How Do Gene Expression Patterns Change in Response to Osmotic Stresses in Kuruma Shrimp (Marsupenaeus japonicus)?
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Effects of Acute Salinity Changes on Post-Larvae Shrimp
2.3. Salinity Acclimation Experiment of Post-Larvae Shrimp
2.4. Acute Salinity Exposure Experiment of Adult Shrimp
2.5. Salinity Acclimation Experiment of Adult Shrimp
2.6. RNA Isolation and cDNA Synthesis
2.7. Identification and Characterization of Osmoregulatory Genes
2.8. Expression Analysis of Osmoregulatory Genes
3. Results
3.1. Molecular Characterization of Osmoregulatory Genes
3.2. Tissue Expression Patterns of Genes in Adult Shrimp
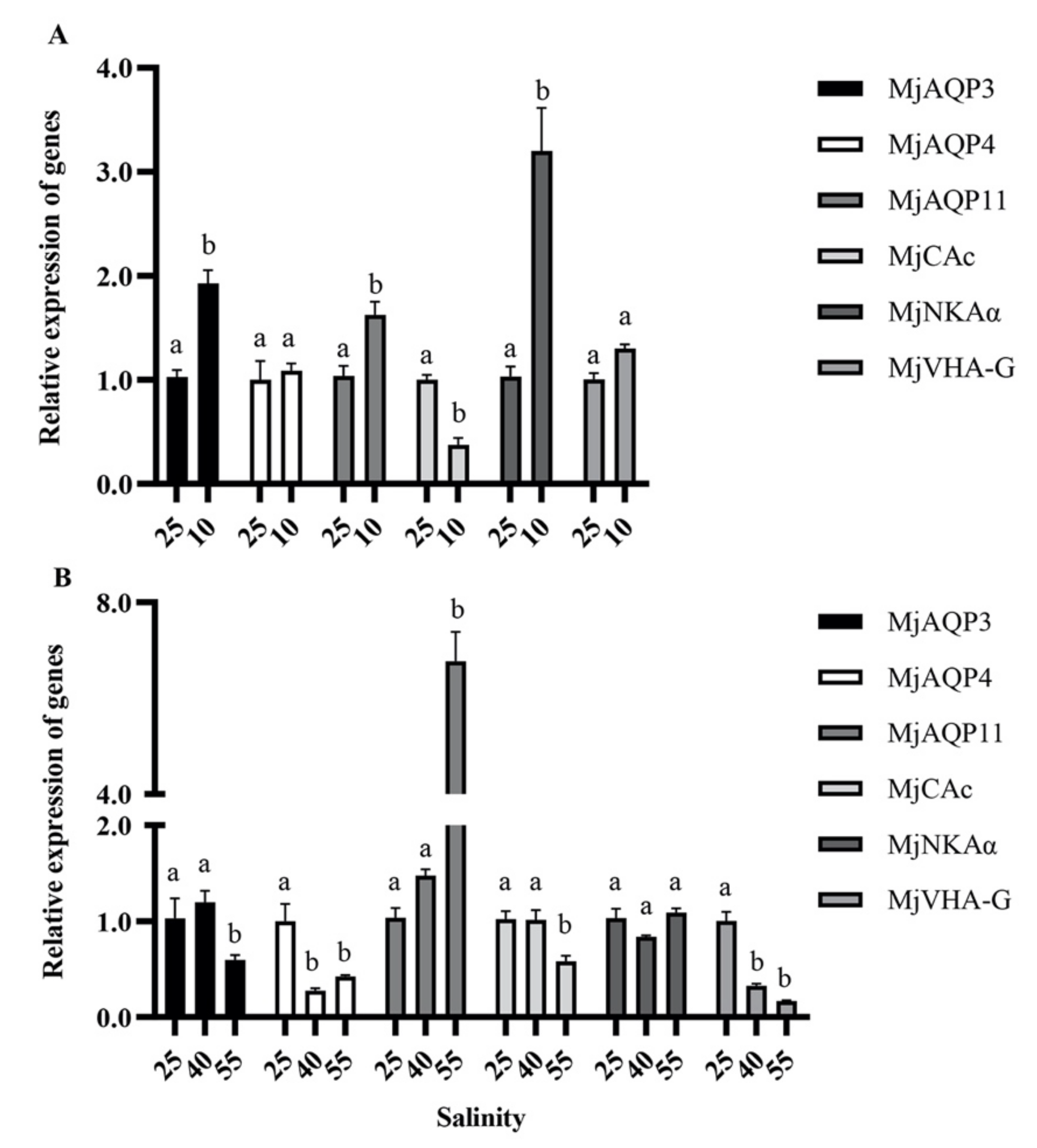
3.3. Effects of Acute Salinity Stress on Post-Larvae Shrimp
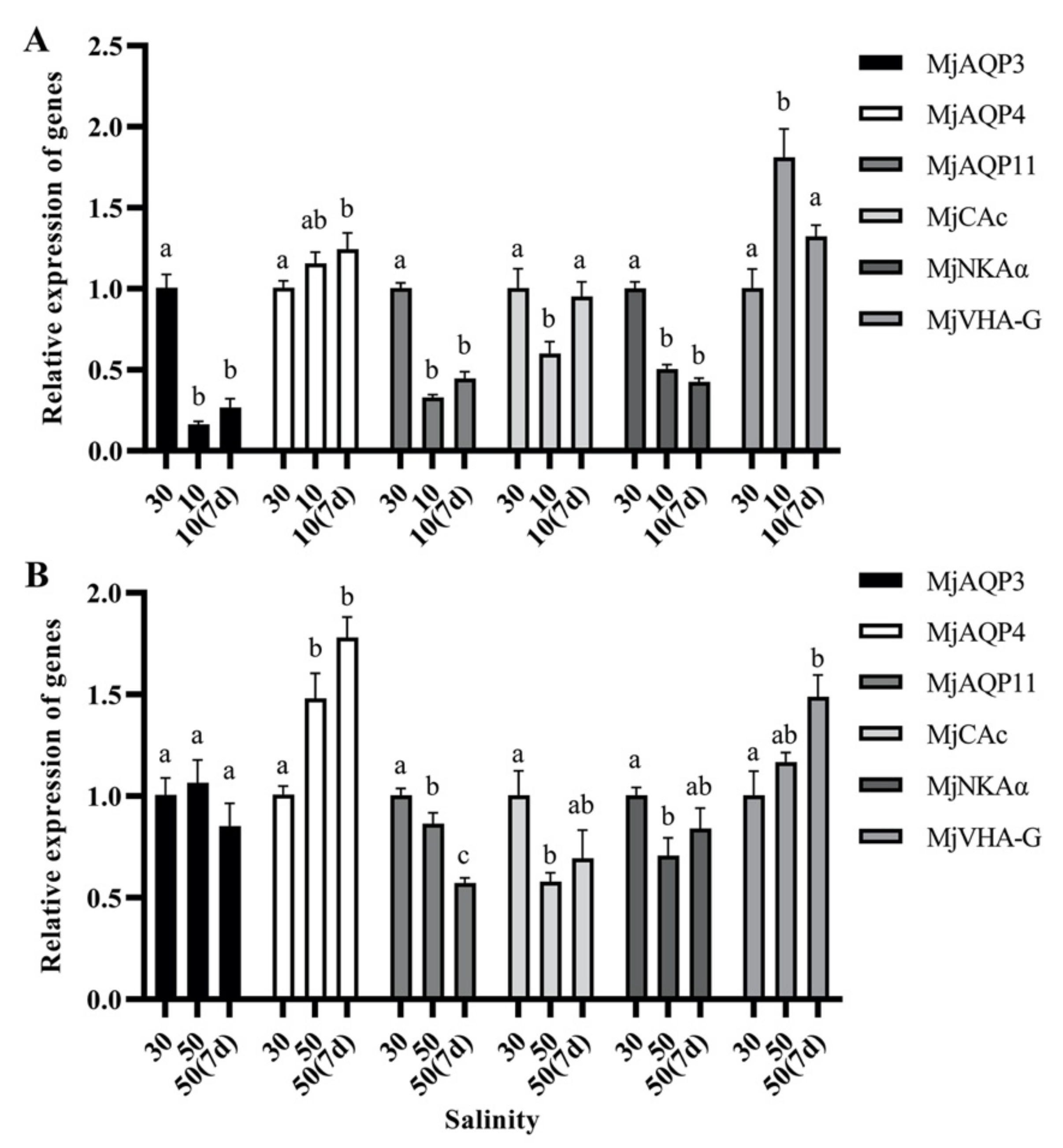
3.4. Gene Expression in Post-Larvae Shrimp during the Salinity Acclimation
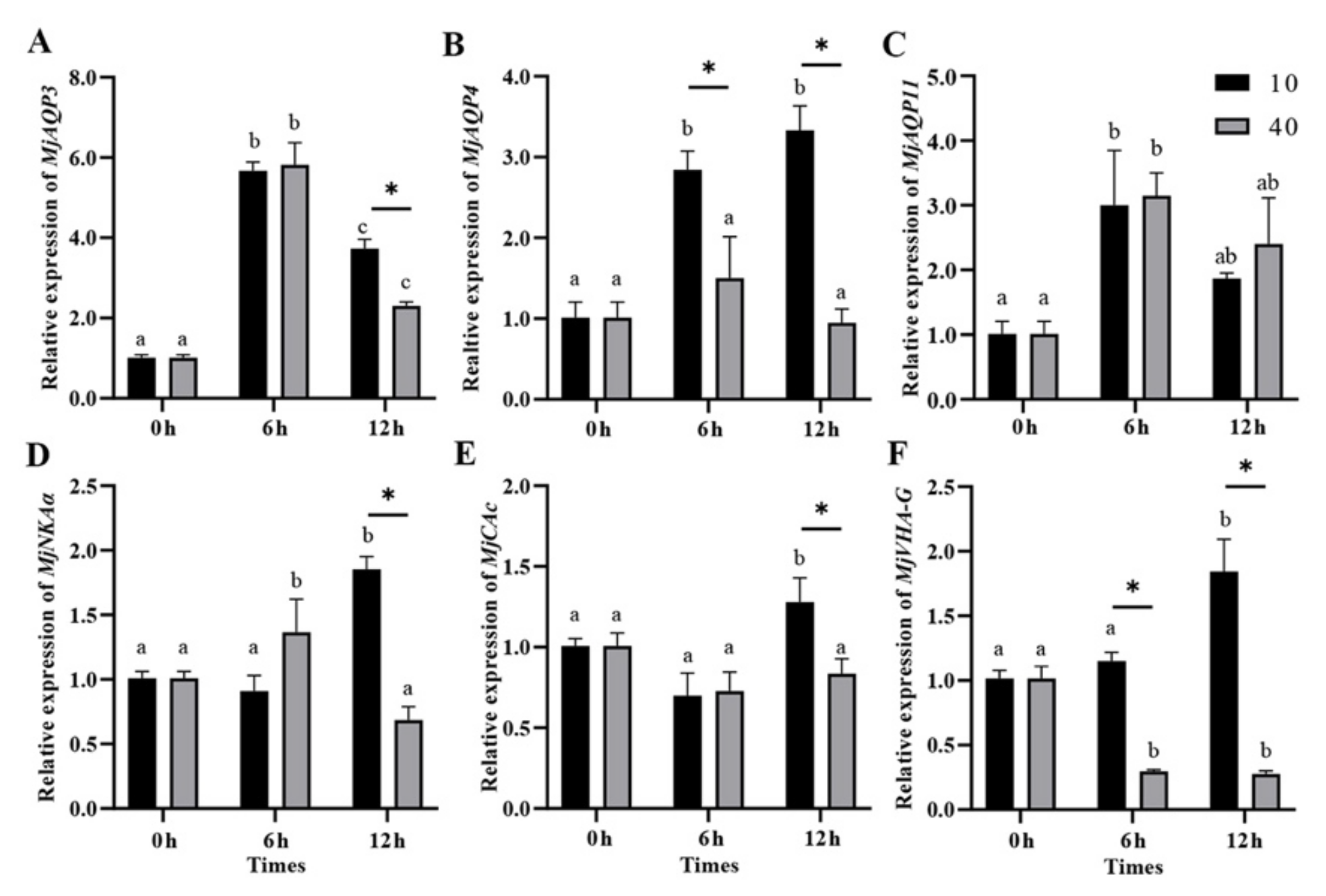
3.5. Gene Expression in Response to Acute Salinity Stresses in the Gills
3.6. Gene Expression in Response to Salinity Acclimation in the Gills
4. Discussion
4.1. Tolerance of M. japonicus to Salinity Challenges
4.2. Expression of MjAQPs in Response to Salinity Challenges
4.3. Expression of Ion-Transportation-Related Genes in Response to Salinity Challenges
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Chen, Y.; Cui, Y.; Shen, M.; Wang, R.; Wang, Z. Transcriptome Analysis of Pacific White Shrimp (Litopenaeus vannamei) Under Prolonged High-Salinity Stress. J. Ocean Univ. China 2022, 21, 430–444. [Google Scholar] [CrossRef]
- McNamara, J.C.; Faria, S.C. Evolution of Osmoregulatory Patterns and Gill Ion Transport Mechanisms in the Decapod Crustacea: A Review. J. Comp. Physiol. B 2012, 182, 997–1014. [Google Scholar] [CrossRef]
- Rahi, M.L.; Moshtaghi, A.; Mather, P.B.; Hurwood, D.A. Osmoregulation in Decapod Crustaceans: Physiological and Genomic Perspectives. Hydrobiologia 2018, 825, 177–188. [Google Scholar] [CrossRef]
- Rahi, M.L.; Amin, S.; Mather, P.B.; Hurwood, D.A. Candidate Genes That Have Facilitated Freshwater Adaptation by Palaemonid Prawns in the Genus Macrobrachium: Identification and Expression Validation in a Model Species (M. Koombooloomba). PeerJ 2017, 5, e2977. [Google Scholar] [CrossRef][Green Version]
- McNamara, J.C.; Freire, C.A.; Torres, A.H., Jr.; Faria, S.C. The Conquest of Fresh Water by the Palaemonid Shrimps: An Evolutionary History Scripted in the Osmoregulatory Epithelia of the Gills and Antennal Glands. Biol. J. Linn. Soc. 2015, 114, 673–688. [Google Scholar] [CrossRef]
- Thabet, R.; Ayadi, H.; Koken, M.; Leignel, V. Homeostatic Responses of Crustaceans to Salinity Changes. Hydrobiologia 2017, 799, 1–20. [Google Scholar] [CrossRef]
- Lucu, Č.; Towle, D.W. Na++K+-ATPase in Gills of Aquatic Crustacea. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2003, 135, 195–214. [Google Scholar] [CrossRef]
- Boudour-Boucheker, N.; Boulo, V.; Charmantier-Daures, M.; Anger, K.; Charmantier, G.; Lorin-Nebel, C. Osmoregulation in Larvae and Juveniles of Two Recently Separated Macrobrachium Species: Expression Patterns of Ion Transporter Genes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 195, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.-R.; Lin, H.-C. V-Type H+-ATPase and Na+, K+-ATPase in the Gills of 13 Euryhaline Crabs during Salinity Acclimation. J. Exp. Biol. 2007, 210, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Havird, J.C.; Henry, R.P.; Wilson, A.E. Altered Expression of Na+/K+–ATPase and Other Osmoregulatory Genes in the Gills of Euryhaline Animals in Response to Salinity Transfer: A Meta-Analysis of 59 Quantitative PCR Studies over 10 years. Comp. Biochem. Physiol. Part D Genom. Proteom. 2013, 8, 131–140. [Google Scholar] [CrossRef]
- Leone, F.A.; Lucena, M.N.; Garçon, D.P.; Pinto, M.R.; McNamara, J.C. Gill Ion Transport ATPases and Ammonia Excretion in Aquatic Crustaceans. In Acid-Base Balance and Nitrogen Excretion in Invertebrates; Springer: Cham, Switzerland, 2017; pp. 61–107. [Google Scholar]
- Weihrauch, D.; Ziegler, A.; Siebers, D.; Towle, D.W. Molecular Characterization of V-Type H(+)-ATPase (B-Subunit) in Gills of Euryhaline Crabs and Its Physiological Role in Osmoregulatory Ion Uptake. J. Exp. Biol. 2001, 204, 25–37. [Google Scholar] [CrossRef]
- Burnett, L.E.; Dunn, T.N.; Infantino, R.L. The Function of Carbonic Anhydrase in Crustacean Gills. In Proceedings of the Transport Processes, Iono- and Osmoregulation; Gilles, R., Gilles-Baillien, M., Eds.; Springer: Berlin/Heidelberg, Germany, 1985; pp. 159–168. [Google Scholar]
- Ge, Q.; Li, J.; Wang, J.; Li, Z.; Li, J. Characterization, Functional Analysis, and Expression Levels of Three Carbonic Anhydrases in Response to PH and Saline–Alkaline Stresses in the Ridgetail White Prawn Exopalaemon carinicauda. Cell Stress Chaperones 2019, 24, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Irisarri, I.; Zardoya, R. Diversity and Evolution of Membrane Intrinsic Proteins. Biochim. Biophys. Acta BBA Gen. Subj. 2014, 1840, 1468–1481. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.N.; Cerdà, J. Evolution and Functional Diversity of Aquaporins. Biol. Bull. 2015, 229, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Benga, G. On the Definition, Nomenclature and Classification of Water Channel Proteins (Aquaporins and Relatives). Mol. Asp. Med. 2012, 33, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Morishita, Y.; Tanaka, Y. The Evolutionary Aspects of Aquaporin Family. In Aquaporins; Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2017; Volume 969, pp. 35–50. [Google Scholar] [CrossRef]
- Boyle, R.T.; Oliveira, L.F.; Bianchini, A.; Souza, M.M. The Effects of Copper on Na+/K--ATPase and Aquaporin Expression in Two Euryhaline Invertebrates. Bull. Environ. Contam. Toxicol. 2013, 90, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Foguesatto, K.; Boyle, R.T.; Rovani, M.T.; Freire, C.A.; Souza, M.M. Aquaporin in Different Moult Stages of a Freshwater Decapod Crustacean: Expression and Participation in Muscle Hydration Control. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2017, 208, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Lind, U.; Järvå, M.; Alm Rosenblad, M.; Pingitore, P.; Karlsson, E.; Wrange, A.-L.; Kamdal, E.; Sundell, K.; André, C.; Jonsson, P.R. Analysis of Aquaporins from the Euryhaline Barnacle Balanus improvisus Reveals Differential Expression in Response to Changes in Salinity. PLoS ONE 2017, 12, e0181192. [Google Scholar] [CrossRef]
- Lv, J.; Liu, P.; Wang, Y.; Gao, B.; Chen, P.; Li, J. Transcriptome Analysis of Portunus trituberculatus in Response to Salinity Stress Provides Insights into the Molecular Basis of Osmoregulation. PLoS ONE 2013, 8, e82155. [Google Scholar] [CrossRef] [PubMed]
- Moshtaghi, A.; Rahi, M.L.; Mather, P.B.; Hurwood, D.A. An Investigation of Gene Expression Patterns That Contribute to Osmoregulation in Macrobrachium australiense: Assessment of Adaptive Responses to Different Osmotic Niches. Gene Rep. 2018, 13, 76–83. [Google Scholar] [CrossRef]
- Stavang, J.A.; Chauvigné, F.; Kongshaug, H.; Cerdà, J.; Nilsen, F.; Finn, R.N. Phylogenomic and Functional Analyses of Salmon Lice Aquaporins Uncover the Molecular Diversity of the Superfamily in Arthropoda. BMC Genom. 2015, 16, 618. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, Z.; Deng, Z.; Sun, J.; Zhao, R.; Cui, Y.; Wang, R.; Li, Y.; Wang, Z. Classical Aquaporins from Pacific White Shrimp (Litopenaeus vannamei): Molecular Characterization and Expression Analysis in Hypersalinity. Aquac. Rep. 2022, 23, 101016. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, Y.; Wang, C.; Zhao, N.; Zhang, Z.; Deng, Z.; Cui, Y.; Wang, R.; Li, Y. Aquaporins in Pacific White Shrimp (Litopenaeus vannamei): Molecular Characterization, Expression Patterns, and Transcriptome Analysis in Response to Salinity Stress. Front. Mar. Sci. 2022, 9, 817868. [Google Scholar] [CrossRef]
- Zhong, S.; Mao, Y.; Wang, J.; Liu, M.; Zhang, M.; Su, Y. Transcriptome Analysis of Kuruma Shrimp (MarsuPenaeus japonicus) Hepatopancreas in Response to White Spot Syndrome Virus (WSSV) under Experimental Infection. Fish Shellfish Immunol. 2017, 70, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, L.; Li, W.; Wu, J. Acute Toxicity of Salinity to Sinogastromyzon szechuanensis. J. Phys. Conf. Ser. 2020, 1629, 012025. [Google Scholar] [CrossRef]
- He, W.; Yu, T.; Cai, Y. Analysis of Control and Use of Alkaline Soil. J. Anshan Univ. Sci. Technol. 2010, 29, 158–160. [Google Scholar]
- Li, C.; Li, N.; Dong, T.; Fu, Q.; Cui, Y.; Li, Y. Analysis of Differential Gene Expression in Litopenaeus vannamei under High Salinity Stress. Aquac. Rep. 2020, 18, 100423. [Google Scholar] [CrossRef]
- Lv, X.; Xu, H.; Li, L.; Zhao, Y. Agricultural Sustainable Utilization and Evaluation of Saline-Alkali Land. Soils 2012, 44, 203–207. [Google Scholar]
- Boyd, C.E.; Fast, A.W. Pond Monitoring and Management: Marine Shrimp Culture Principles an Practices; Elsevier Science Publishing Comp. Inc.: New York, NY, USA, 1992. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Chen, T.; Li, Z.; Liu, J.; Liang, C.; Yuan, L. Transcriptome of Hepatopancreas in Kuruma Shrimp MarsuPenaeus japonicus under Low-Salinity Stress. J. Oceanol. Limnol. 2022, 40, 745–765. [Google Scholar] [CrossRef]
- Cheng, S.-Y.; Lee, W.-C.; Chen, J.-C. Increase of Uricogenesis in the Kuruma Shrimp MarsuPenaeus japonicus Reared under Hyper-Osmotic Conditions. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2004, 138, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Koyama, H.; Kuniyoshi, H.; Piyapattanakorn, S.; Watabe, S. Changes in Metabolite Concentrations in the Abdominal Muscle of the Kuruma Shrimp MarsuPenaeus japonicus in Response to Different Salinities. Fish. Sci. 2021, 87, 383–401. [Google Scholar] [CrossRef]
- Koyama, H.; Mizusawa, N.; Hoashi, M.; Tan, E.; Yasumoto, K.; Jimbo, M.; Ikeda, D.; Yokoyama, T.; Asakawa, S.; Piyapattanakorn, S. Changes in Free Amino Acid Concentrations and Associated Gene Expression Profiles in the Abdominal Muscle of Kuruma Shrimp (MarsuPenaeus japonicus) Acclimated at Different Salinities. J. Exp. Biol. 2018, 221, jeb168997. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-C.; Chen, J.-C. Hemolymph Ammonia, Urea and Uric Acid Levels and Nitrogenous Excretion of MarsuPenaeus japonicus at Different Salinity Levels. J. Exp. Mar. Biol. Ecol. 2003, 288, 39–49. [Google Scholar] [CrossRef]
- Nash, M.T.; Quijada-Rodriguez, A.R.; Allen, G.J.P.; Wilson, J.M.; Weihrauch, D. Characterization of 3 Different Types of Aquaporins in Carcinus maenas and Their Potential Role in Osmoregulation. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2022, 272, 111281. [Google Scholar] [CrossRef]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. Chapter One - Perspectives on the Evolution of Aquaporin Superfamily. In Vitamins and Hormones; Elsevier Inc.: Amsterdam, The Netherlands, 2020; Volume 112, pp. 1–27. [Google Scholar] [CrossRef]
- Kim, Y.K.; Lee, S.Y.; Kim, B.S.; Kim, D.S.; Nam, Y.K. Isolation and MRNA Expression Analysis of Aquaporin Isoforms in Marine Medaka Oryzias dancena, a Euryhaline Teleost. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 171, 1–8. [Google Scholar] [CrossRef]
- Tingaud-Sequeira, A.; Calusinska, M.; Finn, R.N.; Chauvigné, F.; Lozano, J.; Cerdà, J. The Zebrafish Genome Encodes the Largest Vertebrate Repertoire of Functional Aquaporins with Dual Paralogy and Substrate Specificities Similar to Mammals. BMC Evol. Biol. 2010, 10, 38. [Google Scholar] [CrossRef]
- Chung, J.S.; Maurer, L.; Bratcher, M.; Pitula, J.S.; Ogburn, M.B. Cloning of Aquaporin-1 of the Blue Crab, Callinectes sapidus: Its Expression during the Larval Development in Hyposalinity. Aquat. Biosyst. 2012, 8, 21. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, C.; Ren, C.; Qian, J.; He, X.; Jiang, X.; Huang, W. Molecular Cloning of Aquaporin-4 (AQP4) Gene in the Pacific White Shrimp (Litopenaeus vannamei) and the Effect of Salinity Stress on Its Expression in Hepatopancreas. Mar. Sci. 2017, 41, 61–70. [Google Scholar]
- Breves, J.P.; Inokuchi, M.; Yamaguchi, Y.; Seale, A.P.; Hunt, B.L.; Watanabe, S.; Lerner, D.T.; Kaneko, T.; Grau, E.G. Hormonal Regulation of Aquaporin 3: Opposing Actions of Prolactin and Cortisol in Tilapia Gill. J. Endocrinol. 2016, 230, 325–337. [Google Scholar] [CrossRef]
- Cutler, C.P.; Phillips, C.; Hazon, N.; Cramb, G. Cortisol Regulates Eel (Anguilla anguilla) Aquaporin 3 (AQP3) MRNA Expression Levels in Gill. Gen. Comp. Endocrinol. 2007, 152, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.V.; Bollinger, R.J.; Weber, H.M.; Madsen, S.S.; Tipsmark, C.K. Differential Expression and Localization of Branchial AQP1 and AQP3 in Japanese Medaka (Oryzias latipes). Cells 2019, 8, 422. [Google Scholar] [CrossRef]
- Giffard-Mena, I.; Boulo, V.; Aujoulat, F.; Fowden, H.; Castille, R.; Charmantier, G.; Cramb, G. Aquaporin Molecular Characterization in the Sea-Bass (Dicentrarchus labrax): The Effect of Salinity on AQP1 and AQP3 Expression. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 148, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Tipsmark, C.K.; Sørensen, K.J.; Madsen, S. Aquaporin Expression Dynamics in Osmoregulatory Tissues of Atlantic Salmon during Smoltification and Seawater Acclimation. J. Exp. Biol. 2010, 213, 368–379. [Google Scholar] [CrossRef]
- Moshtaghi, A.; Rahi, M.L.; Mather, P.B.; Hurwood, D.A. Understanding the Genomic Basis of Adaptive Response to Variable Osmotic Niches in Freshwater Prawns: A Comparative Intraspecific RNA-Seq Analysis of Macrobrachium australiense. J. Hered. 2017, 108, 544–552. [Google Scholar] [CrossRef]
- Towle, D.W.; Henry, R.P.; Terwilliger, N.B. Microarray-Detected Changes in Gene Expression in Gills of Green Crabs (Carcinus maenas) upon Dilution of Environmental Salinity. Comp. Biochem. Physiol. Part D Genom. Proteom. 2011, 6, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Parpura, V.; Wang, Y.-F. Astroglial Regulation of Magnocellular Neuroendocrine Cell Activities in the Supraoptic Nucleus. Neurochem. Res. 2021, 46, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Geering, K. Functional Roles of Na, K-ATPase Subunits. Curr. Opin. Nephrol. Hypertens. 2008, 17, 526–532. [Google Scholar] [CrossRef]
- Li, J.; Ma, P.; Liu, P.; Chen, P.; Li, J. The Roles of Na+/K+-ATPase α-Subunit Gene from the Ridgetail White Prawn Exopalaemon carinicauda in Response to Salinity Stresses. Fish Shellfish Immunol. 2015, 42, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Lovett, D.L.; Verzi, M.P.; Burgents, J.E.; Tanner, C.A.; Glomski, K.; Lee, J.J.; Towle, D.W. Expression Profiles of Na+, K+-ATPase during Acute and Chronic Hypo-Osmotic Stress in the Blue Crab Callinectes sapidus. Biol. Bull. 2006, 211, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, N.; Masui, D.; McNamara, J.; Leone, F.; Furriel, R. Long-Term Exposure of the Freshwater Shrimp Macrobrachium olfersii to Elevated Salinity: Effects on Gill (Na+, K+)-ATPase α-Subunit Expression and K+-Phosphatase Activity. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 146, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, R.; Weihrauch, D.; Lignot, J.; Towle, D. ATPase in Gills of the Blue Crab Callinectes sapidus: CDNA Sequencing and Salinit-Related Expression of Alpha Subunit MRNA and Protein; American Society of Zoologists: Lawrence, KS, USA, 2000; Volume 40, pp. 1166–1167. [Google Scholar]
- Sun, H.; Zhang, L.; Ren, C.; Chen, C.; Fan, S.; Xia, J.J.; Lin, H.; Hu, C. The Expression of Na, K-ATPase in Litopenaeus vannamei under Salinity Stress. Mar. Biol. Res. 2011, 7, 623–628. [Google Scholar] [CrossRef]
- Masui, D.C.; Mantelatto, F.L.; McNamara, J.C.; Furriel, R.P.; Leone, F.A. Na+, K+-ATPase Activity in Gill Microsomes from the Blue Crab, Callinectes danae, Acclimated to Low Salinity: Novel Perspectives on Ammonia Excretion. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 153, 141–148. [Google Scholar] [CrossRef]
- Wilder, M.N.; Atmomarsono, M.; Hien, T.T.T.; Phu, T.Q.; Yang, W.-J. Characterization of Na/K-ATPase in Macrobrachium rosenbergii and the Effects of Changing Salinity on Enzymatic Activity. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2000, 125, 377–388. [Google Scholar] [CrossRef]
- Bouaricha, N.; Thuet, P.; Charmantier, G.; Charmantier-Daures, M.; Trilles, J.-P. Na+-K+ ATPase and Carbonic Anhydrase Activities in Larvae, Postlarvae and Adults of the Shrimp Penaeus japonicus (Decapoda, Penaeidea). Comp. Biochem. Physiol. A Physiol. 1991, 100, 433–437. [Google Scholar] [CrossRef]
- Gilmour, K.M. New Insights into the Many Functions of Carbonic Anhydrase in Fish Gills. Respir. Physiol. Neurobiol. 2012, 184, 223–230. [Google Scholar] [CrossRef]
- Syrjänen, L.; Tolvanen, M.; Hilvo, M.; Olatubosun, A.; Innocenti, A.; Scozzafava, A.; Leppiniemi, J.; Niederhauser, B.; Hytönen, V.P.; Gorr, T.A. Characterization of the First Beta-Class Carbonic Anhydrase from an Arthropod (Drosophila melanogaster) and Phylogenetic Analysis of Beta-Class Carbonic Anhydrases in Invertebrates. BMC Biochem. 2010, 11, 28. [Google Scholar] [CrossRef]
- Henry, R.P.; Garrelts, E.E.; McCarty, M.M.; Towle, D.W. Differential Induction of Branchial Carbonic Anhydrase and Na+/K+ ATPase Activity in the Euryhaline Crab, Carcinus maenas, in Response to Low Salinity Exposure. J. Exp. Zool. 2002, 292, 595–603. [Google Scholar] [CrossRef]
- Henry, R.P.; Thomason, K.L.; Towle, D.W. Quantitative Changes in Branchial Carbonic Anhydrase Activity and Expression in the Euryhaline Green Crab, Carcinus maenas, in Response to Low Salinity Exposure. J. Exp. Zool. Part A Comp. Exp. Biol. 2006, 305, 842–850. [Google Scholar] [CrossRef]
- Liu, M.; Liu, S.; Hu, Y.; Pan, L. Cloning and Expression Analysis of Two Carbonic Anhydrase Genes in White Shrimp Litopenaeus vannamei, Induced by PH and Salinity Stresses. Aquaculture 2015, 448, 391–400. [Google Scholar] [CrossRef]
- Pongsomboon, S.; Udomlertpreecha, S.; Amparyup, P.; Wuthisuthimethavee, S.; Tassanakajon, A. Gene Expression and Activity of Carbonic Anhydrase in Salinity Stressed Penaeus monodon. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009, 152, 225–233. [Google Scholar] [CrossRef]
- Serrano, L.; Halanych, K.M.; Henry, R.P. Salinity-Stimulated Changes in Expression and Activity of Two Carbonic Anhydrase Isoforms in the Blue Crab Callinectes sapidus. J. Exp. Biol. 2007, 210, 2320–2332. [Google Scholar] [CrossRef]
- Serrano, L.; Henry, R.P. Differential Expression and Induction of Two Carbonic Anhydrase Isoforms in the Gills of the Euryhaline Green Crab, Carcinus maenas, in Response to Low Salinity. Comp. Biochem. Physiol. Part D Genom. Proteom. 2008, 3, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Sun, D.; Lv, J.; Ren, X.; Liu, P.; Li, J. Transcriptomic Analysis Provides Insight into the Mechanism of Salinity Adjustment in Swimming Crab Portunus trituberculatus. Genes Genom. 2019, 41, 961–971. [Google Scholar] [CrossRef]
- Toei, M.; Saum, R.; Forgac, M. Regulation and Isoform Function of the V-ATPases. Biochemistry 2010, 49, 4715–4723. [Google Scholar] [CrossRef] [PubMed]
- Beyenbach, K.W.; Wieczorek, H. The V-Type H+ ATPase: Molecular Structure and Function, Physiological Roles and Regulation. J. Exp. Biol. 2006, 209, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Nishi, T.; Forgac, M. The Vacuolar (H+)-ATPases—Nature’s Most Versatile Proton Pumps. Nat. Rev. Mol. Cell Biol. 2002, 3, 94–103. [Google Scholar] [CrossRef]
- Qi, T.; Liu, J.; Zhao, P.; Ge, B.; Liu, Q.; Jiang, S.; Wang, Z.; Zhang, H.; Tang, B.; Ding, G. A Novel Modulation of Physiological Regulation in Cultured Chinese Mitten Crab (Eriocheir japonica sinensis) in Response to Consistent Salinity Changes. Gene 2020, 756, 144914. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.-N.; Liu, Y.; Cai, D.-X.; Li, J.-Z.; Wang, A.-L. Two Types of ATPases from the Pacific White Shrimp, Litopenaeus vannamei in Response to Environmental Stress. Mol. Biol. Rep. 2012, 39, 6427–6438. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, J.; Wei, B.; Cheng, Y.; Zhang, L.; Zhen, X. Comparative Transcriptome Analysis Reveals Osmotic-Regulated Genes in the Gill of Chinese Mitten Crab (Eriocheir sinensis). PLoS ONE 2019, 14, e0210469. [Google Scholar] [CrossRef] [PubMed]
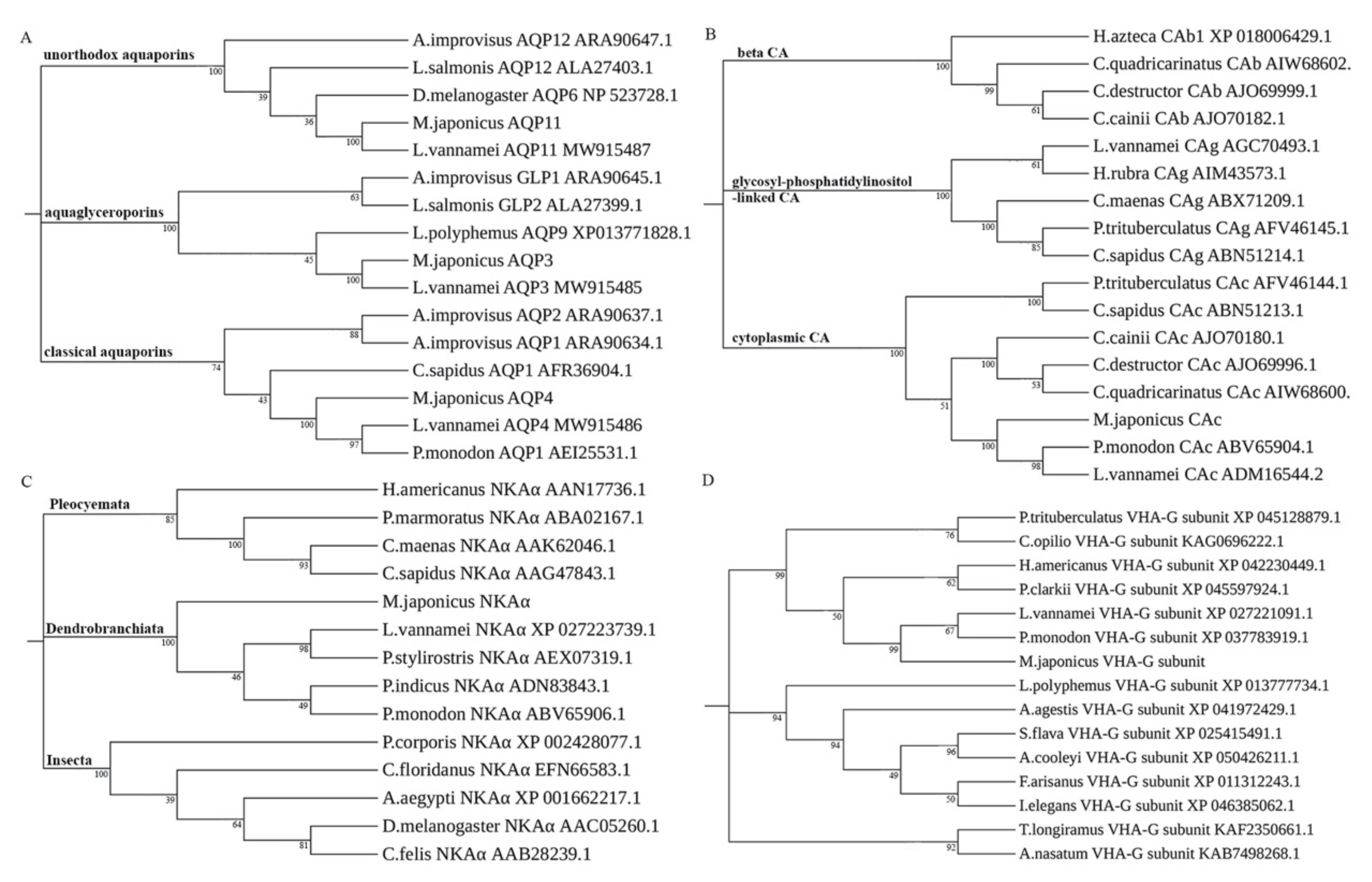
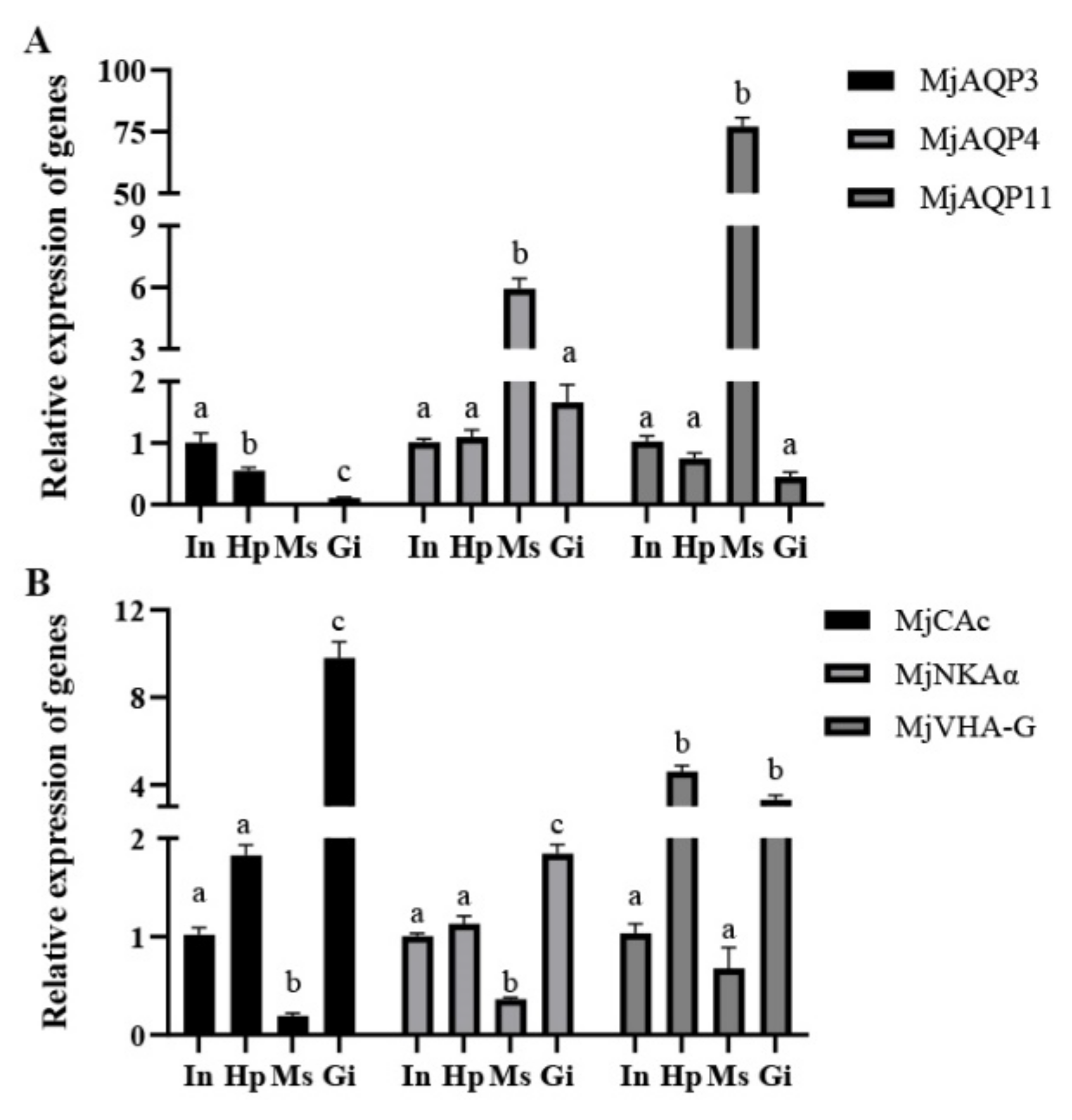
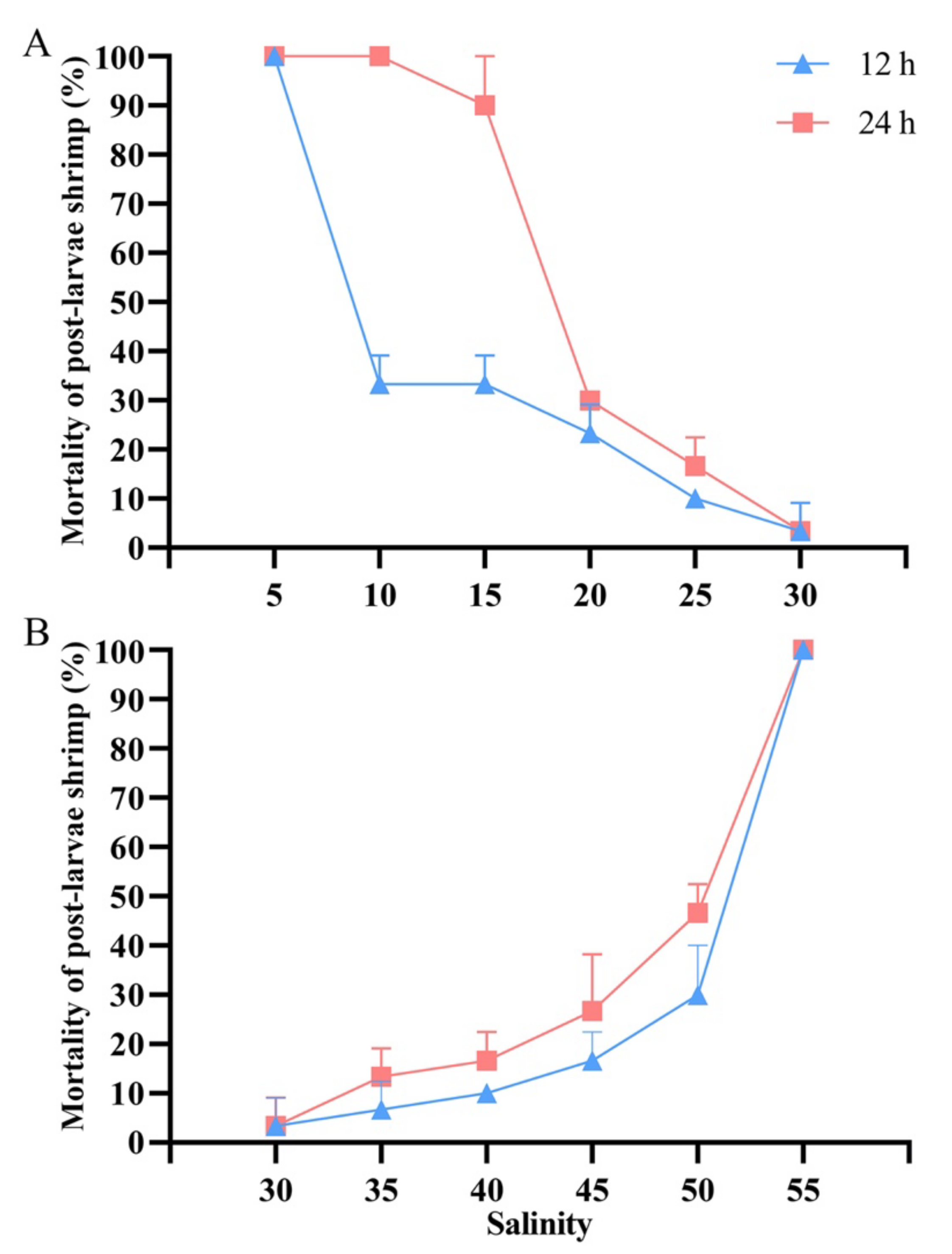
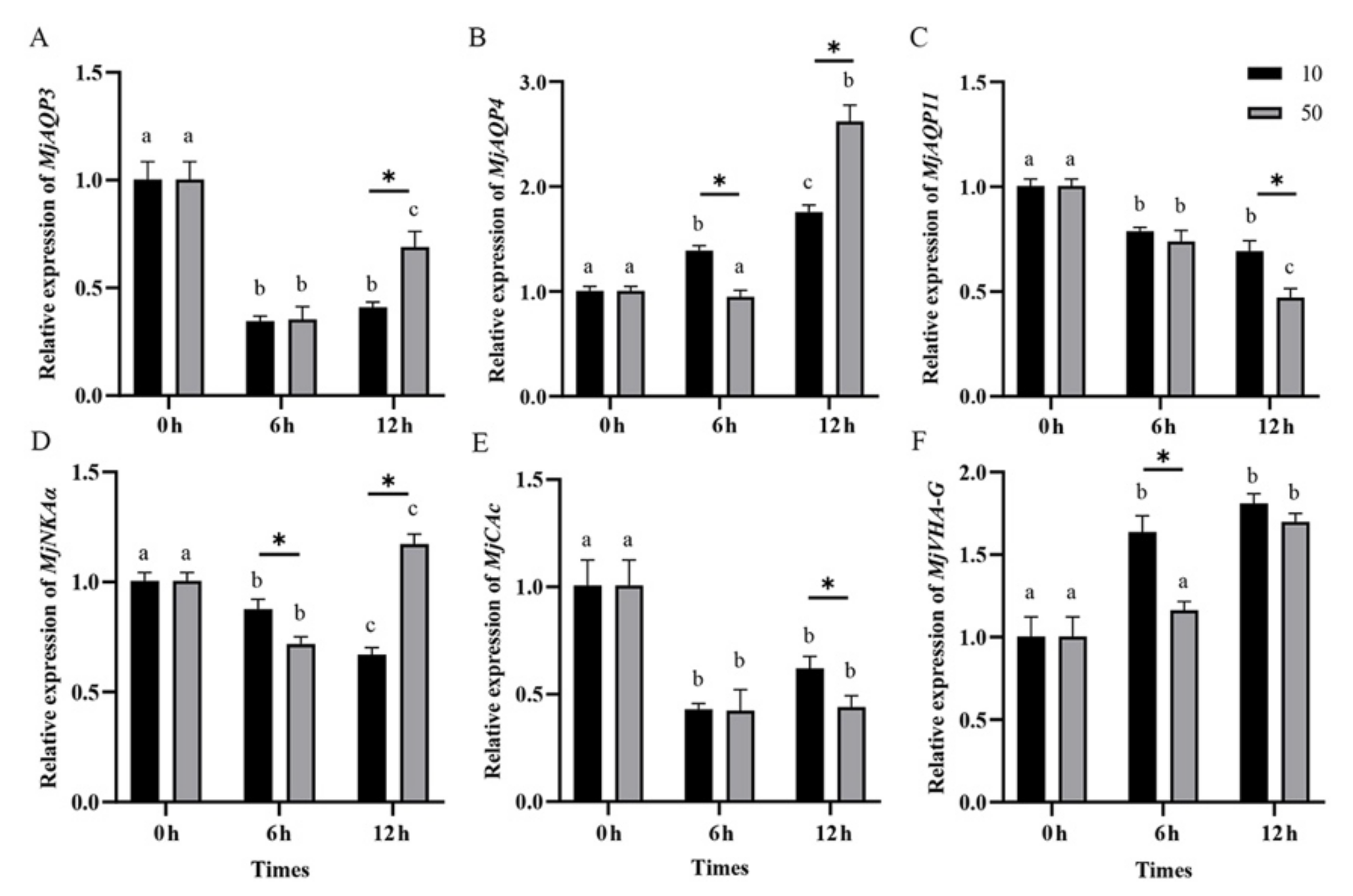
| Gene Name | Primer Type | Sequence (5′-3′) |
|---|---|---|
| MjAQP3 | Forward | AGTGTACAGGAACAAAGGCTTAC |
| Reverse | ACTACTCCTCCTTAGTCGCTCTC | |
| MjAQP4 | Forward | GAAGAGCAAGAAACCACCATC |
| Reverse | CCGAGCCAGTAGATCTTTCAT | |
| MjAQP11 | Forward | CATCTCCGCAACCATGTCGAT |
| Reverse | TGTGACACTACCGAAAATCCTC | |
| MjNKAα | Forward | GCCTAGTGGCTTGTACATAAGTG |
| Reverse | TGAGCTCTAGTAATTCTTGCGC | |
| MjCAc | Forward | GACAGTTAAACATGGTTGGCTG |
| Reverse | GAGTGTCACTTATTTCTCTGGTCA | |
| MjVHA-G | Forward | ACAAGACACACACAGCAGACG |
| Reverse | CAGTTCTACACACTTATCATGGAGT |
| Gene Name | Primer Type | Sequence (5′-3′) | Melting Temperature | Efficiency |
|---|---|---|---|---|
| MjAQP3 | Forward | ACCTTGGGTGTGCTCGTATCT | 86.0 °C | 97.23% |
| Reverse | TGGGCAATAACATACACGGG | |||
| MjAQP4 | Forward | TGGCTGCTGCTCTTATCTACTCC | 81.0 °C | 95.36% |
| Reverse | CGTTGTTGGTTCGCTTATTGTA | |||
| MjAQP11 | Forward | CGACCTCCAAGTGCCTGTAG | 85.0 °C | 96.53% |
| Reverse | AAGCACGGGATTGAAGTAACC | |||
| MjNKAα | Forward | ACCCGCCGTAACTCTATTGTC | 82.5 °C | 96.97% |
| Reverse | TGCCTGGGGTGTAGGAAAG | |||
| MjCAc | Forward | ACATTCATGGAAGGCTCAGGT | 84.0 °C | 95.67% |
| Reverse | AGTAGCAGCGACCATTGACG | |||
| MjVHA-G | Forward | TGAATAAGCAGGTTGCCCAC | 81.5 °C | 96.78% |
| Reverse | CCTTCTTAGCGTTGGTGTGC | |||
| EF1α | Forward | GGAACTGGAGGCAGGACC | 85.0 °C | 98.98% |
| Reverse | AGCCACCGTTTGCTTCAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, Z.; Wang, Z.; Deng, Z.; Zhao, R.; Sun, J.; Hao, P.; Zhang, L.; Wang, X.; Liu, F.; et al. How Do Gene Expression Patterns Change in Response to Osmotic Stresses in Kuruma Shrimp (Marsupenaeus japonicus)? J. Mar. Sci. Eng. 2022, 10, 1870. https://doi.org/10.3390/jmse10121870
Li Y, Zhang Z, Wang Z, Deng Z, Zhao R, Sun J, Hao P, Zhang L, Wang X, Liu F, et al. How Do Gene Expression Patterns Change in Response to Osmotic Stresses in Kuruma Shrimp (Marsupenaeus japonicus)? Journal of Marine Science and Engineering. 2022; 10(12):1870. https://doi.org/10.3390/jmse10121870
Chicago/Turabian StyleLi, Yuquan, Zhihao Zhang, Zhongkai Wang, Zhitong Deng, Ruiyang Zhao, Jinfeng Sun, Pengyuan Hao, Long Zhang, Xiaofan Wang, Fei Liu, and et al. 2022. "How Do Gene Expression Patterns Change in Response to Osmotic Stresses in Kuruma Shrimp (Marsupenaeus japonicus)?" Journal of Marine Science and Engineering 10, no. 12: 1870. https://doi.org/10.3390/jmse10121870
APA StyleLi, Y., Zhang, Z., Wang, Z., Deng, Z., Zhao, R., Sun, J., Hao, P., Zhang, L., Wang, X., Liu, F., Wang, R., & Cui, Y. (2022). How Do Gene Expression Patterns Change in Response to Osmotic Stresses in Kuruma Shrimp (Marsupenaeus japonicus)? Journal of Marine Science and Engineering, 10(12), 1870. https://doi.org/10.3390/jmse10121870







