S.; Takahashi, K.; Masuda, R. Environmental DNA reflects spatial and temporal jellyfish distribution. PLoS ONE 2017, 12, e0173073. [Google Scholar] [CrossRef] [PubMed]
- Takahara, T.; Minamoto, T.; Yamanaka, H.; Doi, H.; Kawabata, Z. Estimation of Fish Biomass Using Environmental DNA. PLoS ONE 2012, 7, e35868. [Google Scholar] [CrossRef] [PubMed]
- Dunn, N.; Priestley, V.; Herraiz, A.; Arnold, R.; Savolainen, V. Behavior and season affect crayfish detection and density inference using environmental DNA. Ecol. Evol. 2017, 7, 7777–7785. [Google Scholar] [CrossRef] [PubMed]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Factors influencing detection of eDNA from a stream-dwelling amphibian. Mol. Ecol. Resour. 2014, 14, 109–116. [Google Scholar] [CrossRef]
- Allan, E.A.; DiBenedetto, M.H.; Lavery, A.C.; Govindarajan, A.F.; Zhang, W.G. Modeling characterization of the vertical and temporal variability of environmental DNA in the mesopelagic ocean. Sci. Rep. 2021, 11, 21273. [Google Scholar] [CrossRef]
- Cowart, D.A.; Murphy, K.R.; Cheng, C.H.C. Metagenomic sequencing of environmental DNA reveals marine faunal assemblages from the West Antarctic Peninsula. Mar. Genom. 2018, 37, 148–160. [Google Scholar] [CrossRef]
- Jo, T.; Minamoto, T. Complex interactions between environmental DNA (eDNA) state and water chemistries on eDNA persistence suggested by meta-analyses. Mol. Ecol. Resour. 2021, 21, 1490–1503. [Google Scholar] [CrossRef] [PubMed]
- Driessen, R.P.C.; Sitters, G.; Laurens, N.; Moolenaar, G.F.; Wuite, G.J.L.; Goosen, N.; Dame, R.T. Effect of temperature on the intrinsic flexibility of DNA and its interaction with architectural proteins. Biochemistry 2014, 53, 6430–6438. [Google Scholar] [CrossRef] [PubMed]
- Eichmiller, J.J.; Best, S.E.; Sorensen, P.W. Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 2016, 50, 1859–1867. [Google Scholar] [CrossRef] [PubMed]
- Lance, R.F.; Klymus, K.E.; Richter, C.A.; Guan, X.; Farrington, H.L.; Carr, M.R.; Thompson, N.; Chapman, D.C.; Baerwaldt, K.L. Experimental observations on the decay of environmental DNA from bighead and silver carps. Manag. Biol. Invasions 2017, 8, 343–359. [Google Scholar] [CrossRef]
- Tsuji, S.; Ushio, M.; Sakurai, S.; Minamoto, T.; Yamanaka, H. Water temperature-dependent degradation of environmental DNA and its relation to bacterial abundance. PLoS ONE 2017, 12, e0176608. [Google Scholar] [CrossRef]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of environmental DNA in marine systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef]
- Salter, I. Seasonal variability in the persistence of dissolved environmental DNA (eDNA) in a marine system: The role of microbial nutrient limitation. PLoS ONE 2018, 13, e0192409. [Google Scholar] [CrossRef]
- Carraro, L.; Stauffer, J.B.; Altermatt, F. How to design optimal eDNA sampling strategies for biomonitoring in river networks. Environ. DNA 2021, 3, 157–172. [Google Scholar] [CrossRef]
- Marshall, N.T.; Vanderploeg, H.A.; Changanti, S.R. Environmenta (e)RNA advances the reliability of eDNA by predicting its age. Sci. Rep. 2021, 11, 2769. [Google Scholar] [CrossRef]
- Andruszkiewicz, E.A.; Koseff, J.R.; Fringer, O.B.; Ouellette, N.T.; Lowe, A.B.; Edwards, C.; Boehm, A.B. Modeling Environmental DNA Transport in the Coastal Ocean Using Lagrangian Particle Tracking. Front. Mar. Sci. 2019, 6, 477. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport Distance of Invertebrate Environmental DNA in a Natural River. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef] [PubMed]
- Deiner, K.; Fronhofer, E.A.; Machler, E.; Walser, J.C.; Altermatt, F. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sansom, B.J.; Sassoubre, L.M. Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 2017, 51, 14244–14253. [Google Scholar] [CrossRef] [PubMed]
- Shogren, A.J.; Tank, J.L.; Andruszkiewicz, E.; Olds, B.; Mahon, A.R.; Jerde, C.L.; Bolster, D. Controls on eDNA movement in streams: Transport, Retention, and Resuspension. Sci. Rep. 2017, 7, 5065. [Google Scholar] [CrossRef]
- Jerde, C.L.; Olds, B.P.; Shogren, A.J.; Andruszkiewicz, E.A.; Mahon, A.R.; Bolster, D.; Tank, J. Influence of Stream Bottom Substrate on Retention and Transport of Vertebrate Environmental DNA. Environ. Sci. Technol. 2016, 50, 8770–8779. [Google Scholar] [CrossRef]
- O’Donnell, J.L.; Kelly, R.P.; Shelton, A.O.; Samhouri, J.F.; Lowell, N.C.; Williams, G.D. Spatial distribution of environmental DNA in a nearshore marine habitat. PeerJ 2017, 5, e3044. [Google Scholar] [CrossRef]
- Yamamoto, S.; Masuda, R.; Sato, Y.; Sado, T.; Araki, H.; Kondoh, M.; Minamoto, T.; Miya, M. Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 2017, 7, 40368. [Google Scholar] [CrossRef]
- Jeunen, G.J.; Knapp, M.; Spencer, H.G.; Lamare, M.D.; Taylor, H.R.; Stat, M.; Bunce, M.; Gemmell, N.J. Environmental DNA (eDNA) metabarcoding reveals strong discrimination among diverse marine habitats connected by water movement. Mol. Ecol. Resour. 2019, 19, 426–438. [Google Scholar] [CrossRef]
- Friebertshauser, R.; Shollenberger, K.; Janosik, A.; Garner, J.T.; Johnston, C. The effect of bivalve filtration on eDNA-based detection of aquatic organisms. PLoS ONE 2019, 14, e0222830. [Google Scholar] [CrossRef]
- Pont, D.; Rocle, M.; Valentini, A.; Civade, R.; Jean, P.; Maire, A.; Roset, N.; Schabuss, M.; Zornig, H.; Dejean, T. Environmental DNA reveals quantitative patterns of fish biodiversity in large rivers despite its downstream transportation. Sci. Rep. 2018, 8, 10361. [Google Scholar] [CrossRef]
- Andruszkiewicz, E.A.; Sassoubre, L.M.; Boehm, A.B. Persistence of marine fish environmental DNA and the influence of sunlight. PLoS ONE 2017, 12, e0185043. [Google Scholar] [CrossRef] [PubMed]
- Lacoursiere-Roussel, A.; Howland, K.; Normandeau, E.; Grey, E.K.; Archambault, P.; Deiner, K.; Lodge, D.M.; Hernandez, C.; Leduc, N.; Bernatchez, L. eDNA metabarcoding as a new surveillance approach for coastal Arctic biodiversity. Ecol. Evol. 2018, 8, 7763–7777. [Google Scholar] [CrossRef] [PubMed]
- Uthicke, S.; Lamare, M.; Doyle, J.R. eDNA detection of corallivorous seastar (Acanthaster cf. solaris) outbreaks on the Great Barrier Reef using digital droplet PCR. Coral Reefs 2018, 37, 1229–1239. [Google Scholar] [CrossRef]
- Paul, J.; Kellogg, C.; Jiang, S. Viruses and DNA in marine environments. In Microbial Diversity in Time and Space; Colwell, R.R., Simidu, U., Ohwada, K., Eds.; Springer (Plenum Press): New York, NY, USA, 1996; pp. 115–124, Chapter 14. [Google Scholar] [CrossRef]
- Moushomi, R.; Wilgar, G.; Carvalho, G.; Creer, S.; Seymour, M. Environmental DNA size sorting and degradation experiment indicates the state of Daphnia magna mitochondrial and nuclear eDNA is subcellular. Sci. Rep. 2019, 9, 12500. [Google Scholar] [CrossRef]
- Diaz-Ferguson, E.E.; Moyer, G.R. History, applications, methodological issues and perspectives for the use of environmental DNA (eDNA) in marine and freshwater environments. Rev. Biol. Trop. 2014, 62, 1273–1284. [Google Scholar] [CrossRef]
- Laroche, O.; Kersten, O.; Smith, C.R.; Goetze, E. From Sea Surface to Seafloor: A Benthic Allochthonous eDNA Survey for the Abyssal Ocean. Front. Mar. Sci. 2020, 7, 682. [Google Scholar] [CrossRef]
- Willerslev, E.; Hansen, A.J.; Binladen, J.; Brand, T.B.; Gilbert, M.T.P.; Shapiro, B.; Bunce, M.; Wiuf, C.; Gilichinsky, D.A.; Cooper, A. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 2003, 300, 791–795. [Google Scholar] [CrossRef]
- Anglès d’Auriac, M.; Strand, D.; Mjelde, M.; Demars, B.; Thaulow, J. Detection of an invasive aquatic plant in natural water bodies using environmental DNA. PLoS ONE 2019, 14, e0219700. [Google Scholar] [CrossRef]
- Turner, C.R.; Uy, K.L.; Everhart, R.C. Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol. Conserv. 2015, 183, 93–102. [Google Scholar] [CrossRef]
- Guardiola, M.; Wangensteen, O.S.; Taberlet, P.; Coissac, E.; Jesus Uriz, M.; Turon, X. Spatio-temporal monitoring of deep-sea communities using metabarcoding of sediment DNA and RNA. PeerJ 2016, 4, e2807. [Google Scholar] [CrossRef]
- Lejzerowicz, F.; Gooday, A.J.; Angeles, I.B.; Cordier, T.; Morard, R.; Apotheloz-Perret-Gentil, L.; Lins, L.; Menot, L.; Brandt, A.; Levin, L.A.; et al. Eukaryotic Biodiversity and Spatial Patterns in the Clarion-Clipperton Zone and Other Abyssal Regions: Insights From Sediment DNA and RNA Metabarcoding. Front. Mar. Sci. 2021, 8, 536. [Google Scholar] [CrossRef]
- Won, N.I.; Kim, K.H.; Kang, J.H.; Park, S.R.; Lee, H.J. Exploring the Impacts of Anthropogenic Disturbance on Seawater and Sediment Microbial Communities in Korean Coastal Waters Using Metagenomics Analysis. Int. J. Environ. Res. Public Health 2017, 14, 130. [Google Scholar] [CrossRef] [PubMed]
- Garel, E.; Rey, C.C.; Ferreira, O.; van Koningsveld, M. Applicability of the “Frame of Reference” approach for environmental monitoring of offshore renewable energy projects. J. Environ. Manag. 2014, 141, 16–28. [Google Scholar] [CrossRef] [PubMed]
- Coston-Guarini, J.; Guarini, J.M.; Hinz, S.; Wilson, J.; Chauvaud, L. A roadmap for a quantitative ecosystem-based environmental impact assessment. ICES J. Mar. Sci. 2017, 74, 2012–2023. [Google Scholar] [CrossRef]
- Beentjes, K.K.; Speksnijder, A.G.; Schilthuizen, M.; Hoogeveen, M.; van der Hoorn, B.B. The effects of spatial and temporal replicate sampling on eDNA metabarcoding. PeerJ 2019, 7, e7335. [Google Scholar] [CrossRef] [PubMed]
- Koziol, A.; Stat, M.; Simpson, T.; Jarman, S.; DiBattista, J.; Harvey, E.S.; Marnane, M.; McDonald, J.; Bunce, M. Environmental DNA metabarcoding studies are critically affected by substrate selection. Mol. Ecol. Resour. 2019, 19, 366–376. [Google Scholar] [CrossRef]
- Cashmore, M. The role of science in environmental impact assessment: Process and procedure versus purpose in the development of theory. Environ. Impact Assess. Rev. 2004, 24, 403–426. [Google Scholar] [CrossRef]
- Cowart, D.A.; Matabos, M.; Brandt, M.I.; Marticorena, J.; Sarrazin, J. Exploring Environmental DNA (eDNA) to Assess Biodiversity of Hard Substratum Faunal Communities on the Lucky Strike Vent Field (Mid-Atlantic Ridge) and Investigate Recolonization Dynamics After an Induced Disturbance. Front. Mar. Sci. 2020, 6, 783. [Google Scholar] [CrossRef]
- Pinfield, R.; Dillane, E.; Runge, A.K.W.; Evans, A.; Mirimin, L.; Niemann, J.; Reed, T.E.; Reid, D.G.; Rogan, E.; Samarra, F.I.P.; et al. False-negative detections from environmental DNA collected in the presence of large numbers of killer whales (Orcinus orca). Environ. DNA 2019, 1, 316–328. [Google Scholar] [CrossRef]
- Doi, H.; Akamatsu, Y.; Watanabe, Y.; Goto, M.; Inui, R.; Katano, I.; Nagano, M.; Takahara, T.; Minamoto, T. Water sampling for environmental DNA surveys by using an unmanned aerial vehicle. Limnol. Oceanogr.-Methods 2017, 15, 939–944. [Google Scholar] [CrossRef]
- Doi, H.; Uchii, K.; Matsuhashi, S.; Takahara, T.; Yamanaka, H.; Minamoto, T. Isopropanol precipitation method for collecting fish environmental DNA. Limnol. Oceanogr.-Methods 2017, 15, 212–218. [Google Scholar] [CrossRef]
- Forster, D.; Filker, S.; Kochems, R.; Breiner, H.W.; Cordier, T.; Pawlowski, J.; Stoeck, T. A Comparison of Different Ciliate Metabarcode Genes as Bioindicators for Environmental Impact Assessments of Salmon Aquaculture. J. Eukaryot. Microbiol. 2019, 66, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Yamahara, K.; Preston, C.; Birch, J.; Walz, K.; Marin, R., III; Jensen, S.; Pargett, D.; Roman, B.; Ussler, W., III; Zhang, Y.; et al. In situ Autonomous Acquisition and Preservation of Marine Environmental DNA Using an Autonomous Underwater Vehicle. Front. Mar. Sci. 2019, 6, 1–14. [Google Scholar] [CrossRef]
- Schmidt, B.R.; Kery, M.; Ursenbacher, S.; Hyman, O.J.; Collins, J.P. Site occupancy models in the analysis of environmental DNA presence/absence surveys: A case study of an emerging amphibian pathogen. Methods Ecol. Evol. 2013, 4, 646–653. [Google Scholar] [CrossRef]
- MacKenzie, D.; Nichols, J.; Lachman, G.; Droege, S.; Royle, J.; Langtimm, C. Estimating site occupancy rates when detection probabilities are less than one. Ecology 2002, 83, 2248–2255. [Google Scholar] [CrossRef]
- Lahoz-Monfort, J.J.; Guillera-Arroita, G.; Tingley, R. Statistical approaches to account for false-positive errors in environmental DNA samples. Mol. Ecol. Resour. 2016, 16, 673–685. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Taberlet, P.; Coissac, E. How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 2016, 16, 604–607. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Pansu, J.; Bonin, A.; Coissac, E.; Giguet-Covex, C.; De Barba, M.; Gielly, L.; Lopes, C.M.; Boyer, F.; Pompanon, F.; et al. Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 2015, 15, 543–556. [Google Scholar] [CrossRef]
- Griffin, J.E.; Matechou, E.; Buxton, A.S.; Bormpoudakis, D.; Griffiths, R.A. Modelling environmental DNA data; Bayesian variable selection accounting for false positive and false negative errors. J. R. Stat. Soc. Ser. C-Appl. Stat. 2020, 69, 377–392. [Google Scholar] [CrossRef]
- Leese, F.; Elbrecht, V. Can DNA-Based Ecosystem Assessments Quantify Species Abundance? Testing Primer Bias and Biomass—Sequence Relationships with an Innovative Metabarcoding Protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef]
- Bylemans, J.; Gleeson, D.M.; Duncan, R.P.; Hardy, C.M.; Furlan, E.M. A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes. Environ. DNA 2019, 1, 402–414. [Google Scholar] [CrossRef]
- Harper, L.R.; Handley, L.; Hahn, C.; Boonham, N.; Rees, H.C.; Gough, K.C.; Lewis, E.; Adams, I.P.; Brotherton, P.; Phillips, S.; et al. Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol. Evol. 2018, 8, 6330–6341. [Google Scholar] [CrossRef] [PubMed]
- Boero, F. The Study of Species in the Era of Biodiversity: A Tale of Stupidity. Diversity 2010, 2, 115–126. [Google Scholar] [CrossRef]
- Darling, J.A. How to learn to stop worrying and love environmental DNA monitoring. Aquat. Ecosyst. Health Manag. 2019, 22, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Glaubrecht, M. On “Darwinian Mysteries” or Molluscs as Models in Evolutionary Biology: From Local Speciation to Global Radiation. Am. Malacol. Bull. 2009, 27, 3–23. [Google Scholar] [CrossRef]
- Emerson, J.; Adams, R.; Betancourt Román, C.; Brooks, B.; Coil, D.; Dahlhausen, K.; Ganz, H.; Hartmann, E.; Hsu, T.; Justice, N.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Engelhard, G.H.; Thurstan, R.H.; MacKenzie, B.R.; Alleway, H.K.; Bannister, R.C.A.; Massimiliano, C.; Clarke, M.W.; Currie, J.C.; Fortibuoni, R.; Holm, P.; et al. CES meets marine historical ecology: Placing the history of fish and fisheries in current policy context. ICES J. Mar. Sci. 2015, 73, 1386–1403. [Google Scholar] [CrossRef]
- Reinholdt Jensen, M.; Egelyng Sigsgaard, E.; Agersnap, S.; Jessen Rasmussen, J.; Baattrup-Pedersen, A.; Wiberg-Larsen, P.; Francis Thomsen, P. Seasonal turnover in community composition of stream-associated macroinvertebrates inferred from freshwater environmental DNA metabarcoding. Environ. DNA 2021, 3, 861–876. [Google Scholar] [CrossRef]
- Gontier, M.; Balfors, B.; Mörtberg, U. Biodiversity in environmental assessment—current practice and tools for prediction. Environ. Impact Assess. Rev. 2006, 26, 268–286. [Google Scholar] [CrossRef]
- White, N.E.; Guzik, M.T.; Austin, A.D.; Moore, G.I.; Humphreys, W.F.; Alexander, J.; Bunce, M. Detection of the rare Australian endemic blind cave eel (Ophisternon candidum) with environmental DNA: Implications for threatened species management in subterranean environments. Hydrobiologia 2020, 847, 3201–3211. [Google Scholar] [CrossRef]
- Yang, C.; Bohmann, K.; Wang, X.; Cai, W.; Wales, N.; Ding, Z.; Gopalakrishnan, S.; Yu, D.W. Biodiversity Soup II: A bulk-sample metabarcoding pipeline emphasizing error reduction. Methods Ecol. Evol. 2021, 12, 1252–1264. [Google Scholar] [CrossRef]
- Cordier, T.; Forster, D.; Dufresne, Y.; Martins, C.I.M.; Stoeck, T.; Pawlowski, J. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour. 2018, 18, 1381–1391. [Google Scholar] [CrossRef] [PubMed]
- Seymour, M.; Edwards, F.K.; Cosby, B.J.; Kelly, M.G.; de Bruyn, M.; Carvalho, G.R.; Creer, S. Executing multi-taxa eDNA ecological assessment via traditional metrics and interactive networks. Sci. Total. Environ. 2020, 729, 138801. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Fruehe, L.; Forster, D.; Cordier, T.; Martins, C.I.M.; Pawlowski, J. Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Mar. Pollut. Bull. 2018, 127, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Sinniger, F.; Pawlowski, J.; Harii, S.; Gooday, A.J.; Yamamoto, H.; Chevaldonne, P.; Cedhagen, T.; Carvalho, G.; Creer, S. Worldwide Analysis of Sedimentary DNA Reveals Major Gaps in Taxonomic Knowledge of Deep-Sea Benthos. Front. Mar. Sci. 2016, 3, 92. [Google Scholar] [CrossRef]
- Brandt, M.I.; Trouche, B.; Henry, N.; Liautard-Haag, C.; Maignien, L.; de Vargas, C.; Wincker, P.; Poulain, J.; Zeppilli, D.; Arnaud-Haond, S. An Assessment of Environmental Metabarcoding Protocols Aiming at Favoring Contemporary Biodiversity in Inventories of Deep-Sea Communities. Front. Mar. Sci. 2020, 7, 234. [Google Scholar] [CrossRef]
- Boschen, R.E.; Collins, P.C.; Tunnicliffe, V.; Carlsson, J.; Gardner, J.P.A.; Lowe, J.; McCrone, A.; Metaxas, A.; Sinniger, F.; Swaddling, A. A primer for use of genetic tools in selecting and testing the suitability of set-aside sites protected from deep-sea seafloor massive sulfide mining activities. Ocean. Coast. Manag. 2016, 122, 37–48. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (E)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total. Environ. 2018, 637, 1295–1310. [Google Scholar] [CrossRef]
- Cordier, T.; Esling, P.; Lejzerowicz, F.; Visco, J.; Ouadahi, A.; Martins, C.; Cedhagen, T.; Pawlowski, J. Predicting the Ecological Quality Status of Marine Environments from eDNA Metabarcoding Data Using Supervised Machine Learning. Environ. Sci. Technol. 2017, 51, 9118–9126. [Google Scholar] [CrossRef]
- Harrison, J.B.; Chronopoulou, P.M.; Salone, I.S.; Jilbert, T.; Koho, K.A. 16S and 18S rRNA Gene metabarcoding provide congruent information on the responses of sediment communities to eutrophication. Front. Mar. Sci. 2021, 8, 708716. [Google Scholar] [CrossRef]
- Mauffrey, F.; Cordier, T.; Apotheloz-Perret-Gentil, L.; Cermakova, K.; Merzi, T.; Delefosse, M.; Blanc, P.; Pawlowski, J. Benthic monitoring of oil and gas offshore platforms in the North Sea using environmental DNA metabarcoding. Mol. Ecol. 2021, 30, 3007–3022. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.P.; O’Donnell, J.L.; Lowell, N.C.; Shelton, A.O.; Samhouri, J.F.; Hennessey, S.M.; Feist, B.E.; Williams, G.D. Genetic signatures of ecological diversity along an urbanization gradient. PeerJ 2016, 4, e2444. [Google Scholar] [CrossRef] [PubMed]
- Rojahn, J.; Gleeson, D.M.; Furlan, E.; Haeusler, T.; Bylemans, J. Improving the detection of rare native fish species in environmental DNA metabarcoding surveys. Aquat.-Conserv.-Mar. Freshw. Ecosyst. 2021, 31, 990–997. [Google Scholar] [CrossRef]
- Sigsgaard, E.E.; Jensen, M.R.; Winkelmann, I.E.; Moller, P.R.; Hansen, M.M.; Thomsen, P.F. Population-level inferences from environmental DNA-Current status and future perspectives. Evol. Appl. 2020, 13, 245–262. [Google Scholar] [CrossRef]
- Taguchi, M.; King, J.R.; Wetklo, M.; Withler, R.E.; Yokawa, K. Population genetic structure and demographic history of Pacific blue sharks (Prionace glauca) inferred from mitochondrial DNA analysis. Mar. Freshw. Res. 2014, 66, 267–275. [Google Scholar] [CrossRef]
- Ciborowski, K.L.; Consuegra, S.; Garcia de Leaniz, C.; Beaumont, M.A.; Wang, J.; Jordan, W.C. Rare and fleeting: An example of interspecific recombination in animal mitochondrial DNA. Biol. Lett. 2007, 3, 554–557. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ujvari, B.; Dowton, M.; Madsen, T. Mitochondrial DNA recombination in a free-ranging Australian lizard. Biol. Lett. 2007, 3, 189–192. [Google Scholar] [CrossRef]
- Parsons, K.M.; Everett, M.; Dahlheim, M.; Park, L. Water, water everywhere: Environmental DNA can unlock population structure in elusive marine species. R. Soc. Open Sci. 2018, 5, 180537. [Google Scholar] [CrossRef]
- Stat, M.; Huggett, M.J.; Bernasconi, R.; DiBattista, J.D.; Berry, T.E.; Newman, S.J.; Harvey, E.S.; Brunce, M. Ecosystem biomonitoring with eDNA: Metabarcoding across the tree of life in a tropical marine environment. Sci. Rep. 2017, 7, 12240. [Google Scholar] [CrossRef]
- Stepien, C.A.; Snyder, M.R.; Etz, A.E. Invasion genetics of the silver carp Hypophthalmichthys molitrix across North America: Differentiation of fronts, introgression, and eDNA metabarcode detection. PLoS ONE 2019, 14, e0203012. [Google Scholar] [CrossRef]
- Andres, K.J.; Sethi, S.A.; Lodge, D.M.; Andres, J. Nuclear eDNA estimates population allele frequencies and abundance in experimental mesocosms and field samples. Mol. Ecol. 2021, 30, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.; Worm, B. How many species are there on Earth and in the ocean? PLoS Biol. 2011, 9, e1001127. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.B.; Miller, E.C.; Rhodes, M.K.; Wiens, J.J. Inordinate fondness multiplied and redstributed: The number of species on Earth and the new pie of life. Q. Rev. Biol. 2017, 92, 230–265. [Google Scholar] [CrossRef]
- Weigand, H.; Beerman, A.J.; Ciampor, F.; Costa, F.O.; Csabai, Z.; Duarte, S.; Geiger, M.F.; Grabowski, M.; Rimet, F.; Rulik, B.; et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total. Environ. 2019, 678, 499–524. [Google Scholar] [CrossRef]
- Marques, V.; Milhau, T.; Albouy, C.; Dejean, T.; Manel, S.; Mouillot, D.; Juhel, J.B. GAPeDNA: Assessing and mapping global species gaps in genetic databases for eDNA metabarcoding. Divers. Distrib. 2021, 27, 1880–1892. [Google Scholar] [CrossRef]
- Moritz, C.; Cicero, C. Correspondence: DNA barcoding: Promise and pitfalls. PLoS Biol. 2004, 2, 1520–1531. [Google Scholar] [CrossRef]
- Ardura, A. Species-specific markers for early detection of marine invertebrate invaders through eDNA methods: Gaps and priorities in GenBank as database example. J. Nat. Conserv. 2019, 47, 51–57. [Google Scholar] [CrossRef]
- Kim, J.I.; Linton, E.W.; Shin, W. Morphological and genetic diversity of Euglena deses group (Euglenophyceae) with emphasis on cryptic species. Algae 2016, 31, 219–230. [Google Scholar] [CrossRef]
- Mathieu, C.; Hermans, S.M.; Lear, G.; Buckley, T.R.; Lee, K.C.; Buckley, H.L. A Systematic Review of Sources of Variability and Uncertainty in eDNA Data for Environmental Monitoring. Front. Ecol. Evol. 2020, 8, 135. [Google Scholar] [CrossRef]
- Foote, A.D.; Thomsen, P.F.; Sveegaard, S.; Wahlberg, M.; Kielgast, J.; Kyhn, L.A.; Salling, A.B.; Galatius, A.; Orlando, L.; Gilbert, M.T.P. Investigating the Potential Use of Environmental DNA (eDNA) for Genetic Monitoring of Marine Mammals. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Kawk, I.S.; Park, Y.S.; Chang, K.H. Application and Utilization of Environmental DNA Technology for Biodiversity in Water Ecosystems. Korean J. Ecol. Environ. 2021, 54, 151–155. [Google Scholar] [CrossRef]
- Johnston, E.L.; Roberts, D.A. Contaminants reduce the richness and evenness of marine communities: A review and meta-analysis. Environ. Pollut. 2009, 157, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Borja, A.; Franco, J.; Perez, V. A marine biotic index to establish the ecological quality of soft-bottom benthos within European estuarine and coastal environments. Mar. Pollut. Bull. 2000, 40, 1100–1114. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Rodríguez-Ezpeleta, N. Environmental status assessment using DNA metabarcoding: Towards a genetics based marine biotic index (gAMBI). PLoS ONE 2014, 9, e90529. [Google Scholar] [CrossRef]
- Darling, J.A.; Galil, B.S.; Carvalho, G.R.; Rius, M.; Viard, F.; Piraino, S. Recommendations for developing and applying genetic tools to assess and manage biological invasions in marine ecosystems. Mar. Policy 2017, 85, 54–64. [Google Scholar] [CrossRef]
- Yu, O.H.; Lee, H.G.; Lee, J.H. Influence of environmental variables on the distribution of macrobenthos in the Han River Estuary. Korean Oceanogr. Sci. J. 2012, 47, 519–528. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Muxika, I.; Rodriguez-Ezpeleta, N. Adapting metabarcoding-based benthic biomonitoring into routine marine ecological status assessment networks. Ecol. Indic. 2018, 95, 194–202. [Google Scholar] [CrossRef]
- Popper, A.N.; Hawkins, A.D. An overview of fish bioacoustics and the impacts of anthropogenic sounds on fishes. J. Fish Biol. 2019, 94, 692–713. [Google Scholar] [CrossRef]
- Popper, A.N.; Hawkins, A.D.; Thomsen, F. Taking the Animals’ Perspective Regarding Anthropogenic Underwater Sound. Trends Ecol. Evol. 2020, 35, 787–794. [Google Scholar] [CrossRef]
- Stat, M.; John, J.; DiBattista, J.D.; Newman, S.J.; Bunce, M.; Harvey, E.S. Combined use of eDNA metabarcoding and video surveillance for the assessment of fish biodiversity. Conserv. Biol. 2019, 33, 196–205. [Google Scholar] [CrossRef]
- Mirimin, L.; Desmet, S.; Romero, D.L.; Fernandez, S.F.; Miller, D.L.; Mynott, S.; Brincau, A.G.; Stefanni, S.; Berry, A.; Gaughan, P.; et al. Don’t catch me if you can—Using cabled observatories as multidisciplinary platforms for marine fish community monitoring: An in situ case study combining Underwater Video and environmental DNA data. Sci. Total. Environ. 2021, 773, 145351. [Google Scholar] [CrossRef] [PubMed]
- Aglieri, G.; Baillie, C.; Mariani, S.; Cattano, C.; Calo, A.; Turco, G.; Spatafora, D.; Di Franco, A.; Di Lorenzo, M.; Guidetti, P.; et al. Environmental DNA effectively captures functional diversity of coastal fish communities. Mol. Ecol. 2021, 30, 3127–3139. [Google Scholar] [CrossRef] [PubMed]
- Lodge, D.M.; Turner, C.R.; Jerde, C.L.; Barnes, M.A.; Chadderton, L.; Egan, S.P.; Feder, J.L.; Mahon, A.R.; Pfrender, M.E. Conservation in a cup of water: Estimating biodiversity and population abundance from environmental DNA. Mol. Ecol. 2012, 21, 2555–2558. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Balasubramanian, S.; Church, G.M.; Gilbert, W.; Rogers, J.; Schloss, J.A.; Waterston, R.H. DNA sequencing at 40: Past, present and future. Nature 2017, 550, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Gasol, J.M.; Kirchman, D.L. Introduction: The evolution of microbial ecology of the ocean. In Microbial Ecology of the Oceans, 3rd ed.; Gasol, J.M., Kirchman, D.L., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2018; Chapter 1; pp. 1–46. ISBN 978-1-119-10718-7. [Google Scholar]
| Steps | Description |
|---|---|
| Screening | Preliminary identification of potential impacts created by a proposed project and description of alternative propositions, can be an iterative process |
| Scoping | Potential impact receptors are specified, determination of the extent and scales of the impact assessment study, includes definition of a project’s zone of influence and identification of data gaps |
| Baseline | Part of Scoping, Describes baseline measurements needed to estimate impacts |
| Impact Assessment | Evaluation of significance and consequences of the potential impacts on receptors identified during Scoping |
| Monitoring | Measurements done during project installation and operation to compare the predicted and actual impacts and the effectiveness of the mitigation measures taken |
| EIA Stage | New Capabilities Introduced |
|---|---|
| Screening | Rare, endangered, or protected species presence detections |
| Scoping | Enables objective comparisons with earlier studies on project area, changes notion of cumulative impact |
| Baselines | Potential for high frequency sampling of sensitive receptor(s) throughout project area, high-throughput screening methods could produce lists of groups potentially present with less effort, possible to store information on organisms present in situ for analysis post-project |
| Impact assessment | More sensitive detection of changes allowing more responsive management; ability to assess changes at an assemblage level |
| Monitoring | Potential to increase speed and reduce the cost of biological surveys, could use smaller field teams, could collect information on wider range of species simultaneously |
| EIA Stage | Opportunities | Limitations | Challenges |
|---|---|---|---|
| Screening | More ecological components (e.g., species, assemblages) detected | Conditioned by availability in data libraries | When unknown, eDNA shedding dynamics should be characterized with preparatory studies |
| Scoping | Better characterization of ecological system structure | Requires access to complementary information about receptor(s) | Developing a quantitative index of cumulative impact accounting |
| Baselines | Increased spatio-temporal resolution of species presence; more efficient detection and databanking of species present | Need for reference samples of known compositions for inter-laboratory comparisons | Standarization and normalization of analysis protocols; ensure backwards compatibility; develop formal sampling plans using statistical principles |
| Impact assessment | Probabilistic prediction of impact on species assemblages; sensitive detection of rare species presence changes | Need to link discrepancies between eDNA and receptor(s) changes | Integrate quantification of eDNA fate and transport over region of interest |
| Monitoring | Cost effective field surveys with higher taxonomic resolution and coverage | Could require long-term access to cold storage and analysis facilities | Determination of mitigation effectiveness could require formal sampling plans using statistical principles |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinz, S.; Coston-Guarini, J.; Marnane, M.; Guarini, J.-M. Evaluating eDNA for Use within Marine Environmental Impact Assessments. J. Mar. Sci. Eng. 2022, 10, 375. https://doi.org/10.3390/jmse10030375
Hinz S, Coston-Guarini J, Marnane M, Guarini J-M. Evaluating eDNA for Use within Marine Environmental Impact Assessments. Journal of Marine Science and Engineering. 2022; 10(3):375. https://doi.org/10.3390/jmse10030375
Chicago/Turabian StyleHinz, Shawn, Jennifer Coston-Guarini, Michael Marnane, and Jean-Marc Guarini. 2022. "Evaluating eDNA for Use within Marine Environmental Impact Assessments" Journal of Marine Science and Engineering 10, no. 3: 375. https://doi.org/10.3390/jmse10030375
APA StyleHinz, S., Coston-Guarini, J., Marnane, M., & Guarini, J.-M. (2022). Evaluating eDNA for Use within Marine Environmental Impact Assessments. Journal of Marine Science and Engineering, 10(3), 375. https://doi.org/10.3390/jmse10030375






