Abstract
Two deep-sea eels collected from the East Mariana Basin in the western Pacific Ocean are described in this study. Based on their morphological features, two eel specimens were assumed to belong to the Gray cutthroat eel family, Synaphobranchidae. Mitochondrial DNA (mtDNA) genes have been widely used as genetic markers to identify fish species. To accurately identify the species of the two eel specimens, we sequenced the mitochondrial cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA (16S rRNA) genes from the two eel specimens. The sequences from the specimens were 100% identical. The molecular phylogenetic tree confirmed that the two eel specimens were closely related to Synaphobranchus affinis with a bootstrap value of 100%. This is the first study to report new records of S. affinis from the East Mariana Basin in the western Pacific Ocean.
1. Introduction
The deep sea (>200 m depth) is the Earth’s largest ecosystem and a largely unexplored deep marine biosphere [1,2]. Deep-sea environments are extremely harsh, and deep-sea organisms survive with scarce food resources and under extreme pressure, cold, and constant darkness [3,4]. Deep-sea fishes are critical components of deep-water ecosystems, and their habitat range varies above a depth of 8200 m [5,6,7]. The identification and characterization of deep-sea fish species are important for understanding deep-sea biodiversity [8,9,10]. However, there is little information about deep-sea fish species because deep-ocean environments are difficult to access for collecting biological samples.
The eel family Synaphobranchidae is an important ecological component of the deep-sea fauna and is globally found in tropical and temperate seas [11]. In particular, Synaphobranchidae is widely distributed in the vertical ocean column from less than 100 m to several thousand meters [12]. The Synaphobranchidae family currently comprises 12 genera (about 40 species) [13,14]. Among them, the cutthroat eel genus Synaphobranchus belongs to the family Synaphobranchidae. Currently, Synaphobranchus eels include six valid species [13]: S. kaupii Johnson, 1862 (Kaup’s arrowtooth eel) [15]; S. affinis Günther, 1877 (Gray cutthroat) [16]; S. brevidorsalis Günther, 1887 (Shortdorsal cutthroat eel) [17]; S. dolichorhynchus (Lea, 1913) [18]; S. oregoni Castle, 1961 [19]; and S. calvus Melo, 2007 [20].
To date, all species of this genus have been recorded in the North Atlantic Ocean, except for S. calvus [11,18]. In particular, S. affinis has been recorded in the Atlantic (western North Atlantic, eastern South Atlantic, eastern Atlantic, and eastern-central Atlantic), Pacific (western North Pacific, western South Pacific, eastern North Pacific, western South Pacific) and Indian Oceans (southwestern Indian Ocean and eastern Indian Ocean) [1,21,22]. However, there was no evidence of the presence of S. affinis from the East Mariana Basin in the western Pacific Ocean so far. Nevertheless, we collected two cutthroat eels from off Mariana Basin in the western Pacific and identified them as S. affinis, based both on morphological and molecular characteristics.
Recently, DNA-based markers, especially mtDNA markers, have been extensively studied in fish taxonomy and phylogenetics. Mitochondrial COI and 16S rRNA genes are good genetic markers for identifying and differentiating deep-sea species, including fish [23,24,25]. DNA barcoding is used for the species identification of some deep-sea species, such as the squat lobsters, Munida distiza and M. militaris [26], and sea cucumber, Benthodytes marianensis [27]. Indeed, species identification of two deep-sea eels (Bassozetus sp. and Synaphobranchus sp.) has been described based on mitochondrial COI and 16S rRNA genes [28]. In addition, mtDNA genes were used to describe the molecular phylogenetics and evolution of 77 deep-sea fishes [29], indicating that DNA barcoding is very effective in identifying deep-sea species, including deep-sea eels. Therefore, mtDNA markers were considered a useful tool for the accurate identification of the two eel specimens from the East Mariana Basin in the western Pacific Ocean.
This study represents the first reliable record of S. affinis from the Mariana Basin. It provides species identification based on mitochondrial cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA (16S rRNA) sequences and brief morphological information.
2. Materials and Methods
2.1. Sampling Collection
During an oceanographic cruise of the Korea Institute of Ocean Science and Technology (KIOST) to the western Pacific Ocean in May 2020, two eel specimens were collected in a baited trap (composed of meat and fish) from different areas. The sampling areas are shown in Figure 1 and Table 1.
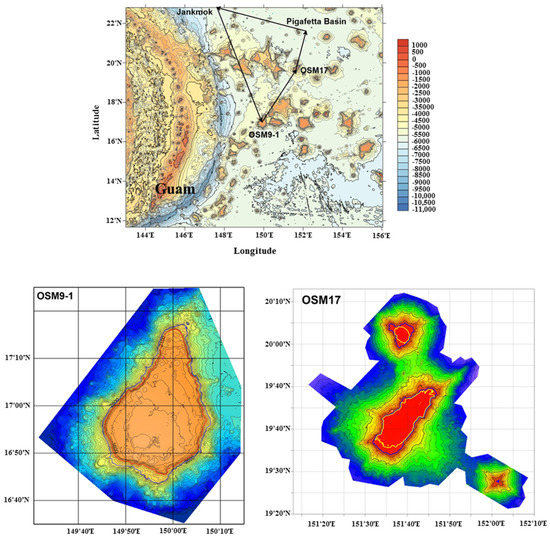
Figure 1.
Map of the western Pacific Ocean and geographic distribution of sampling sites.

Table 1.
List of samples and collection locations.
The pictures of the two eel specimens (sample IDs: BT02 and BT05) are shown in Figure 2. This study did not include live fish, and the samples had naturally died when they were collected. The dead fish were preserved and stored at −20 °C until morphological analysis and DNA isolation. For taxonomical examination, they were fixed in 10% formalin and transferred into 75% ethyl alcohol for long-term preservation. Counts, measurements, and terminology of the cephalic lateral canal were performed following the guideline reported in previous studies [11,30]. Vertebral count was based on radiographs (Softex, CMB-2, Japan). Morphometric data are presented in Table 2 and Table 3.

Figure 2.
Picture of the two deep-sea Synaphobranchus eels.

Table 2.
Morphometric characters of the two eel specimens collected from the East Mariana Basin, western Pacific Ocean.

Table 3.
Meristic characters of the two eel specimens collected from the East Mariana Basin, western Pacific Ocean. Data based on specimens listed in Table 2.
2.2. DNA Extraction, PCR Amplification and DNA Sequencing
Genomic DNA was extracted from the muscle with DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. The quantity and quality of the isolated DNA were analyzed and measured at 230, 260, and 280 nm using a spectrophotometer (NanoDrop One, Thermo Fisher Scientific Inc., Madison, WI, USA). The PCR amplification was carried out in a 50 μL reaction mixture containing 32.875 μL of sterilized distilled water, 6 μL of 10X Ex Taq Buffer (TaKaRa, Japan), 5 μL of dNTP mixture (2.5 mM each), 1 μL of each primer (5 μM), 0.125 μL of EX Taq DNA polymerase (5 units/μL), and 4 μL of DNA template. Two mitochondrial DNA genes (COI and 16S rDNA), as the barcoding marker genes for animals, were amplified with forward and reverse primers (Table 4) [31]. PCR cycling was performed using a thermal cycler PCR machine (C1000 Touch Thermal Cycler, Bio-Rad). The amplification conditions were as follows: initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing for 30 s at 65 °C, and extension at 72 °C for 45 s. The final extension was performed at 72 °C for 5 min. PCR products were confirmed by 1.0% agarose gel electrophoresis and visualized using FluoroBox (Blue LED Gel doc, NeoScience, Gyeonngi-do, Seoul, South Korea). All PCR products were sequenced by Macrogen (Seoul, South Korea).

Table 4.
Primer set used in this study.
2.3. Sequence Alignment and Phylogenetic Analysis
To determine the phylogenetic relationships of the two eel species, mitochondrial COI and 16S rRNA sequences from the eel family Synaphobranchidae (genus: Simenchelys, llyophis, Histiobranchus, Dysomma, Dysommina, Diastobrachus, and Synaphobranchus) were obtained from GenBank database. A total of 30 and 12 sequences for COI and 16S rRNA were selected and aligned using ‘MAFFT’ v7.450 with the default setting [32]. The most suitable models for each sequence were estimated using a Modeltest-NG [33] in raxmlGUI 2.0 software for Linux [34]. According to the Akaike information criterion (AIC), the best-fit model STMTREV + I + G4 + F of amino-acid and GTR + I of nucleotide substitution were selected for COI and 16S rRNA sequences, respectively. Based on the best-fit model for each sequence, a maximum likelihood (ML) phylogenetic tree was constructed using RAxML-NG in raxmlGUI 2.0 software with 1000 bootstrap replicates and inferring the best scoring tree [35]. The Simenchelys species in the subfamily Simenchelyinae was used as an outgroup, as previously described [36,37].
3. Results and Discussion
The specimens collected from the Mariana Basin were assigned as a member of the genus Synaphobranchus based on the presence of two of the following features: gill slits confluent along the ventral midline; a dorsal fin originating from far behind the tip of the pectoral fin; 12 pores in the preoperculo-mandibular canal, 6 in the supraorbital branch of the supraorbital canal, and 2 in the supratemporal cross-commissure canal; the absence of temporal canal pore series; a pectoral fin longer than one-half of the gape length; interspace between the posterior margin of the orbit and a first trunk lateral-line pore shorter than the snout length; completed in paving stone array; a uniserial vomerine tooth patch, except irregularly biserial only in the anterior portion; a premaxillary-ethmoid tooth patch that is short and oval with teeth irregularly placed; and lateral-line pores to vertical of anus 29–32 [11].
The specimens used in this study are most similar to S. affinis among the six valid species of the genus Synaphobranchus, based on the sharing of the diagnostic characters of the discriminating species, such as the elongated oval scales on the body, 135–139 vertebral numbers, and uniserial vomerine teeth (Figure 3 and Table 2 and Table 3). In addition, they are easily differentiated from both S. calvus and S. oligolepis (without scales) due to the presence of a scaly head, and from both S. brevidorsalis (oval) and S. oregoni (rounded) because of the elongated oval scales on the body. The present specimens, in accordance with that of S. kaupii, show a dorsal fin origin located well behind from a vertical at the vent. In addition, the vertebral number of S. kaupii (143–153) is higher than that of S. affinis (128–140). However, Synaphobranchus eels are difficult to distinguish at the species levels because of the broad range of vertebral values and similar external morphologies [38,39]. To date, morphological characteristics have been used as basic tools for identifying species and distinguishing among them. However, distinguishing between similar species and identifying new species based on morphological characteristics is challenging because of the diversity in morphological characteristics and the ontogenetic changes in morphology during fish development [23,40,41]. Therefore, although our two eels are closely related to S. affinis, it is difficult to accurately identify the species solely based on morphological characteristics.
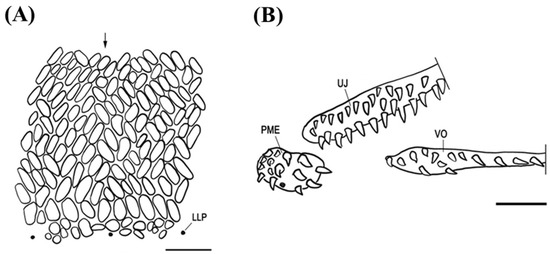
Figure 3.
Scale pattern (A) below origin of dorsal fin (arrow) and teeth (B) on premaxillary-ethmoid maxillary (PME), upper jaw (UJ), and vomer (VO) of Synaphobranchus affinis, BT05, 1023 mm in total length. LLP, lateral line pore. Posterior parts of UJ and VO omitted. Scales bars indicate 5 mm.
In this study, the mitochondrial COI and 16S rRNA genes of the two Synaphobranchus eels were 655 base pairs (bp) and 611 bp in length, respectively. The partial mtDNA COI and 16S rRNA gene sequences were sequenced and deposited at GenBank (BT02 [COI; MZ221136 and16S rRNA; MZ227814] and BT05 [COI; MZ221137 and16S rRNA; MZ227815]). The mitochondrial COI and 16S rRNA sequences from the two eel specimens showed 100% identity, indicating that the specimens were from the same species. Based on sequence analysis of the amplified mitochondrial COI and 16S, molecular phylogenetic trees were constructed according using the maximum likelihood (ML) (Figure 4).
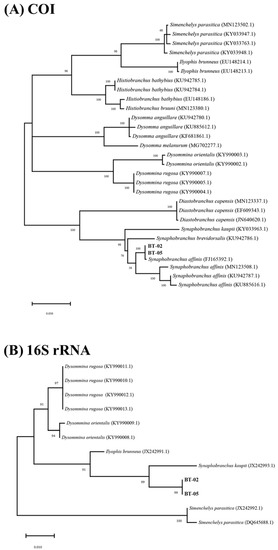
Figure 4.
Phylogenic tree based on (A) mitochondrial CO1 and (B) 16S rRNA gene sequences using RAxML-NG in raxmlGUI 2.0 software.
The two eel specimens genetically belonged to the genus Synaphobranchus, based on the mitochondrial COI and 16S rRNA sequences. The phylogenetic tree of mitochondrial COI and 16S rRNA showed that the two eel species were deeply nested within Synaphobranchus eels. The phylogenetic tree of the mitochondrial COI showed that the two eel specimens were closely related to S. affinis with a 100% bootstrap value. The sequences from the two eel specimens converged with the one branch containing one individual S. affinis. Despite the non-monophyletic relationship among the four S. affinis, the two specimens (e.g., BT02 and BT05) clearly formed a separate clade from S. brevidorsalis, suggesting that the two newly sampled species are congeneric species to S. affinis. Taken together, supported by the phylogenetic tree of mitochondrial COI and 16S rRNA, the two eel specimens are S. affinis.
In summary, S. affinis was reported for the first time from the East Mariana Basin in the western Pacific Ocean based on molecular and morphological characteristics. The mitochondrial COI and 16S rRNA sequences were sufficient to identify the Synaphobranchus species collected from the East Mariana Basin in the western Pacific Ocean, supporting the usefulness of the mtDNA-based method in fish species identification. This study provides insights into the deep-sea fish species diversity in the western Pacific Ocean. However, replicate samples from the western Pacific Ocean are needed for accurate species identification and a better understanding of species distribution.
Author Contributions
Data curation, formal analysis, writing—original draft, J.H.; investigation, H.-J.K. and B.-J.K.; project administration, funding acquisition, K.H. and C.N.; conceptualization, formal analysis, writing—original draft, Y.-U.C. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the KIOST projects “Acquisition of marine bioresources from the high seas and screening of biological potency” (PM62830).
Institutional Review Board Statement
All experiments were conducted in compliance with the guidelines of the Institutional Animal Care and Experimental Committee of Korea Institute of Ocean Science and Technology (KIOST)“ that approved the experimental protocol.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are available via the data repository of the KIOST. Requests for material should be made to the corresponding author.
Acknowledgments
The authors would like to thank the support of KIOST projects “Acquisition of marine bioresources from the high seas and screening of biological potency” (PM62830). Finally, we thank the editor and the anonymous reviewers whose comments greatly improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ramirez-Llodra, E.; Brandt, A.; Danovaro, R.; Mol, B.D.; Escobar, E.; German, C.R.; Levin, L.A.; Arbizu, P.M.; Menot, L.; Buhl-Mortensen, P.; et al. Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Rex, M.A. Community structure in the deep-sea Benthos. Annu. Rev. Ecol. Syst. 1981, 12, 331–353. [Google Scholar] [CrossRef]
- Robison, B.H. Deep pelagic biology. J. Exp. Mar. Biol. Ecol. 2004, 300, 253–272. [Google Scholar] [CrossRef]
- Sanders, H.L.; Hessler, R.R. Ecology of the deep-sea benthos. Science 1969, 163, 1419–1424. [Google Scholar] [CrossRef]
- Drazen, J.C.; Sutton, T.T. Dining in the Deep: The Feeding Ecology of Deep-Sea Fishes. Annu. Rev. Mar. Sci. 2017, 9, 337–366. [Google Scholar] [CrossRef]
- Linley, T.D.; Gerringer, M.E.; Yancey, P.H.; Drazen, J.C.; Weinstock, C.L.; Jamieson, A.J. Fishesof the hadal zone including new species, in situ observations and depth records of Liparidae. Deep Sea Res. Part I Oceanogr. Res. Pap. 2016, 114, 99–110. [Google Scholar] [CrossRef]
- Orlov, A.M.; Tokranov, A.M. Checklist of deep-sea fishes of the Russian northwestern Pacific Ocean found at depths below 1000 m. Prog. Oceanogr. 2019, 176, 102143. [Google Scholar] [CrossRef]
- Austen, G.E.; Bindemann, M.; Griffiths, R.A.; Roberts, D.L. Species identification by experts and non-experts: Comparing images from field guides. Sci. Rep. 2016, 20, 33634. [Google Scholar] [CrossRef]
- Elphick, C.S. How you count counts: The importance of methods research in applied ecology. J. Appl. Ecol. 2008, 45, 1313–1320. [Google Scholar] [CrossRef]
- Farnsworth, E.J.; Chu, M.; Kress, W.J.; Neill, A.K.; Best, J.H.; Pickering, J.; Stevenson, R.D.; Courtney, G.W.; VanDyk, J.K.; Ellison, A.M. Next-Generation Field Guides. Bioscience 2013, 63, 891–899. [Google Scholar] [CrossRef]
- Sulak, K.J.; Shcherbachev, Y.N. Zoogeography and systematics of six deep-living genera of synaphobranchid eels, with a key to taxa and description of two new species of Ilyophis. Bull. Mar. Sci. 1997, 60, 1158–1194. [Google Scholar]
- Priede, I.G.; Froese, R. Colonization of the deep sea by fishes. J. Fish Biol. 2013, 83, 1528–1550. [Google Scholar] [CrossRef]
- Eschmeyer, W.N. Catalog of Fishes. Electronic Version. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 4 April 2018).
- Ho, H.-C.; Smith, D.G.; McCosker, J.E.; Hibino, Y.; Loh, K.-H.; Tighe, K.A.; Shao, K.-T. Annotated checklist of eels (orders Anguilliformes and Saccopharyngiformes) from Taiwan. Zootaxa 2015, 4060, 140–189. [Google Scholar] [CrossRef]
- Johnson, J.Y. Descriptions of some new genera and species of fishes obtained at Madeira. Proc. Zool. Soc. Lond. 1862, 30, 167–180. [Google Scholar]
- Günther, A. Preliminary notes on new fishes collected in Japan during the Expedition of H.M.S. “Challenger”. Ann. Mag. Nat. Hist. 1877, 20, 433–446. [Google Scholar] [CrossRef]
- Günther, A. Report on the deep-sea fishes collected by H.M.S. Challenger during the years 1873–1876. Zoology 1887, 5, 1–335. [Google Scholar]
- Lea, E. Muraenoid Larvae: From the “Michael Sars” North Atlantic Deep-Sea Expedition, 1910. Rep. Sci. Res. “Michael Sars” N. Atl. Deep-Sea Exped. 1913, 3, 1–59. [Google Scholar]
- Castle, P.H.J. Deep-Water Eels from Cook Strait, New Zealand; Zoology Publications from Victoria University Wellington: Wellington, New Zealand, 1961; Volume 27, pp. 1–30. [Google Scholar]
- Melo, M.R.S. A new synaphobranchid eel (Anguilliformes: Synaphobranchidae) from Brazil, with comments on the species from the Western South Atlantic. Copeia 2007, 2, 315–323. [Google Scholar] [CrossRef]
- Almeida, A.J.; Biscoito, M. New records of Synaphobranchus (Anguilliformes, Synaphobranchidae) from the Azores, (eastern Atlantic Ocean). Cybium 2007, 31, 391–392. [Google Scholar]
- Almeida, A.J.; Biscoito, M.; Santana, J.I.; González, J.A. New records of grey cutthroat, Synaphobranchus affinis (Actinopterygii: Anguilliformes: Synaphobranchidae), from the eastern-central Atlantic Ocean. Acta Ichthyol. Piscat. 2010, 40, 66–70. [Google Scholar] [CrossRef]
- Zhang, J.B.; Hanner, R. DNA barcoding is a useful tool for the identification of marine fishes from Japan. Biochem. Syst. Ecol. 2011, 39, 31–42. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D.N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zhang, H.; Liu, J.; Niu, S.; Xiao, Y.; Chen, Y. DNA barcoding of the family Sparidae along the coast of China and revelation of potential cryptic diversity in the Indo-West Pacific oceans based on COI and 16S rRNA genes. J. Oceanol. Limnol. 2018, 36, 1753–1770. [Google Scholar] [CrossRef]
- Cabezas, P.; Lin, C.W.; Chan, T.Y. Two new species of the deep-sea squat lobster genus Munida Leach, 1820 (Crustacea: Decapoda: Munididae) from Taiwan: Morphological and molecular evidence. Zootaxa 2011, 3036, 26–38. [Google Scholar] [CrossRef]
- Mu, W.; Liu, J.; Zhang, H. Complete mitochondrial genome of Benthodytes marianensis (Holothuroidea: Elasipodida: Psychropotidae): Insight into deep sea adaptation in the sea cucumber. PLoS ONE 2018, 13, e0208051. [Google Scholar] [CrossRef]
- Kim, H.-J.; Han, J.; Kim, B.-J.; Lee, K.-W.; Hyeong, K.; Choi, Y.-U. New records of two deep-sea eels collected from the Western Pacific Ocean based on COI and 16S rRNA genes. Mol. Biol. Rep. 2021, 48, 5795–5801. [Google Scholar] [CrossRef]
- Shen, Y.; Hubert, N.; Huang, Y.; Wang, X.; Gan, X.; Peng, Z.; He, S. DNA barcoding the ichthyofauna of the Yangtze River: Insights from the molecular inventory of a mega-diverse temperate fauna. Mol. Ecol. Resour. 2019, 19, 1278–1291. [Google Scholar] [CrossRef]
- Böhlke, E.B. Fishes of the Western North Atlantic. Number 1. Memoirs of the Sears Foundation of Marine Research Part 9. In Orders Anguilliformes and Saccopharyngiformes; Yale University Press: New Haven, CT, USA, 1989; Volume 1. [Google Scholar]
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Tang, K.L.; Fielitz, C. Phylogeny of moray eels (Anguilliformes: Muraenidae), with a revised classification of true eels (Teleostei: Elopomorpha: Anguilliformes). Mitochondrial DNA 2013, 24, 55–66. [Google Scholar] [CrossRef]
- Zhang, K.; Zhu, K.; Liu, Y.; Zhang, H.; Gong, L.; Jiang, L.; Liu, L.; Lü, Z.; Liu, B. Novel gene rearrangement in the mitochondrial genome of Muraenesox cinereus and the phylogenetic relationship of Anguilliformes. Sci. Rep. 2021, 11, 2411. [Google Scholar] [CrossRef]
- Robins, C.H. The Comparative Morphology of the Synaphobranchid Eels of the Straits of Florida. Proc. Acad. Nat. Sci. Phila. 1971, 123, 153–204. [Google Scholar]
- Smith, M.A.; Wood, D.M.; Janzen, D.H.; Hallwachs, W.; Hebert, P.D.N. DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. Proc. Natl. Acad. Sci. USA 2007, 104, 4967–4972. [Google Scholar] [CrossRef]
- Matarese, A.C.; Spies, I.B.; Busby, M.S.; Orr, J.W. Early larvae of Zesticelus profundorum (family Cottidae) identified using DNA barcoding. Ichthyol. Res. 2011, 58, 170–174. [Google Scholar] [CrossRef]
- Triantafyllidis, A.; Bobori, D.; Koliamitra, C.; Gbandi, E.; Mpanti, M.; Petriki, O.; Karaiskou, N. DNA barcoding analysis of fish species diversity in four north Greek lakes. Mitochondrial DNA 2011, 22, 37–42. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).