Locating QTL Controlling the Yield-Related Traits in Perennial Chinese Rice “Shendao3#”
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotyping Experiment
2.3. Trait Measurement
2.4. Data Analysis
2.5. Linkage Map and QTL Analysis
3. Results
3.1. Phenotype of 16 Yield-Related Traits of SD3# in Both MC and RC of 2024
3.2. Correlations of 16 Yield-Related Traits of SD3# in Both MC and RC of 2024
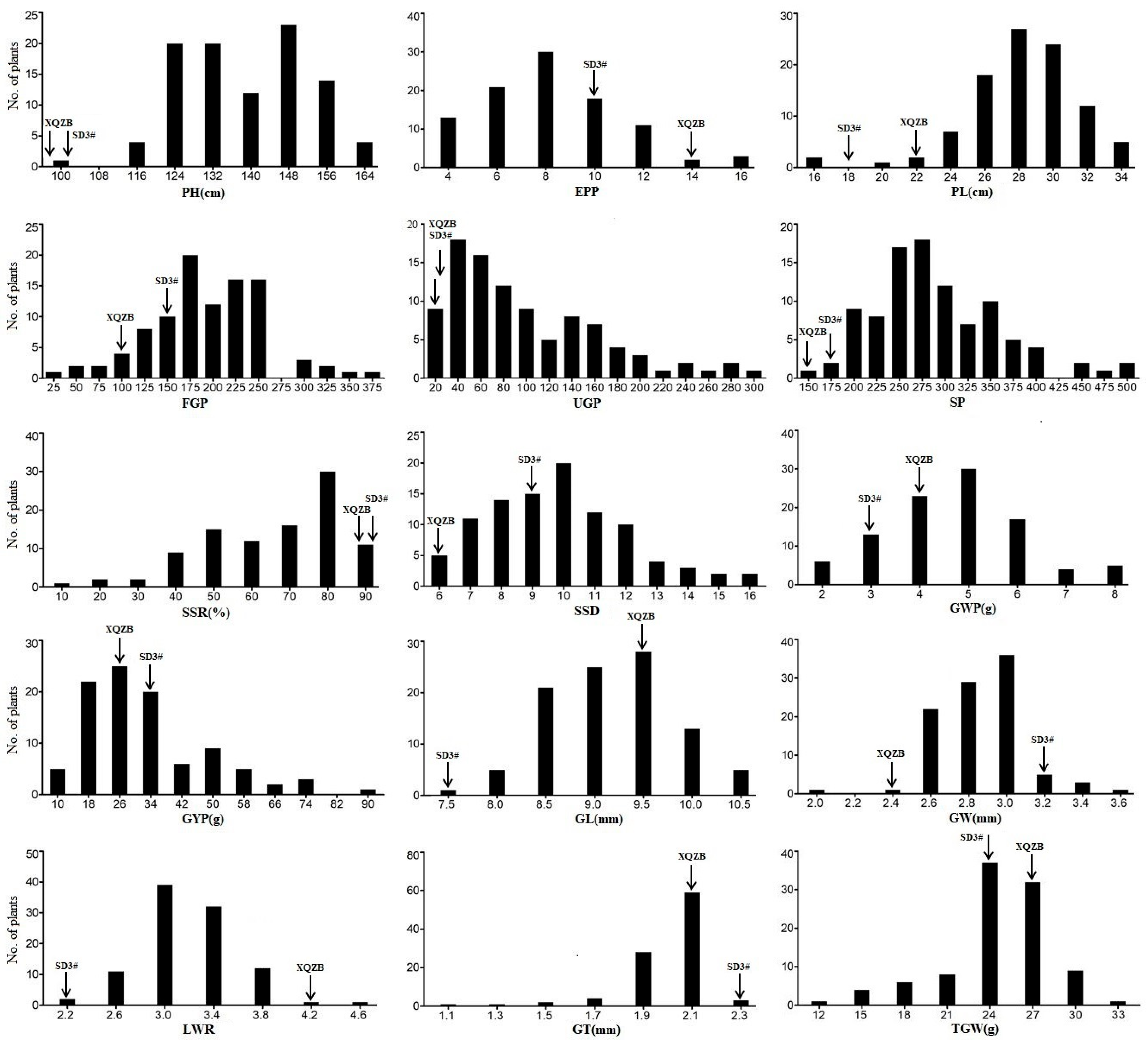
3.3. Phenotypic Variation for Yield-Related Traits in SD3#-Population and Its Bi-Parents
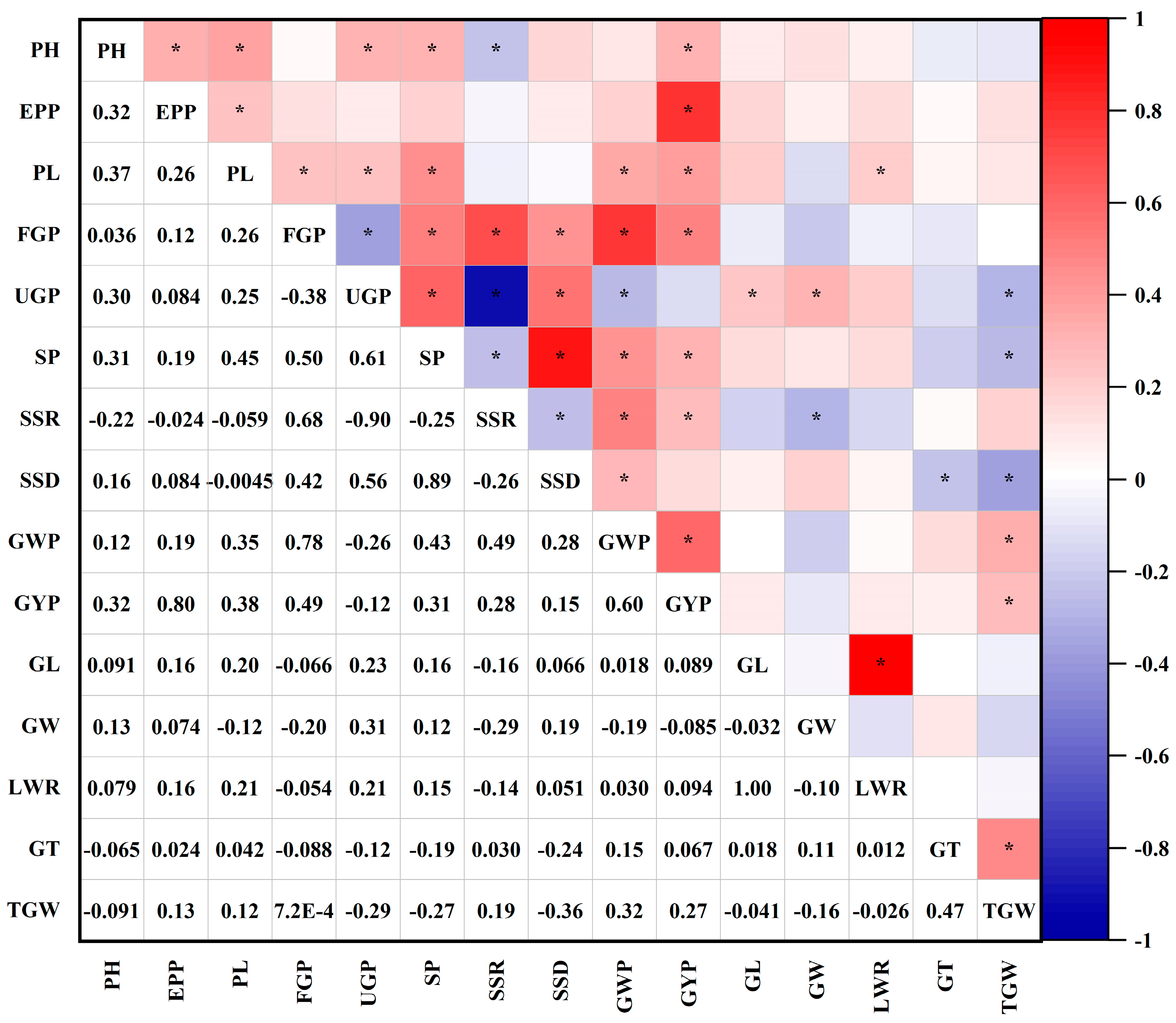
3.4. Correlation of the 15 Yield-Related Traits in the SD3#-Population
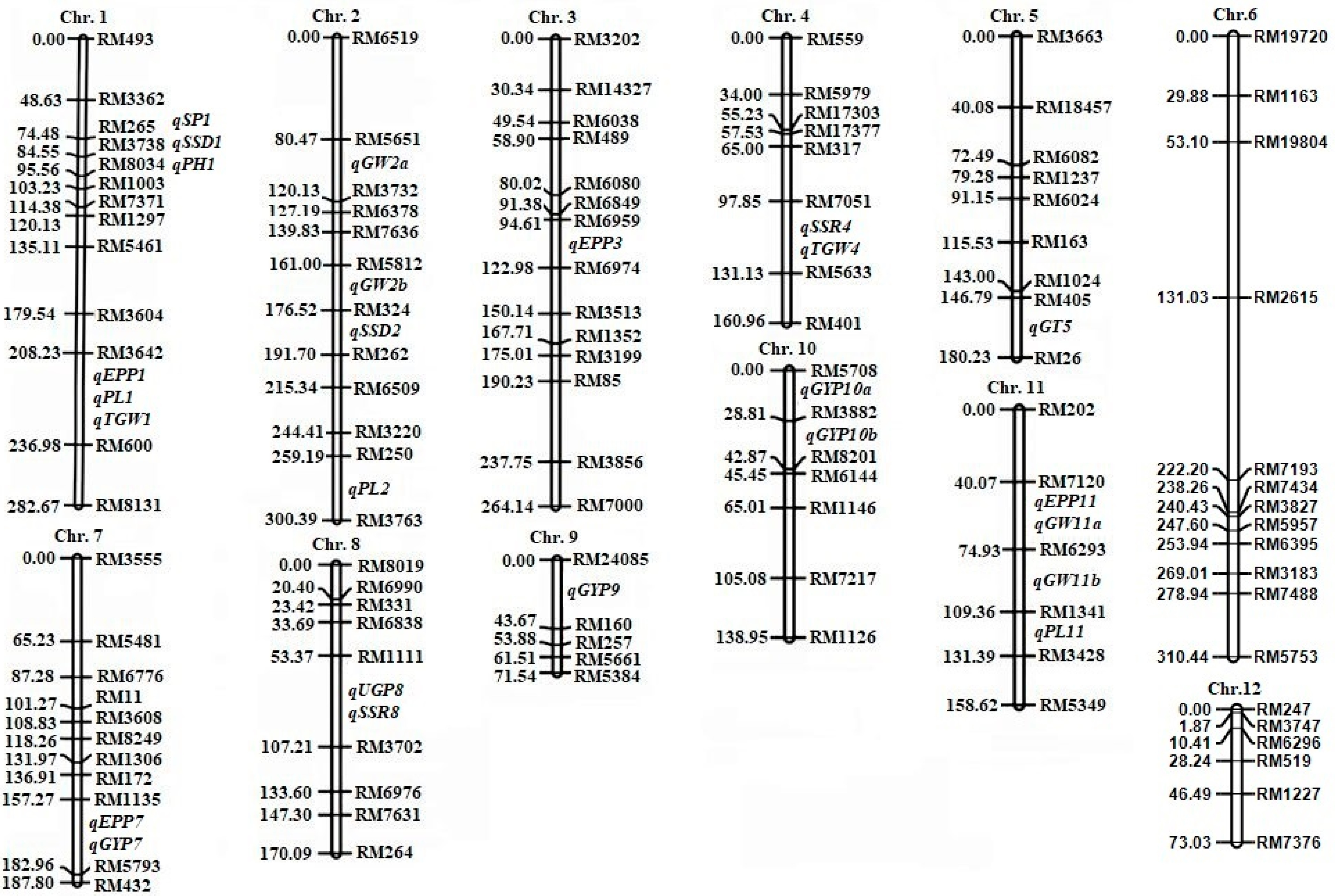
3.5. QTLs for Yield-Related Traits
3.6. Pleiotropic QTLs for Yield-Related Traits
3.7. Digenic Epistatic QTLs for Yield-Related Traits
4. Discussion
4.1. The Significance of the Research on Perennial Rice Germplasm
4.2. Comparison with Previous QTL Mapping
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Takahashi, Y.; Shomura, A.; Sasaki, T.; Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes a subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 2001, 98, 7922–7927. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Ashikari, M.; Ueguchi-Tanaka, M.; Itoh, H.; Nishimura, A.; Swapan, D.; Ishiyama, K.; Saito, T.; Kobayashi, M.; Khush, G.S.; et al. Green revolution: A mutant gibberellin synthesis gene in rice. Nature 2002, 416, 701–702. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Qian, Q.; Fu, Z.M.; Wang, Y.H.; Xiong, G.S.; Zeng, D.L.; Wang, X.Q.; Liu, X.F.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.Y.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- He, G.M.; Luo, X.J.; Tian, F.; Li, K.G.; Zhu, Z.F.; Su, W.; Qian, X.Y.; Fu, Y.C.; Wang, X.K.; Sun, C.Q.; et al. Haplotype variation in structure and expression of a gene cluster associated with a quantitative trait locus for improved yield in rice. Genome Res. 2006, 16, 618–626. [Google Scholar] [CrossRef]
- Fan, C.C.; Xing, Y.Z.; Mao, H.L.; Lu, T.T.; Han, B.; Xu, C.G.; Li, X.H.; Zhang, Q.F. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Xue, W.Y.; Xing, Y.Z.; Weng, X.Y.; Zhao, Y.; Tang, W.J.; Wang, L.; Zhou, H.J.; Yu, S.B.; Xu, C.G.; Li, X.H.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Huang, X.Z.; Qian, Q.; Liu, Z.B.; Sun, H.Y.; He, S.Y.; Luo, D.; Xia, G.M.; Chu, C.C.; Li, J.Y.; Fu, X.D. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Jiao, Y.Q.; Wang, Y.H.; Xue, D.X.; Wang, J.; Yan, M.X.; Liu, G.F.; Dong, G.J.; Zeng, D.L.; Lu, Z.F.; Zhu, X.D.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Wang, S.K.; Wu, K.; Yun, Q.B.; Liu, X.Y.; Liu, Z.B.; Lin, X.Y.; Zeng, R.Z.; Zhu, H.T.; Dong, G.J.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef]
- Zhang, X.X.; Meng, W.J.; Liu, D.P.; Pan, D.Z.; Yang, Y.Z.; Chen, Z.; Ma, X.D.; Yin, W.C.; Niu, M.; Dong, N.N.; et al. Enhancing rice panicle branching and grain yield through tissue-specific brassinosteroid inhibition. Science 2024, 383, eadk8838. [Google Scholar] [CrossRef]
- Yu, J.M.; Zhou, W.; Shao, G.N.; Hu, P.S. Gene mining and breeding utilization of important agronomic traits in rice. Chin. Sci. Bull. 2025, 70, 3126–3148. [Google Scholar] [CrossRef]
- Liang, Y.S.; Nan, W.B.; Qin, X.J.; Zhang, H.M. Field performance on grain yield and quality and genetic diversity of overwintering cultivated rice (Oryza sativa L.) in southwest China. Sci. Rep. 2021, 11, 1846. [Google Scholar] [CrossRef]
- Huang, G.F.; Qin, S.W.; Zhang, S.L.; Cai, X.L.; Wu, S.K.; Dao, J.R.; Zhang, J.; Huang, L.Y.; Harnpichitvitays, D.; Wade, L.J.; et al. Performance, Economics and Potential Impact of Perennial Rice PR23 Relative to Annual Rice Cultivars at Multiple Locations in Yunnan Province of China. Sustainability 2018, 10, 1086. [Google Scholar] [CrossRef]
- Zhang, S.L.; Huang, G.F.; Zhang, Y.J.; Lv, X.T.; Wan, K.J.; Liang, J.; Feng, Y.P.; Dao, J.R.; Wu, S.K.; Zhang, L.; et al. Sustained productivity and agronomic potential of perennial rice. Nat. Sustain. 2022, 6, 28–38. [Google Scholar] [CrossRef]
- Liang, Y.S.; Gong, J.Y.; Yan, Y.X.; Wang, B.B.; Gong, W.A.; Wen, H.; Wu, Q.; Nan, W.B.; Qin, X.J.; Zhang, H.M. Survey of overwintering trait in Chinese rice cultivars (Oryza sativa L.). Euphytica 2022, 218, 94. [Google Scholar] [CrossRef]
- Liu, L.; Chen, T.; Wang, Z.; Zhang, H.; Yang, J.; Zhang, J. Combination of site-specific nitrogen management and alternate wetting and drying irrigation increases grain yield and nitrogen and water use efficiency in super rice. Field Crops Res. 2013, 154, 226–235. [Google Scholar] [CrossRef]
- Shen, Z.D. Crop Breeding Experiment; Chinese Agricultural Press: Beijing, China, 1995; pp. 112–114. [Google Scholar]
- Motulsky, H.J. Prism 5 Statistics Guide; GraphPad Software Inc.: San Diego, CA, USA, 2007; Available online: www.graphpad.com (accessed on 20 September 2025).
- Meng, L.; Li, H.H.; Zhang, L.Y.; Wang, J.K. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- McCouch, S.R.; Cho, Y.G.; Yang, M. Report on QTL nomenclature. Rice Genet. Newsl. 1997, 14, 11–13. [Google Scholar]
- Peng, J.; Richards, D.E.; Hartley, N.M.; Murphy, G.P.; Devos, K.M.; Flintham, J.E.; Beales, J.; Fish, L.J.; Worland, A.J.; Pelica, F.; et al. ‘Green revolution’ genes encode mutant gibberellin response modulatores. Nature 1999, 400, 256–261. [Google Scholar]
- Yuan, L.P. A preliminary report on male sterility in rice, Oryza sativa L. Chin. Sci. Bull. 1966, 4, 185–188. [Google Scholar]
- Shi, M.S. The discovery and preliminary studies of the photoperiod-sensitive recessive male sterile rice (Oryza sativa L. subsp. japonica). Sci. Agric. Sin. 1985, 2, 44–48. [Google Scholar]
- Xie, H.A.; Luo, J.M.; Zhang, S.G.; Zheng, J.T.; Lin, M.J.; Zhang, J.X.; Jiang, Z.H.; Xu, X.M.; Yu, Y.A. Breeding of restorer lines in indica hybrid rice. Hybrid Rice 1994, 3, 31–34. [Google Scholar]
- Deng, Y.W.; Zhai, K.R.; Xie, Z.; Yang, D.Y.; Zhu, X.D.; Liu, J.Z.; Wang, X.; Qin, P.; Yang, Y.Z.; Zhang, G.M.; et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 2017, 355, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.L.; Zhang, X.P.; Fan, Y.L.; Gao, Y.; Zhu, Q.L.; Zheng, C.K.; Qin, T.F.; Li, Y.Q.; Che, J.Y.; Zhang, M.W.; et al. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 2015, 8, 290–302. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, J.; Wang, Z.Z.; Jing, S.L.; Wang, Y.; Ouyang, Y.D.; Cai, B.D.; Xin, X.F.; Liu, X.; Zhang, C.X.; et al. Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc. Natl. Acad. Sci. USA 2016, 113, 12850–12855. [Google Scholar] [CrossRef]
- Li, W.; Yang, K.; Hu, C.F.; Abbas, W.; Zhang, J.; Xu, P.K.; Cheng, B.; Zhang, J.C.; Yin, W.J.; Shalmani, A.; et al. A natural gene on-off system confers field thermo tolerance for grain quality and yield in rice. Cell 2025, 188, 3661–3678. [Google Scholar] [CrossRef]
- Zhang, S.L.; Huang, G.F.; Zhang, J.; Huang, L.Y.; Cheng, M.; Wang, Z.L.; Zhang, Y.N.; Wang, C.L.; Zhu, P.F.; Yu, X.L.; et al. Genotype by environment interactions for performance of perennial rice genotypes (Oryza sativa L./Oryza longistaminata) relative to annual rice genotypes over regrowth cycles and locations in southern China. Field Crops Res. 2019, 241, 107556. [Google Scholar] [CrossRef]
- Wang, J.K.; Li, H.H.; Zhang, X.C.; Yin, C.B.; Li, Y.; Ma, Y.Z.; Li, X.H.; Qiu, L.J.; Wan, J.M. Molecular design breeding in crops in China. Acta Agron. Sin. 2011, 37, 191–201. [Google Scholar] [CrossRef]
- Xu, Y.B.; Zhu, L.H.; Huang, N.; McCouch, S.R. Chromosomal regions associated with segregation distortion of molecular markers in F2, backcross, doubled haploid, and recombinant inbred populations in rice (Oryza sativa L.). Mol. Genet. Genom. 1997, 253, 535–545. [Google Scholar] [CrossRef]
- Han, L.F.; Zhou, R.; Zhou, T.; Lin, C.X.; Gan, Q.; Ni, D.H.; Shi, Y.Y.; Song, F.H. Correlation analysis and QTLs mapping of lodging resistance and yield traits in rice. J. Biol. 2023, 40, 65–70. [Google Scholar]
- Sheng, Z.H.; Zhu, Z.L.; Ma, N.; Li, W.; He, J.W.; Wei, X.J.; Shao, G.N.; Wang, J.L.; Hu, P.S.; Tan, S.Q. QTL mapping of yield related traits in super rice variety Zhongjiazao 17. Chin. J. Rice Sci. 2016, 30, 35–43. [Google Scholar]
- Tan, Z.B.; Shen, L.S.; Yuan, Z.L.; Lu, C.F.; Chen, Y.; Zhou, K.D.; Zhu, L.H. Identification of QTLs for ratooning ability and grain yield traits of rice and analysis of their genetic effects. Acta Agron. Sin. 1997, 23, 289–295. [Google Scholar]
- Moncada, P.; Martinez, C.P.; Borrero, J.; Chatel, M.; Gauch, H.; Guimaraes, E.; Tohme, J.; McCouch, S.R. Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 2001, 102, 41–42. [Google Scholar] [CrossRef]
- Hu, D.W.; Sheng, Z.H.; Chen, W.; Li, Q.L.; Wei, X.J.; Shao, G.N.; Jiao, G.A.; Wang, J.L.; Hu, P.S.; Xie, L.H.; et al. Identification of QTLs associated with high yield of super rice variety Zhongjiazao17. Acta Agron. Sin. 2017, 43, 1434–1447. [Google Scholar] [CrossRef]
- Thomson, M.J.; Tai, T.H.; McClung, A.M.; Lai, X.H.; Hinga, M.E.; Lobos, K.B.; Xu, Y.B.; Martinez, C.P.; McCouch, S.R. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 2003, 107, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.L.; Xue, Q.Z.; Luo, L.J.; Li, Z.K. QTL dissection of panicle number per plant and spikelet number per panicle in rice (Oryza sativa L.). Acta Genet. Sin. 2001, 28, 752–759. [Google Scholar]
- Lu, C.F.; Shen, L.H.; Tan, Z.B.; Xu, Y.B.; He, P.; Chen, Y.; Zhu, L.H. Comparative mapping of QTLs for agronomic traits of rice across environments by using a doubled-haploid population. Theor. Appl. Genet. 1997, 94, 145–150. [Google Scholar] [CrossRef]
- Zhuang, J.Y.; Lin, H.X.; Lu, J.; Qian, H.R.; Hittalmani, S.; Huang, N.; Zheng, K.L. Analysis of QTL x environment interaction for yield components and plant height in rice. Theor. Appl. Genet. 1997, 95, 799–808. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Luo, R.J.; Sheng, Z.H.; Jiao, G.A.; Tang, S.Q.; Hu, P.S.; Wei, X.J. QTL mapping of yield associated traits of Nipponbare/Zhongjiazao17 RIL population. Sci. Agric. Sin. 2017, 50, 3640–3651. [Google Scholar]
- Aluko, G.; Martinez, C.; Tohme, J.; Castano, C.; Bergman, C.; Oard, J.H. QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor. Appl. Genet. 2004, 109, 630–639. [Google Scholar] [CrossRef]
- Huang, N.; Parco, A.; Mew, T.; Magpantay, G.; McCouch, S.R.; Guiderdoni, E.; Xu, J.C.; Subudhi, P.; Enrique, R.; Khush, G.S. RFLP mapping of isozymes, RAPD and QTLs for grain shape, brown planthopper resistance in a doubled haploid rice population. Mol. Breed. 1997, 3, 105–113. [Google Scholar] [CrossRef]
- Xiao, J.H.; Li, J.; Grandillo, S.; Ahn, S.N.; Yuan, L.; Tanksley, S.D.; McCouch, S.R. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 1998, 150, 899–909. [Google Scholar] [CrossRef]
- Tian, F.; Li, D.J.; Fu, Q.; Zhu, Z.F.; Fu, Y.C.; Wang, X.K.; Sun, C.Q. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 2005, 112, 570–580. [Google Scholar] [CrossRef]
- You, A.Q.; Lu, X.G.; Jin, H.J.; Ren, X.; Liu, K.; Yang, G.C.; Yang, H.Y.; Zhu, L.L.; He, G.C. Identification of quantitative trait loci across recombinant inbred lines and testcross populations for traits of agronomic importance in rice. Genetics 2006, 105, 1287–1300. [Google Scholar] [CrossRef]
- Tahmasebi, S.; Heidari, B.; Pakniyat, H.; McIntyre, C.L. Mapping QTLs associated with agronomic and physiological traits under terminal drought and heat stress conditions in wheat (Triticum aestivum L.). Genome 2017, 60, 26–45. [Google Scholar] [CrossRef]



| Traits | Shendao3# | t-Test Value | |||
|---|---|---|---|---|---|
| Major Crop | Ratooning Crop | ||||
| Means ± SD | CV (%) | Means ± SD | CV (%) | ||
| HD (d) | 108.50 ± 1.71 | 1.58 | 127 ± 1.41 | 1.11 | 13.86 ** |
| PH (cm) | 121.42 ± 2.71 | 2.23 | 110.40 ± 5.31 | 4.81 | 7.08 ** |
| EPP | 18.50 ± 5.12 | 27.68 | 19.00 ± 4.77 | 25.11 | 0.37 |
| PL (cm) | 19.07 ± 1.09 | 5.72 | 20.52 ± 0.67 | 3.27 | 2.66 |
| FGP | 180.83 ± 5.55 | 3.07 | 205.20 ± 27.44 | 13.37 | 1.88 |
| UGP | 13.33 ± 2.29 | 17.18 | 48.80 ± 7.91 | 16.21 | 8.67 ** |
| SP | 167.50 ± 4.54 | 2.71 | 156.40 ± 27.49 | 17.58 | 0.82 |
| SSR (%) | 92.64 ± 1.14 | 1.23 | 75.86 ± 4.97 | 6.55 | 5.96 |
| SSD | 9.52 ± 0.66 | 6.93 | 9.99 ± 1.24 | 12.41 | 0.49 |
| GWP (g) | 3.63 ± 0.18 | 4.96 | 3.77 ± 0.48 | 12.73 | 0.91 |
| GYP (g) | 49.83 ± 18.56 | 37.25 | 43.74 ± 9.71 | 22.20 | 0.98 |
| GL (mm) | 7.05 ± 0.24 | 3.40 | 7.34 ± 0.17 | 2.32 | 1.73 |
| GW (mm) | 3.08 ± 0.13 | 4.22 | 3.03 ± 0.14 | 4.62 | 0.20 |
| LWR | 2.29 ± 0.06 | 2.62 | 2.43 ± 0.07 | 2.88 | 0.06 |
| GT (mm) | 2.25 ± 0.06 | 2.67 | 2.12 ± 0.09 | 4.25 | 1.30 |
| TGW (g) | 24.83 ± 1.07 | 4.31 | 24.20 ± 2.14 | 8.84 | 0.16 |
| Traits | HD | PH | EPP | PL | FGP | UGP | SP | SSR | SSD | GWP | GYP | GL | GW | LWR | GT | TGW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HD | 1.00 | 0.97 * | 0.45 | 0.55 | −0.93 * | 0.22 | −0.90 * | 0.55 | 0.86 | 0.87 | 0.34 | 0.77 | 0.84 | 0.84 | 0.86 | 0.67 |
| PH | 0.01 | 1.00 | 0.55 | 0.63 | −0.90 * | 0.26 | −0.91 * | 0.45 | −0.92 * | −0.92 * | 0.45 | 0.85 | −0.90 * | −0.90 * | −0.92 * | 0.56 |
| EPP | 0.29 | 0.85 | 1.00 | −0.97 * | 0.53 | 0.04 | 0.46 | 0.09 | 0.73 | 0.72 | 0.87 | 0.80 | 0.77 | 0.74 | 0.75 | 0.59 |
| PL | 0.31 | 0.03 | 0.09 | 1.00 | 0.56 | 0.14 | 0.61 | 0.04 | 0.83 | 0.82 | −0.92 * | 0.89 * | 0.87 | 0.84 | 0.84 | 0.71 |
| FGP | 0.64 | 0.14 | 0.03 | 0.12 | 1.00 | 0.15 | 0.69 | 0.77 | 0.74 | 0.75 | 0.25 | 0.65 | 0.72 | 0.72 | 0.75 | 0.64 |
| UGP | 0.52 | 0.16 | 0.07 | 0.28 | 0.88 | 1.00 | 0.61 | 0.64 | 0.44 | 0.44 | 0.41 | 0.46 | 0.42 | 0.46 | 0.42 | 0.10 |
| SP | 0.29 | 0.43 | 0.40 | 0.67 | 0.62 | 0.26 | 1.00 | 0.12 | 0.94 * | 0.95 * | 0.57 | 0.89 * | 0.92 * | 0.94 * | 0.94 * | 0.61 |
| SSR | 0.64 | 0.66 | 0.59 | 0.25 | 0.75 | 0.76 | 0.01 | 1.00 | 0.15 | 0.15 | 0.35 | 0.02 | 0.12 | 0.10 | 0.15 | 0.29 |
| SSD | 0.10 | 0.72 | 0.83 | 0.34 | 0.48 | 0.37 | 0.82 | 0.20 | 1.00 | 1.00 * | 0.75 | 0.99 * | 1.00 * | 1.00 * | 1.00 * | 0.66 |
| GWP | 0.08 | 0.72 | 0.79 | 0.46 | 0.44 | 0.27 | 0.87 | 0.26 | 0.99 * | 1.00 | 0.74 | 0.98 * | 1.00 * | 1.00 * | 1.00 * | 0.68 |
| GYP | 0.07 | 0.69 | 0.78 | 0.48 | 0.46 | 0.28 | −0.88 * | 0.24 | −0.99 * | −1.00 * | 1.00 | 0.85 | 0.79 | 0.78 | 0.76 | 0.55 |
| GL | 0.10 | 0.67 | 0.74 | 0.52 | 0.48 | 0.27 | 0.91 * | 0.22 | 0.97 * | 1.00 * | −1.00 * | 1.00 | 0.99 * | 0.99 * | 0.99 * | 0.65 |
| GW | 0.05 | 0.67 | 0.78 | 0.49 | 0.47 | 0.30 | 0.88 * | 0.22 | 0.99 * | 1.00 * | −1.00 * | 0.99 * | 1.00 | 1.00 * | 1.00 * | 0.70 |
| LWR | 0.08 | 0.72 | 0.81 | 0.42 | 0.45 | 0.31 | 0.86 | 0.24 | 0.99 * | 1.00 * | −1.00 * | 0.99 * | 1.00 * | 1.00 | 1.00 * | 0.68 |
| GT | 0.04 | 0.68 | 0.80 | 0.44 | 0.47 | 0.33 | 0.85 | 0.22 | 0.99 * | 1.00 * | −1.00 * | 0.99 * | 1.00 * | 1.00 * | 1.00 | 0.69 |
| TGW | 0.10 | 0.77 | 0.35 | 0.11 | 0.43 | 0.61 | 0.25 | 0.69 | 0.24 | 0.31 | 0.29 | 0.30 | 0.24 | 0.29 | 0.23 | 1.00 |
| Traits | Bi-Parents | SD3#-Population | ||||
|---|---|---|---|---|---|---|
| SD3# | XQZB | t-Test Value | Means ± SD | Range | CV (%) | |
| PH (cm) | 102.80 | 82.40 | 13.07 ** | 137.96 ± 16.36 | 100.00–167.00 | 9.78 |
| EPP | 9.80 | 13.20 | 3.30 * | 7.73 ± 2.86 | 3.00–17.00 | 37.80 |
| PL (cm) | 17.60 | 22.04 | 7.89 ** | 28.33 ± 3.15 | 16.20–34.00 | 11.78 |
| FGP | 157.20 | 96.60 | 6.13 ** | 193.29 ± 62.46 | 27.00–379.00 | 32.12 |
| UGP | 6.20 | 10.00 | 1.26 | 99.02 ± 68.02 | 12.00–296.00 | 67.52 |
| SP | 163.40 | 106.60 | 4.56 ** | 292.32 ± 72.86 | 143.00–502.00 | 24.17 |
| SSR (%) | 96.25 | 90.88 | 2.74 * | 67.00 ± 0.19 | 8.36–94.59 | 27.74 |
| SSD | 9.28 | 4.83 | 7.85 ** | 10.32 ± 2.25 | 6.27–17.16 | 22.07 |
| GWP (g) | 3.99 | 2.71 | 6.29 ** | 5.26 ± 1.47 | 2.32–8.94 | 27.88 |
| GYP (g) | 33.04 | 24.27 | 4.43 * | 34.38 ± 15.84 | 5.57–88.84 | 47.17 |
| GL (mm) | 6.77 | 9.75 | 25.49 ** | 10.16 ± 9.49 | 7.60–10.72 | 7.02 |
| GW (mm) | 3.09 | 2.43 | 11.63 ** | 2.87 ± 0.23 | 2.17–3.71 | 7.81 |
| LWR | 2.19 | 4.00 | 30.35 ** | 3.56 ± 3.35 | 2.18–4.54 | 11.78 |
| GT (mm) | 2.19 | 2.07 | 3.36 * | 1.99 ± 0.18 | 1.17–2.21 | 8.70 |
| TGW (g) | 24.60 | 26.80 | 2.42 | 24.70 ± 0.37 | 13.50–33.90 | 15.15 |
| Traits | PH | EPP | PL | FGP | UGP | SP | SSR | SSD | GWP | GYP | GL | GW | LWR | GT | TGW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | 1.00 | ||||||||||||||
| EPP | 0.32 * | 1.00 | |||||||||||||
| PL | 0.37 * | 0.26 * | 1.00 | ||||||||||||
| FGP | 0.04 | 0.12 | 0.26 * | 1.00 | |||||||||||
| UGP | 0.30 * | 0.08 | 0.25 * | −0.38 * | 1.00 | ||||||||||
| SP | 0.31 * | 0.19 | 0.45 * | 0.50 * | 0.61 * | 1.00 | |||||||||
| SSR | −0.22 * | −0.02 | −0.06 | 0.68 * | −0.90 * | −0.25 * | 1.00 | ||||||||
| SSD | 0.16 | 0.08 | −0.00 | 0.42 * | 0.56 * | 0.89 * | −0.26 * | 1.00 | |||||||
| GWP | 0.12 | 0.19 | 0.35 * | 0.78 * | −0.26 * | 0.43 * | 0.49 * | 0.28 * | 1.00 | ||||||
| GYP | 0.32 * | 0.80 * | 0.38 * | 0.49 * | −0.12 | 0.31 * | 0.28 * | 0.15 | 0.60 * | 1.00 | |||||
| GL | 0.09 | 0.16 | 0.20 | −0.07 | 0.23 * | 0.16 | −0.16 | 0.07 | 0.02 | 0.09 | 1.00 | ||||
| GW | 0.13 | 0.07 | −0.12 | −0.20 | 0.31 * | 0.12 | −0.29 * | 0.19 | −0.19 | −0.09 | −0.03 | 1.00 | |||
| LWR | 0.08 | 0.16 | 0.21 * | −0.05 | 0.21 | 0.15 | −0.14 | 0.05 | 0.03 | 0.09 | 1.00 * | −0.10 | 1.00 | ||
| GT | −0.07 | 0.02 | 0.04 | −0.09 | −0.12 | −0.19 | 0.03 | −0.24 * | 0.15 | 0.07 | 0.02 | 0.11 | 0.01 | 1.00 | |
| TGW | −0.09 | 0.13 | 0.12 | 0.00 | −0.29 * | −0.27 * | 0.19 | −0.36 * | 0.32 * | 0.27 * | −0.04 | −0.16 | −0.03 | 0.47 * | 1.00 |
| Trait | QTL | Chromosome | Genomic Position | Marker Interval | LOD | Additive | Dominant | R2 (%) | Favorable Allele |
|---|---|---|---|---|---|---|---|---|---|
| PH | qPH1 | 1 | 34902085-37261443 | RM3738-RM8084 | 6.06 | 9.93 | 7.21 | 27.30 | SD3# |
| EPP | qEPP1 | 1 | 9463544-24866202 | RM3642-RM600 | 3.06 | −3.02 | −4.80 | 8.06 | XQZB |
| qEPP3 | 3 | 4333680-13933574 | RM489-RM6080 | 3.02 | 3.84 | −5.56 | 7.33 | SD3# | |
| qEPP7 | 7 | 16932001-17489638 | RM1135-RM5793 | 5.23 | −3.58 | −6.52 | 10.82 | XQZB | |
| qEPP11 | 11 | 11763775-2888052 | RM7120-RM6293 | 4.28 | −3.04 | −5.09 | 8.53 | XQZB | |
| PL | qPL1 | 1 | 27925715-32774365 | RM3642-RM600 | 3.00 | 6.05 | 4.19 | 6.07 | SD3# |
| qPL2 | 2 | 13481661-19677083 | RM250-RM3763 | 4.67 | −6.67 | 3.94 | 8.05 | XQZB | |
| qPL11 | 11 | 1124242-4773752 | RM1341-RM3428 | 3.96 | −3.11 | 5.53 | 8.23 | XQZB | |
| UGP | qUGP8 | 8 | 35196573-37261443 | RM1111-RM3702 | 3.10 | 49.48 | −132.62 | 14.32 | SD3# |
| SP | qSP1 | 1 | 13059580-24116775 | RM265-RM3738 | 5.52 | 48.12 | 33.14 | 26.56 | SD3# |
| SSR | qSSR4 | 4 | 11389704-20800963 | RM7051-RM5633 | 3.88 | 0.02 | −0.32 | 4.61 | SD3# |
| qSSR8 | 8 | 16932001-17489638 | RM1111-RM3702 | 5.90 | −0.17 | 0.46 | 5.40 | XQZB | |
| SSD | qSSD1 | 1 | 10811135-19788247 | RM265-RM3738 | 4.53 | 0.47 | 1.82 | 14.11 | SD3# |
| qSSD2 | 2 | 2722348-14527760 | RM324-RM262 | 5.25 | 1.49 | −3.80 | 20.65 | SD3# | |
| GYP | qGYP7 | 7 | 2722348-13761888 | RM1135-RM5793 | 3.32 | −14.32 | −23.54 | 7.25 | XQZB |
| qGYP9 | 9 | 4407860-23568212 | RM24085-RM160 | 3.41 | 21.40 | −14.19 | 5.79 | SD3# | |
| qGYP10a | 10 | 11389704-15894177 | RM5708-RM3882 | 4.24 | 17.86 | −13.64 | 9.07 | SD3# | |
| qGYP10b | 10 | 19677083-28788052 | RM3882-RM8201 | 3.74 | 19.60 | −23.56 | 5.16 | SD3# | |
| GW | qGW2a | 2 | 3073406-27342022 | RM5651-RM3732 | 3.04 | 0.34 | −0.37 | 4.21 | SD3# |
| qGW2b | 2 | 11389704-15894177 | RM5812-RM324 | 4.07 | 0.28 | −0.26 | 5.38 | SD3# | |
| qGW11a | 11 | 11763775-28788053 | RM7120-RM6293 | 3.82 | 0.27 | −0.35 | 5.34 | SD3# | |
| qGW11b | 11 | 19677083-28788052 | RM6293-RM1341 | 3.46 | 0.28 | −0.35 | 5.03 | SD3# | |
| GT | qGT5 | 5 | 3073406-27342022 | RM405-RM26 | 3.98 | −0.16 | 0.28 | 9.30 | XQZB |
| TGW | qTGW1 | 1 | 9463544-24866202 | RM3642-RM600 | 4.15 | 0.44 | 0.32 | 4.76 | SD3# |
| qTGW4 | 4 | 13059580-24116775 | RM7051-RM5633 | 4.43 | 0.38 | 0.57 | 5.08 | SD3# |
| Traits | QTL | Chromosome | Marker Interval | LOD Value | Additive | Dominant | R2 (%) |
|---|---|---|---|---|---|---|---|
| SP | qSP1 | 1 | RM265-RM3738 | 5.52 | 48.12 | 33.14 | 26.56 |
| SSD | qSSD1 | 1 | RM265-RM3738 | 4.53 | 0.47 | 1.82 | 14.11 |
| EPP | qEPP1 | 1 | RM3642-RM600 | 3.06 | −3.02 | −4.80 | 8.06 |
| PL | qPL1 | 1 | RM3642-RM600 | 3.00 | 6.05 | 4.19 | 6.07 |
| SSR | qSSR4 | 4 | RM7051-RM5633 | 3.88 | 0.02 | −0.32 | 4.61 |
| TGW | qTGW4 | 4 | RM7051-RM5633 | 4.43 | 0.38 | 0.57 | 5.08 |
| EPP | qEPP7 | 7 | RM1135-RM5793 | 5.23 | −3.58 | −6.52 | 10.82 |
| GYP | qGYP7 | 7 | RM1135-RM5793 | 3.32 | −14.32 | −23.54 | 7.25 |
| UGP | qUGP8 | 8 | RM1111-RM3702 | 3.10 | 49.48 | −132.62 | 14.32 |
| SSR | qSSR8 | 8 | RM1111-RM3702 | 5.90 | −0.17 | 0.46 | 5.40 |
| EPP | qEPP11 | 11 | RM7120-RM6293 | 4.28 | −3.04 | −5.09 | 8.53 |
| GW | qGW11a | 11 | RM7120-RM6293 | 3.82 | 0.27 | −0.35 | 5.34 |
| Traits | Chr a | Marker Interval | Chr a | Marker Interval | LOD Value | Add b | Add | Dom c | Dom | Add × Add | Add × Dom | Dom × Add | Dom × Dom | R2 (%) d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | 1 | RM3738-RM8084 | 11 | RM202-RM7120 | 5.01 | 4.82 | −8.37 | 0.49 | −28.13 | 8.08 | −21.17 | −5.63 | 30.51 | 19.30 |
| EPP | 1 | RM3642-RM600 | 2 | RM6519-RM5651 | 7.08 | −1.97 | 1.90 | −3.21 | 1.56 | −3.38 | 3.30 | 0.42 | −0.59 | 2.21 |
| 3 | RM3513-RM1352 | 5 | RM405-RM26 | 5.03 | 0.91 | −1.03 | 3.50 | 1.48 | −0.70 | −2.76 | 5.63 | −7.37 | 1.49 | |
| 3 | RM85-RM3856 | 10 | RM5708-RM3882 | 6.40 | −0.34 | 1.58 | 4.55 | 0.98 | −1.78 | 2.01 | 2.20 | −8.64 | 1.98 | |
| 7 | RM3555-RM5481 | 7 | RM1135-RM5793 | 6.13 | −3.07 | −1.71 | −2.01 | −6.46 | 0.69 | 2.89 | 1.55 | 4.18 | 2.19 | |
| 7 | RM5793-RM432 | 8 | RM1111-RM3702 | 5.08 | 0.41 | 0.36 | 0.67 | 1.55 | 0.39 | −5.50 | 4.95 | −6.19 | 1.61 | |
| 7 | RM1135-RM5793 | 11 | RM202-RM7120 | 6.84 | 0.55 | −2.71 | −1.06 | 3.88 | 0.89 | −4.19 | 3.52 | −7.08 | 2.20 | |
| PL | 1 | RM3642-RM600 | 8 | RM8019-RM6990 | 5.48 | −4.58 | −3.57 | −2.01 | −2.00 | −2.56 | 6.12 | 1.03 | 7.14 | 6.28 |
| 2 | RM250-RM3763 | 3 | RM1352-RM3199 | 5.10 | −5.30 | 0.77 | 4.32 | 6.97 | 1.58 | 5.93 | −1.27 | −9.50 | 4.94 | |
| 2 | RM7637-RM5812 | 4 | RM7051-RM5633 | 5.49 | 0.91 | 0.34 | 3.40 | 3.33 | −2.59 | −3.69 | −0.77 | −5.13 | 5.13 | |
| UGP | 8 | RM1111-RM3702 | 9 | RM257-RM5661 | 5.11 | 7.29 | −9.84 | −88.52 | −27.85 | −52.25 | −51.59 | 45.48 | −29.79 | 1.74 |
| SSR | 4 | RM317-RM7051 | 4 | RM7051-RM5633 | 5.94 | −0.01 | 0.07 | 0.07 | −0.10 | 0.04 | −0.36 | 0.09 | −0.14 | 1.53 |
| 4 | RM5633-RM401 | 8 | RM1111-RM3702 | 7.42 | 0.07 | 0.00 | −0.33 | 0.21 | −0.01 | −0.04 | −0.15 | 0.34 | 1.45 | |
| 4 | RM7051-RM5633 | 9 | RM24085-RM160 | 5.69 | 0.00 | 0.02 | −0.41 | −0.13 | 0.02 | 0.13 | −0.10 | 0.56 | 1.24 | |
| 4 | RM317-RM7051 | 10 | RM5708-RM3882 | 7.22 | 0.12 | 0.15 | 0.09 | 0.12 | −0.22 | −0.03 | 0.02 | −0.38 | 1.49 | |
| 4 | RM559-RM5979 | 11 | RM7120-RM6293 | 6.72 | −0.03 | 0.02 | 0.00 | −0.24 | 0.24 | −0.22 | −0.12 | 0.47 | 1.54 | |
| 8 | RM1111-RM3702 | 9 | RM24085-RM160 | 5.32 | −0.04 | 0.16 | −0.12 | −0.09 | 0.04 | 0.16 | −0.43 | 0.34 | 1.30 | |
| 8 | RM1111-RM3702 | 10 | RM5708-RM3882 | 7.95 | −0.06 | 0.20 | 0.28 | −0.01 | 0.18 | 0.18 | −0.17 | −0.01 | 1.50 | |
| 8 | RM1111-RM3702 | 11 | RM6293-RM1341 | 6.80 | −0.05 | 0.16 | 0.26 | −0.06 | 0.19 | 0.12 | −0.13 | 0.07 | 1.52 | |
| GYP | 7 | RM3555-RM5481 | 7 | RM1135-RM5793 | 7.01 | 14.80 | −1.74 | 17.34 | −5.76 | 0.78 | −25.40 | 21.76 | −22.25 | 2.28 |
| 7 | RM1135-RM5793 | 8 | RM1111-RM3702 | 5.68 | 1.06 | −18.79 | −18.91 | −8.21 | −8.94 | 8.78 | 8.10 | 6.61 | 2.52 | |
| 7 | RM1135-RM5793 | 11 | RM202-RM7120 | 5.87 | −2.54 | −3.60 | −3.50 | 12.75 | −12.47 | 24.77 | 7.82 | −26.83 | 2.28 | |
| 9 | RM24085-RM160 | 11 | RM7120-RM6293 | 5.31 | 9.01 | −13.95 | 11.16 | −14.50 | −12.05 | −4.57 | 10.83 | −2.13 | 2.36 | |
| 10 | RM5708-RM3882 | 11 | RM202-RM7120 | 5.40 | 7.41 | −13.27 | 1.72 | 15.35 | −2.44 | 18.95 | 20.25 | −31.15 | 2.94 | |
| GW | 2 | RM250-RM3763 | 4 | RM559-RM5979 | 6.25 | 0.12 | −0.01 | 0.46 | 0.28 | 0.18 | 0.00 | 0.34 | −0.79 | 2.49 |
| GT | 5 | RM405-RM26 | 11 | RM202-RM7120 | 8.74 | −0.04 | 0.11 | 0.17 | −0.25 | 0.15 | 0.18 | −0.10 | 0.26 | 2.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Lu, J.; Wu, M.; Peng, T.; Tan, L.; Nan, W.; Qin, X.; Li, M.; Gong, J.; Liang, Y. Locating QTL Controlling the Yield-Related Traits in Perennial Chinese Rice “Shendao3#”. Agriculture 2025, 15, 2453. https://doi.org/10.3390/agriculture15232453
Yan Y, Lu J, Wu M, Peng T, Tan L, Nan W, Qin X, Li M, Gong J, Liang Y. Locating QTL Controlling the Yield-Related Traits in Perennial Chinese Rice “Shendao3#”. Agriculture. 2025; 15(23):2453. https://doi.org/10.3390/agriculture15232453
Chicago/Turabian StyleYan, Yuxin, Jiuyan Lu, Meilin Wu, Tingshen Peng, Lin Tan, Wenbin Nan, Xiaojian Qin, Ming Li, Junyi Gong, and Yongshu Liang. 2025. "Locating QTL Controlling the Yield-Related Traits in Perennial Chinese Rice “Shendao3#”" Agriculture 15, no. 23: 2453. https://doi.org/10.3390/agriculture15232453
APA StyleYan, Y., Lu, J., Wu, M., Peng, T., Tan, L., Nan, W., Qin, X., Li, M., Gong, J., & Liang, Y. (2025). Locating QTL Controlling the Yield-Related Traits in Perennial Chinese Rice “Shendao3#”. Agriculture, 15(23), 2453. https://doi.org/10.3390/agriculture15232453





